The same nucleotide sequence can encode multiple protein products in different reading frames, and these regions encode higher levels of intrinsic structural disorder than non-overlapping genes. Willis and Masel find that...
Keywords: overprinting, gene age, alternative reading frame, evolutionary constraint, mutation-driven evolution
Abstract
The same nucleotide sequence can encode two protein products in different reading frames. Overlapping gene regions encode higher levels of intrinsic structural disorder (ISD) than nonoverlapping genes (39% vs. 25% in our viral dataset). This might be because of the intrinsic properties of the genetic code, because one member per pair was recently born de novo in a process that favors high ISD, or because high ISD relieves increased evolutionary constraint imposed by dual-coding. Here, we quantify the relative contributions of these three alternative hypotheses. We estimate that the recency of de novo gene birth explains or more of the elevation in ISD in overlapping regions of viral genes. While the two reading frames within a same-strand overlapping gene pair have markedly different ISD tendencies that must be controlled for, their effects cancel out to make no net contribution to ISD. The remaining elevation of ISD in the older members of overlapping gene pairs, presumed due to the need to alleviate evolutionary constraint, was already present prior to the origin of the overlap. Same-strand overlapping gene birth events can occur in two different frames, favoring high ISD either in the ancestral gene or in the novel gene; surprisingly, most de novo gene birth events contained completely within the body of an ancestral gene favor high ISD in the ancestral gene (23 phylogenetically independent events vs. 1). This can be explained by mutation bias favoring the frame with more start codons and fewer stop codons.
PROTEIN-CODING genes sometimes overlap, i.e., the same nucleotide sequence encodes different proteins in different reading frames. Most of the overlapping pairs of genes that have been characterized to date are found in viral, bacterial, and mitochondrial genomes, with emerging research showing that they may be common in eukaryotic genomes as well (Nekrutenko et al. 2005; Chung et al. 2007; Kim et al. 2008; Ribrioux et al. 2008; Neme and Tautz 2013). Studying overlapping genes can shed light on the processes of de novo gene birth (Rancurel et al. 2009).
Overlapping genes tend to encode proteins with higher intrinsic structural disorder (ISD) than those encoded by nonoverlapping genes (Rancurel et al. 2009). The term disorder applies broadly to proteins that, at least in the absence of a binding partner, lack a stable secondary and tertiary structure. There are different degrees of disorder: molten globules, partially unstructured proteins, and random coils with regions of disorder spanning from short (<30 residues in length) to long. Disorder can be shown experimentally or predicted from amino acid sequences using software (Ferron et al. 2006). Rancurel et al. (2009) estimated, using the latter approach, that 48% of amino acids in overlapping regions exhibit disorder, compared to only 23% in nonoverlapping regions.
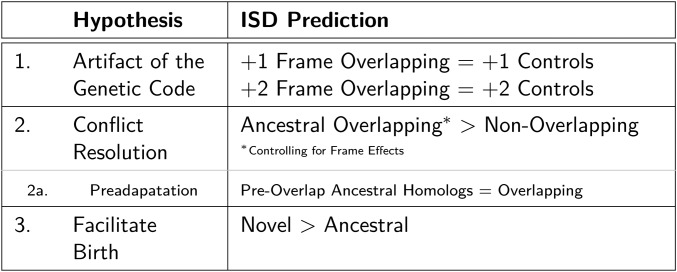
In this work, we explore three nonmutually exclusive hypotheses why this might be, and quantify the extent of each. Two have previously been considered: that elevated ISD in overlapping genes is a mechanism that relieves evolutionary constraint, and that elevated ISD is a holdover from the de novo gene birth process. We add consideration of a third, previously-unexplored, hypothesis—that elevated ISD with dual-coding may be the result of an artifact of the genetic code—to the mix.
Overlapping genes are particularly evolutionarily constrained because a mutation in an overlapping region simultaneously affects both of the two (or occasionally more) genes involved in that overlap. Because 70% of mutations that occur in the third codon position are synonymous, vs. only 5 and 0% of mutations in the first and second codon positions, respectively (Sabath et al. 2008a), a mutation that is synonymous in one reading frame is highly likely to be nonsynonymous in another, so, to permit adaptation, overlapping genes must be relatively tolerant of nonsynonymous changes. Demonstrating the higher constraint on overlapping regions, they have lower genetic diversity and than nonoverlapping regions in RNA viruses (Mizokami et al. 1997; Sabath et al. 2008b, 2012; Simon-Loriere et al. 2013). Further demonstrating their constraint, overlapped sites encode functionally important residues of one gene or the other, but never both, and nonfunctionally important sites vary more in strains in which no overlap is present (Fernandes et al. 2016).
High ISD can alleviate the problem of constraint. Amino acid substitutions that maintain disorder have a reasonable chance of being tolerated, in contrast to the relative fragility of a well-defined three-dimensional structure. This expectation is confirmed by the higher evolutionary rates observed for disordered proteins (Brown et al. 2002; Echave et al. 2016). The known elevation of evolutionary constraint on overlapping genes is usually invoked as the sole (Tokuriki et al. 2009; Xue et al. 2014), or at least dominant (Rancurel et al. 2009), explanation for their high ISD. Given the strength of the evidence for constraint (Mizokami et al. 1997; Sabath et al. 2008b, 2012; Simon-Loriere et al. 2013; Fernandes et al. 2016), we attribute to constraint, by a process of elimination, what cannot be explained by our other two hypotheses.
The second hypothesis that we consider is that high ISD in overlapping genes is an artifact of the process of de novo gene birth (Rancurel et al. 2009). There is no plausible path by which two nonoverlapping genes could re-encode an equivalent protein sequence as overlapping; instead, an overlapping pair arises either when a second gene is born de novo within an existing gene, or when the boundaries of an existing gene are extended to create overlap (Sabath et al. 2008a). In the latter case of “overprinting” (Keese and Gibbs 1992; Rancurel et al. 2009; Carter et al. 2013), the extended portion of that gene, if not the whole gene, is born de novo. One overlapping protein-coding sequence is therefore always evolutionarily younger than the other; we refer to these as “novel” vs. “ancestral” overlapping genes or portions of genes. Genes may eventually lose their overlap through a process of gene duplication followed by subfunctionalization (Keese and Gibbs 1992), enriching overlapping genes for relatively young genes that have not yet been through this process. However, gene duplication may be inaccessible to many viruses (in particular, many RNA, ssDNA, and retroviruses), due to intrinsic geometric constraints on maximum nucleotide length (Miller 1997; Chirico et al. 2010; Campillo-Balderas et al. 2015).
Young genes are known to have higher ISD than old genes, with high ISD at the moment of gene birth facilitating the process (Wilson et al. 2017), perhaps because cells tolerate them better (Tretyachenko et al. 2017). Domains that were more recently born de novo also have higher ISD (Buljan et al. 2010; Ekman and Elofsson 2010; Moore and Bornberg-Bauer 2011; Bornberg-Bauer and Alba 2013). High ISD could be helpful in itself in creating novel function, or it could be a byproduct of a hydrophilic amino acid composition whose function is simply the avoidance of harmful protein aggregation (Liu and Huang 2014; Foy et al. 2017). Regardless of the cause of high ISD in young genes, the “facilitate birth” hypothesis makes a distinct prediction from the constraint hypothesis, namely that the novel overlapping reading frames will tend to encode higher ISD than the ancestral overlapping reading frames.
Under the constraint hypothesis, ancestral overlapping proteins will still have elevated ISD relative to nonoverlapping proteins, even if it is less elevated than that of novel overlapping proteins. Elevated ISD in the ancestral member of the gene pair might have already been there at the moment of gene birth, or it might have subsequently evolved, representing two subhypotheses within the overall hypothesis of constraint. The overlapping gene pairs that we observe are those that have been retained; if either member of the overlapping pair was born with low ISD, then constraint makes it difficult to adapt to a changing environment, and that pair is less likely to be retained. When the ancestral member of the pair already has high ISD at the moment at which the novel gene is functionalized, long-term maintenance of both genes in the face of constraint is more likely. The “already there” or “preadaptation” (Wilson et al. 2017) version of the constraint hypothesis predicts that the preoverlapping ancestors of today’s ancestral overlapping genes had higher ISD than other genes, perhaps because these gene pairs are the ones to have been retained. While these ancestral sequences are not available, as a proxy we use homologous sequences from basal lineages whose most recent common ancestry predates the origin of the overlap. For simplicity, we refer to these sequences as “preoverlapping” to distinguish them from “nonoverlapping” genes never known to have overlap. The preadaptation version of the constraint hypothesis predicts higher ISD in preoverlapping genes than in nonoverlapping genes.
Finally, here we also consider the possibility that the high ISD observed in overlapping genes might simply be an artifact of the genetic code (Kovacs et al. 2010). We perform for the first time the appropriate control, by predicting what the ISD would be if codons were read from alternative reading frames of existing nonoverlapping genes. Any DNA sequence can be read in three reading frames on each of the two strands, for a total of six reading frames. We focus only on same-strand overlap, due to superior availability of reliable data on same-strand overlapping gene pairs. We classify the reading frame of each gene in an overlapping pair relative to its counterpart; if gene A is in the +1 frame with respect to gene B, this means that gene B is in the +2 frame with respect to gene A. We use the +0 frame designation just for nonoverlapping or preoverlapping genes in their original frame. If the high ISD of overlapping genes is driven primarily by the intrinsic properties of the genetic code, then we expect their ISD values to closely match those expected from translation in the +1 vs. +2 frames of nonoverlapping genes.
Here, we test the predictions of all three hypotheses, as summarized in Figure 1, and find that both the birth-facilitation and conflict-resolution hypotheses play a role. The artifact hypothesis plays no appreciable role in elevating the ISD of overlapping regions; while reading frame (+1 vs. +2) strongly affects the ISD of individual genes, each overlapping gene pair has one of each, and the two cancel out such that there is no net contribution to the high ISD found in overlapping regions. Surprisingly, novel genes are more likely to be born in the frame prone to lower ISD; this seems to be a case where mutation bias in the availability of open reading frames (ORFs) is more important than selection favoring higher ISD for novel than ancestral genes.
Figure 1.
Three nonmutually-exclusive hypotheses about why overlapping genes have high ISD. The column on the right describes the ISD patterns we would expect if the hypotheses were true.
Materials and Methods
Overlapping viral genes
A total of 102 viral same-strand overlapping gene pairs were compiled from the literature (Webster et al. 1992; Rancurel et al. 2009; Sabath et al. 2012; Pavesi et al. 2013; Simon-Loriere et al. 2013; Shukla and Hilgenfeld 2015). Of these, 10 were discarded because one or both of the genes involved in the overlap were not found in the National Center for Biotechnology Information (NCBI) databases, either because the accession number had been removed, or because the listed gene could not be located. This left 92 gene pairs for analysis from 80 different species, spanning 33 viral families. Six of these pairs were ssDNA, five were retroviruses, while the remaining 81 were RNA viruses: 7 dsRNA, 61 positive sense RNA and 13 negative sense RNA.
Relative gene age
For 39 of the remaining 92 gene pairs available for analysis, the identity of the older vs. younger member of the pair had been classified in the literature (Morozov and Solovyev 2003; Rancurel et al. 2009; Sabath et al. 2012; Shukla and Hilgenfeld 2015) via phylogenetic analysis. There was disagreement in the literature regarding the TGBp2/TGBp3 overlap; we followed Morozov and Solovyev (2003) rather than Rancurel et al. (2009).
We also used relative levels of codon bias to classify the relative ages of members of each pair. Because all of the overlapping genes are from viral genomes, we can assume that they are highly expressed, leading to a strong expectation of codon bias in general. Novel genes are expected to have less extreme codon bias than ancestral genes due to evolutionary inertia (Sabath et al. 2012; Pavesi et al. 2013).
For each viral species, codon usage data (Nakamura et al. 2000; Zhou et al. 2005) were used to calculate a relative synonymous codon usage (RSCU) value for each codon (Graur 2016):
where is the number of occurrences of codon i in the viral genome, and is the number of synonymous codons that code for the same amino acid as codon i. The relative adaptedness value () for each codon in a viral species was then calculated as:
where RSCU is the RSCU value for the most frequently occurring codon corresponding to the amino acid associated with codon i. The codon adaptation index (CAI) was then calculated for the overlapping portion of each gene, as the geometric mean of the relative adaptedness values:
where L is the number of codons in the overlapping portion of the gene, excluding ATG and TGG codons. This exclusion is because ATG and TGG are the only codons that code for their respective amino acids and so their relative adaptedness values are always 1, thereby introducing no new information. To ensure sufficient statistical power to differentiate between CAI values, we did not analyze CAI for gene pairs with overlapping sections <200 nucleotides long.
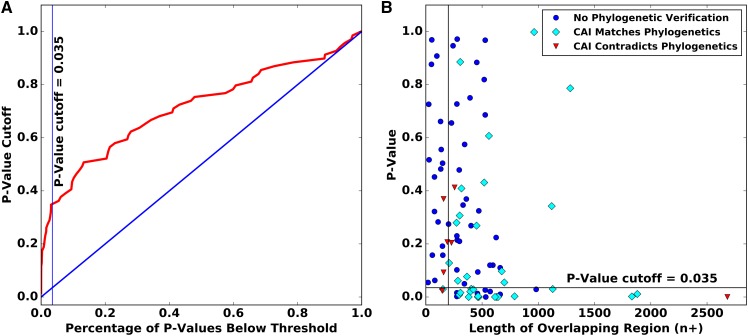
Within each overlapping pair, we provisionally classified the gene with the higher CAI value as ancestral and the gene with lower CAI value as novel. We then compared the two sets of relative adaptedness values using the wilcox.test function in R to perform a Mann-Whitney U Test. We chose a P-value cutoff of 0.035 after analyzing a receiver operating characteristic (ROC) plot (Figure 2A). The combined effects of our length threshold and P-value cutoff are illustrated in Figure 2B. In total, 27 gene pairs were determined to have statistically-significant CAI values, 19 of which had also been classified via phylogenetic analysis.
Figure 2.
Statistical classification of relative ages. (A) The receiver operating characteristic plot for determining which member of an overlapping gene pair has higher CAI, and is hence presumed to be ancestral. Only genes with an overlapping region of at least 200 nucleotides are plotted. (B) CAI classification of the 91 gene pairs for which codon usage data were available was based both on P-value and on the length of the overlapping regions. The vertical line shows the overlapping length cutoff of 200 nucleotides, the horizontal line shows the P-value cutoff; CAI classification was considered informative for the 27 bottom right points.
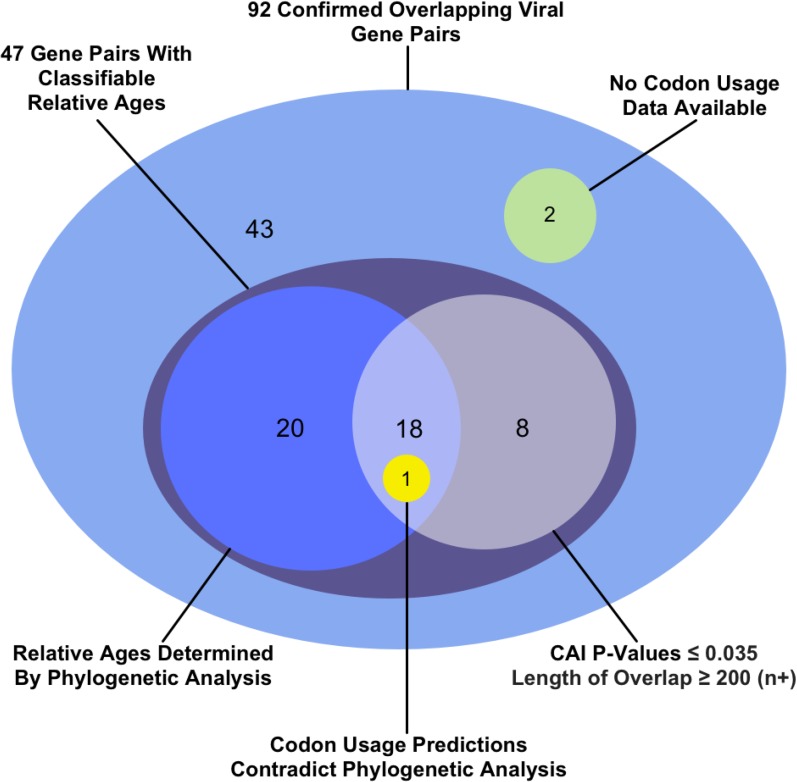
Of the gene pairs whose ancestral vs. novel classification were obtained both by statistically significant CAI differences and by phylogenetics, there was one for which the CAI classification contradicted the phylogenetics. That exception was the p104/p130 overlap in the Providence virus. This overlap is notable because the ancestral member of the pair was acquired through horizontal gene transfer, which renders codon usage an unreliable predictor of relative gene ages (Pavesi et al. 2013). We therefore used the phylogenetic classification and disregarded the CAI results. In total, we were able to classify ancestral vs. novel status for 47 overlapping gene pairs (Figure 3).
Figure 3.
How the relative ages of the genes were classified for 47 out of the 92 overlapping gene pairs for which sequence data were available. Within each shaded region, each gene pair is counted within only one of the subregions shown. Each shaded region’s total is found by summing the individual subtotals within it, some of which are noted outside the shaded regions. For example, the relative ages of 39 genes were classified via phylogenetic analysis: 20 through phylogenetic analysis alone (blue circle), 18 supported via codon analysis (intersection of blue and white circle), and one contradicted by codon analysis (yellow circle).
Nonoverlapping and preoverlapping controls
Both nonoverlapping genes and preoverlapping genes were used as controls; 150 nonoverlapping genes were compiled from the viral species in which the 92 overlapping gene pairs were found. Matching for species helps control for %GC content or other idiosyncrasies of nucleotide composition.
Of the 47 overlapping gene pairs for which we could assign relative ages, we were able to locate preoverlapping homologs for 27 of the ancestral genes in our dataset in the literature (Sabath et al. 2012) and/or NCBI (BLAST search with E-value threshold ).
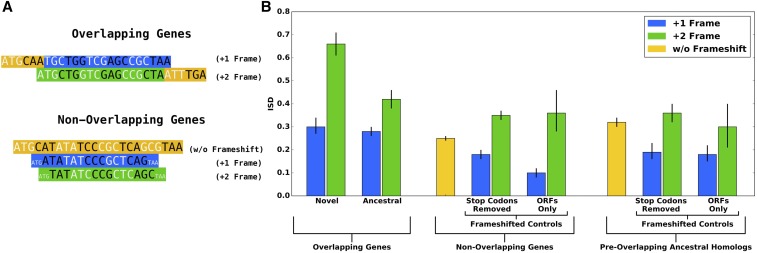
Frameshifting these control sequences was performed in two ways. First, removing one or two nucleotides immediately after the start codon and two or one nucleotides immediately before the stop codon generated +1 and +2 frameshifted non-ORF controls, respectively (Figure 4A). Any stop codons that appeared in the new reading frames were removed.
Figure 4.
ISD results support the birth-facilitation and (preadaptation version of the) conflict resolution hypotheses. (A) Data are from the overlapping sections of the 47 gene pairs whose ages could be classified, from nonoverlapping genes in the species in which those 47 overlapping gene pairs were found, and from the 27 available preoverlapping ancestral homologs. Some numbers in the main text are for the full set of 92 overlapping gene pairs and might not match exactly. (B) While frame significantly impacts disorder content (green is consistently higher than blue), it does not drive the high ISD of overlapping genes. Even controlling for frame, novel ISD ancestral (left), supporting birth facilitation. Supporting conflict resolution, ancestral nonoverlapping either inframe (yellow) or matched frameshifted control. The preadaptation version of the constraint hypothesis is supported by the fact that nonoverlapping ISD preoverlapping (compare the two yellow bars). Means and 66% confidence intervals were calculated from the Box-Cox transformed (with λ = 0.4) means and their SE, and are shown here following back-transformation.
Second, for the frameshifted ORF controls, we took only in situ ORFs within each of the two alternative reading frames. If multiple ORFs terminated at the same stop codon, we used only the longest. We excluded ORFs <25 amino acids in length (after removal of cysteines for analysis by IUPred). For nonoverlapping genes, this yielded 151 and 24 ORFs in the +1 and +2 reading frames, respectively. For the smaller set of preoverlapping genes, it yielded only 25 and 5 ORFs in the +1 and +2 frames, respectively.
Homology groups
Treating each gene as an independent datapoint is a form of pseudoreplication, because homologous genes can share properties via a common ancestor rather than via independent evolution. This problem of phylogenetic confounding can be corrected for by using gene family as a random effect term in a linear model (Wilson et al. 2017), and by counting each gene birth event only once.
We constructed a pHMMer (http://hmmer.org/) database including all overlapping regions, nonoverlapping genes and their frameshifted controls. After an all-against-all search, sequences that were identified as homologous, using an expectation value threshold of , were provisionally assigned the same homology group ID. These provisional groups were used to determine which gene birth events were unique. Two pairs were considered to come from the same gene birth event when both the ancestral and the overlapping sequence were classified as homologous. We also used published phylogenetic analysis to classify the TGBp2/TGBp3 overlap as two birth events (one occurring Virgaviridae, the other occurring in Alpha- and Betaflexiviridae) (Morozov and Solovyev 2003).
Some homologous pairs had such dissimilar protein sequences that ISD values were essentially independent. We therefore manually analyzed sequence similarity within each homology group using the Geneious (Kearse et al. 2012) aligner with free end gaps, using Blosum62 as the cost matrix. The percent similarity using the Blosum62 matrix with similarity threshold 1 was then used as the criterion for whether a protein sequence would remain in its homology group for the ISD analysis. We used protein sequence similarity as the threshold to assign a link between a pair, and then used single-link clustering to assign protein sequences to 561 distinct homology groups. Preoverlapping genes were then assigned to the homology groups of the corresponding ancestral genes.
ISD prediction
We used IUPred (Dosztányi et al. 2005) to calculate ISD values for each sequence. Following Wilson et al. (2017), before running IUPred, we excised all cysteines from each amino acid sequence, because of the uncertainty about their disulfide bond status and hence entropy (Uversky and Dunker 2010). Whether cysteine forms a disulfide bond depends on whether it is in an oxidizing or reducing environment. IUPred implicitly, through the selection of its training data set, assumes most cysteines are in disulfide bonds, which may or may not be accurate for our set of viral proteins. Because cysteines have large effects on ISD (in either direction) depending on disulfide status and hence introduce large inaccuracies, cysteines were dropped from consideration altogether.
IUPred assigns a score between 0 and 1 to each amino acid. To calculate the ISD of an overlapping region, IUPred was run on the complete protein (minus its cysteines), then the average score was taken across only the pertinent subset of amino acids.
Statistical models
Prior to fitting linear models, sequence-level ISD values were transformed using a Box-Cox transform. The optimal value of λ for the combined ancestral, novel and artificially-frameshifted nonoverlapping, non-ORF control group data was 0.41, which we rounded to 0.4 and used throughout all linear models, and for central tendency and confidence intervals in the figures. Simple means and SE are described inline in the text.
We used a multiple regression approach to determine which factors predict ISD values (Sokal and Rohlf 1994). Gene designation (ancestral vs. novel vs. one or more nongenic controls) and relative reading frame (+1 vs. +2) were used as fixed effects. Homologous sequences are not independent; we accounted for this by using a linear mixed model (Oberg and Mahoney 2007), with homology group as a random effect. Species is a stand-in for a number of confounding factors, e.g., %GC content, and so was included as a second random effect. The data were normalized using a Box-Cox transformation prior to analysis. Pairwise comparisons discussed throughout the Results were performed using contrast statements applied to the linear model, using the minimum number of gene designations necessary to make the comparison in question.
We used the lmer and gls functions contained in the nlme and lme4 R packages to generate the linear mixed models. The main model used to calculate the relative effect sizes used frameshifted non-ORF nonoverlapping genes as the nongenic control. In this model, frame, gene designation, species, and homology group terms were retained in the model, with and respectively. All four terms were also easily retained in all other model variants exploring different nongenic controls.
We also considered overlap type (internal vs. terminal) as a fixed effect, but removed it because it did not significantly enhance our model (). To determine the statistical significance of each effect, we used the ANOVA function in R to compare nested models.
Data availability
Scripts and data tables used in this work may be accessed at: https://github.com/MaselLab/Willis_Masel_Overlapping_Genes_Structural_Disorder_Explained.
Results
Confirming elevated ISD in our dataset
Because most verified gene overlaps in the literature, especially longer overlapping sequences, are in viruses (Veeramachaneni et al. 2004; Nakayama et al. 2007; Rancurel et al. 2009), we focused on viral genomes, compiling a list of 92 verified overlapping gene pairs from 80 viral species. The mean predicted ISD of all overlapping regions () was higher than that of the nonoverlapping genes (), confirming previous findings that overlapping genes have elevated ISD.
Frame affects ISD
We artificially frameshifted 150 nonoverlapping viral genes in those 80 species, and found higher ISD in the +2 reading frame () than in the +1 reading frame () for non-ORF controls, and an even more extreme difference for ORF controls ( vs. ). The artifact hypothesis predicts that the +1 and +2 members of the 92 verified overlapping gene pairs will follow suit. In agreement with this, the overlapping regions of genes in the +2 reading frame had higher mean ISD () than those in the +1 reading frame (). While this provides strong evidence that frame shapes ISD as an artifact of the genetic code, average ISD across both ways of frameshifting nonoverlapping genes ( and for non-ORF and ORF frameshifted nonoverlapping genes, respectively) is significantly lower than the ISD of all overlapping sequences (), showing that the artifact hypothesis cannot fully explain elevated ISD in the latter.
The birth-facilitation hypothesis is supported
We find stronger support for the birth-facilitation hypothesis. Of the 92 verified overlapping viral gene pairs, we were able to classify the relative ages of the component genes as ancestral vs. novel for 47 pairs (Table 1). In agreement with the predictions of the birth-facilitation process, and controlling for frame, novel genes have higher ISD than either ancestral members of the same gene pairs or artificially-frameshifted controls (Figure 4B). We confirmed this using a linear mixed model, with frame (+1 vs. +2) as a fixed effect, gene type (novel vs. ancestral) as a fixed effect, species (to control for %GC content and other subtle sequence biases) as a random effect, and homology group (to control for phylogenetic confounding) as a random effect. Within this linear model, the prediction unique to the birth-facilitation hypothesis, namely that ISD in the overlapping regions of novel genes is higher than that in ancestral genes, is supported with . Inspection of Figure 4B suggests that elevation of novel gene ISD above ancestral is specific to the +2 frame; this is confirmed in the analysis of Figure 5B. Running separate statistical models for the two frames, the +2 frame difference is supported with .
Table 1. Gene pairs for which the relative ages could be determined.
| Accession number | Organism | Ancestral gene | Novel gene | Overlap length (n+) | Novel frame |
|---|---|---|---|---|---|
| NC_001401 | Adeno-associated virus 2 | VP2 | AAP | 615 | +1 |
| NC_004285 | Aedes albopictus densovirus | NS1 | NS2 | 1119 | +1 |
| NC_001467 | African cassava mosaic virus | AL1 | AC4 | 423 | +1 |
| NC_009896 | Akabane virusa | N | NSs | 276 | +1 |
| NC_001749 | Apple stem grooving virus | MP | Polyprotein | 963 | +1 |
| NC_001719 | Arctic ground squirrel hepatitis virusb | P | L | 1284 | +1 |
| NC_003481 | Barley stripe mosaic virusc,d,e | TGBp2 | TGBp3 | 191 | +1 |
| NC_003680 | Barley yellow dwarf virusf,g | P5 | MP | 465 | +1 |
| NC_005041 | Blattella germanica densovirus | NS-1 | ORF4 | 789 | +1 |
| NC_001927 | Bunyamwera virusa | N | NSs | 306 | +1 |
| NC_001658 | Cassava common mosaic virusd,e | TGBp2 | TGBp3 | 152 | +1 |
| NC_001427 | Chicken anemia virus | VP2 | Apoptin | 366 | +1 |
| NC_003688 | Cucurbit aphid-born yellowing virusif,g,h | CP | P5 | 572 | +1 |
| NC_005899 | Dendrolimus punctatus Tetravirusf | p71 | p17 | 381 | +1 |
| NC_016561 | hepatitis Bb | P | L | 1128 | +1 |
| NC_003608 | Hibiscus chlorotic ringspot virusf | Coat | p25 | 675 | +1 |
| NC_003608 | Hibiscus chlorotic ringspot virus | Replicase | p23 | 630 | +1 |
| NC_004730 | Indian peanut clump virus | P14 | P17 | 158 | +1 |
| KR732417 | Influenza A virus H5N1 | PB1 | PB1-F2 | 273 | +1 |
| NC_009025 | Israel acute paralysis virus of bees | ORF2 | ORFx | 285 | +1 |
| NC_003627 | Maize chlorotic mottle virus | Coat | 31P | 451 | +1 |
| NC_001498 | Measles virusi | P | C | 561 | +1 |
| NC_005339 | Mossman virusi | P | C | 459 | +1 |
| NC_008311 | Murine norovirus | VP1 | VF1 | 642 | +1 |
| NC_001633 | Mushroom bacilliform virus | ORF1 | Vpg-protease | 533 | +1 |
| NC_001718 | Porcine parvovirus | Capsid | SAT | 207 | +1 |
| NC_001747 | Potato leafroll virus | P0 | P1 | 661 | +1 |
| NC_003725 | Potato mop-top virusc,d | TGBp2 | TGBp2 | 146 | +1 |
| NC_003768 | Rice dwarf virus | Pns12 | OP-ORF | 276 | +1 |
| NC_003771 | Rice ragged stunt virus | P4b | Replicase | 981 | +1 |
| NC_004718 | SARS coronavirus | Nucleocapsid | Protein I | 297 | +1 |
| NC_003809 | Spinach latent virus | Replicase | 2b | 308 | +1 |
| NC_003448 | Striped Jack nervous necrosis virus | Protein A | B2 | 228 | +1 |
| NC_001366 | Theiler’s virus | L | L* | 471 | +1 |
| NC_002199 | Tupaia paramyxovirusi | P | C | 462 | +1 |
| NC_003743 | Turnip yellows virusf,g,h | CP | ORF5 | 528 | +1 |
| NC_001409 | Apple chlorotic leaf spot virus | CP | MP | 317 | +2 |
| NC_001719 | Arctic ground squirrel hepatitis virus | P | Capsid Precursor | 158 | +2 |
| NC_001719 | Arctic ground squirrel hepatitis virus | P | X | 256 | +2 |
| NC_003532 | Cymbidium ringspot virus | MP | p19 | 519 | +2 |
| NC_003093 | Indian citrus ringspot virus | CP | NABP | 301 | +2 |
| NC_004178 | Infectious bursal disease virusj | VP2 | VP5 | 404 | +2 |
| NC_001915 | Infectious pancreatic necrosis virusj | VP2 | VP5 | 395 | +2 |
| NC_001990 | Nudaurelia capensis beta virusf | CP | Replicase | 1832 | +2 |
| NC_014126 | Providence virus | p104 | p130 | 2681 | +2 |
| NC_004366 | Tobacco bushy top virus | MP | RNP | 698 | +2 |
| NC_004063 | Turnip yellows mosaic virus | Replicase | MP | 1880 | +2 |
Genes are phylogenetically independent except as noted in the footnotes.
N/NSs overlaps share sequence similarity.
P/L overlap predicted homologous in HMMer run.
TGBp2 genes share protein sequence similarity.
TGBp2 genes predicted homologous Morozov and Solovyev (2003).
TGBp3 genes predicted homologous Morozov and Solovyev (2003).
Ancestral genes predicted homologous in HMMer run.
Novel genes predicted homologous in HMMer run.
Novel genes predicted homologous in HMMer run.
Novel genes predicted homologous in HMMer run.
Ancestral VP2 genes share protein sequence similarity.
Figure 5.
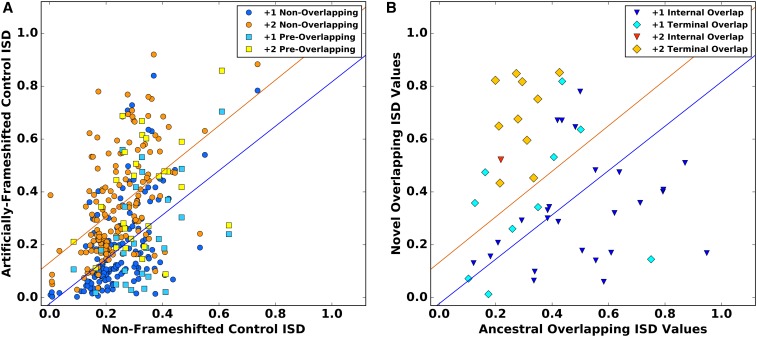
(A) The ISD of a nonoverlapping gene predicts the ISD of the non-ORF frameshifted version of that sequence. Model I regression lines are statistically indistiguishable in our set of 150 nonoverlapping viral genes vs. our set of 27 preoverlapping genes, which are therefore pooled ( and for the +1 and +2 groups, respectively). (B) The overlapping ISD of the 47 gene pairs with classifiable relative ages. Each datapoint represents one overlapping pair. The regression lines from (A) are superimposed to illustrate the elevation of novel gene ISD born into the already intrinsically high-ISD +2 frame, which destroys the correlation ( for +2 in contrast to 0.26 for +1).
These non-ORF frameshifted control sequences do not take into account the fact that ORFs vary in their propensity to appear and disappear, and that this can shape the material available for de novo gene birth, including ISD values (Nielly-Thibault and Landry 2018). However, we did not observe this affecting ISD in our data set. In a linear model comparing ORF vs. non-ORF nonoverlapping control sequences, and one comparing ORF vs. non-ORF preoverlapping controls sequences, there was no significant difference between the two controls ( in both cases, with frame included as a fixed effect, and gene as a random effect). This justifies a focus on the larger non-ORF control dataset, as well as excluding this nonadaptive force as a driver of the birth facilitation hypothesis.
The preadaptation version of the conflict-resolution hypothesis is supported
In agreement with the conflict resolution hypothesis, ancestral overlapping sequences, not just novel ones, have higher ISD than nonoverlapping genes ( vs. ; ). This seems to be because ISD was already high at the time of de novo gene birth; today’s ancestral overlapping genes have indistinguishable ISD from their preoverlapping homologs (), while those preoverlapping homologs have higher ISD than nonoverlapping genes (P = ) (Figure 4B).
It is possible that the overlapping gene pairs for which we are able to identify preoverlapping homologs are significantly younger than other overlapping gene pairs, and that there might therefore simply not have been enough time for the ancestral members of the pair to evolve higher ISD. Contradicting this, these 27 ancestral overlapping genes have indistinguishable ISD from the other 20 ancestral overlapping gene (P = 0.2, controlling for frame and homology group).
The artifact hypothesis is rejected
Nonoverlapping gene ISD is not statistically different from the mean of the +1 and +2 artificially-frameshifted control versions of the same nonoverlapping nucleotide sequences ( contrast statement applied to a linear model with fixed effect of actual nonoverlapping gene sequence vs. +1 artificially-frameshifted version vs. +2 artificially-frameshifted version, and gene identity as a random effect). In other words, despite the enormous effect of +1 vs. +2 reading frame, we find no support for the artifact hypothesis in explaining the elevated ISD of overlapping regions. In each overlapping gene pair, there is always exactly one gene in each of the two reading frames, such that the large effects of each of the two frames cancel each other out when all overlapping genes are considered together. It is nevertheless important to control for the large effect of frame while testing and quantifying other hypotheses.
The relative magnitude of each hypothesis
We calculated the degree to which birth facilitation elevates ISD using a contrast statement, as half the difference between novel and ancestral genes, because exactly half of the genes are novel, and hence elevated above the “normal” ISD level of ancestral genes. By this calculation, birth facilitation accounts for of the estimated total difference in ISD between overlapping and nonoverlapping genes.
Note that frameshifted versions of high-ISD proteins have higher ISD than frameshifted versions of low-ISD proteins (Figure 5A). The two members of an overlapping pair share the same %GC content, and random sequences with higher %GC have substantially higher ISD (Ángyán et al. 2012), so this could be responsible for the trait correlation. A facilitate-birth bias toward high ISD in newborn genes might do so in part via high %GC in the overlapping region at the time of birth, causing overlapping sequences to be biased not just toward high-ISD novel genes, but also toward high-ISD ancestral genes. Our estimate attributes all of the ISD elevation in ancestral overlapping genes to constraint, but given the trait correlation shown in Figure 5A, some of this might also be due to birth facilitation, making an underestimate. Note that novel genes born into the +2 frame have high ISD above and beyond the intrinsic correlation (Figure 5B), and are thus responsible for the statistical difference between ancestral vs. novel when frame is controlled for (Figure 4B).
Mutation bias is responsible for more births in the low-ISD +1 frame
Given the strong influence of frame combined with support for the facilitate-birth hypothesis, we hypothesized that novel genes would be born more often into the +2 frame (Figure 4A, green) because the intrinsically higher ISD of the +2 reading frame would facilitate high ISD in the novel gene and hence birth. Our dataset contained 41 phylogenetically independent overlapping pairs. Surprisingly, we found the opposite of our prediction: 31 of the novel genes were in the +1 frame of their ancestral counterparts, while only 10 were in the +2 frame ( cumulative binomial distribution with trial success probability 0.5).
This unexpected result is stronger for “internal overlaps,” in which one gene is completely contained within its overlapping partner (23 +1 events vs. 1 +2 event, ), and is not found for “terminal overlaps,” in which the end of the downstream gene overlaps with the end of the upstream member of the pair (9 +1 events vs. 9 +2 events). (This double-counts a +1 event for which there were three homologous gene pairs, two of which were internal overlaps, and one of which was a terminal overlap.) Following Belshaw et al. (2007), we interpret the restriction of this finding to internal overlaps as evidence that the cause of the bias applies to complete de novo gene birth, but not to the addition of a sequence to an existing gene.
The unexpected prevalence of +1 gene births, despite birth facilitation favoring +2, can be explained by mutation bias. One artifact of the genetic code is that +1 frameshifts yield more start codons and fewer stop codons, and hence more and longer ORFs (Belshaw et al. 2007). In our control set of 150 nonoverlapping viral genes, we confirm that stop codons are more prevalent in the +2 frame (1 per 11 codons) than the +1 frame (1 per 14), decreasing the mean ORF length, and that start codons are more prevalent in the +1 frame (1 per 27 codons) than the +2 frame (1 per 111). Similar results were found in the preoverlapping ancestral homologs, with more start codons in the +1 frame (1 per 33) than the +2 frame (1 per 169), and fewer stop codons in the +1 frame (1 per 20) than the +2 frame (1 per 13).
This is reflected in the relative numbers and sizes of our frameshifted ORF controls. Prior to implementing a minimum length requirement (see Materials and Methods), we found 465 ORFs in the +1 frame of our nonoverlapping genes, with a mean and maximum length of 24 and 149 amino acids, respectively, while only 92 ORFs were found in the +2 frame with a mean and maximum length of 19 and 92 amino acids. The same pattern was found in the preoverlapping ancestral homologs, with 65 ORFs found in the +1 frame with a mean and maximum length of 36 and 315 amino acids, vs. 13 ORFs in the +2 frame with a mean and maximum length of 36 and 173 amino acids.
Discussion
There is growing interest in the topic of de novo gene birth, but identifying de novo genes is plagued with high rates of both false positives and false negatives (McLysaght and Hurst 2016), with phylostratigraphy tools being particularly controversial due to homology detection biases (Moyers and Zhang 2017). The overlapping viral genes that we study are unlikely either to be nongenes, and must have arisen via de novo gene birth, and so circumvent many of these difficulties. Carvunis et al. (2012) have disputed that young genes have high ISD, in an analysis that was prone to false positives (Wilson et al. 2017); our findings here provide an independent line of evidence, free from the danger of homology detection bias, that younger genes have higher ISD.
The study of overlapping genes has of course its own statistical traps. In particular, the preponderance of novel genes in the +1 frame demonstrates the need to control for the strong effects of frame when testing hypotheses. Ancestral genes are more frequently in the high-ISD +2 frame, while the depressed ISD of the +1 frame lowers the ISD of the novel. As a result, when frame is not considered, ancestral and novel overlapping sequences encode very similar levels of disorder ( vs. respectively), making it easy to miss the evidence for the facilitate-birth hypothesis.
More broadly, our results are consistent with a major role for mutational availability in shaping adaptive evolution. Rare adaptive changes happen at a rate given by the product of mutation and the probability of fixation, with the latter approximately proportional to the selection coefficient (McCandlish and Stoltzfus 2014). This means that differences in the beneficial mutation rate are just as important as differences in the selection coefficient in determining which path adaptive evolution takes (Yampolsky and Stoltzfus 2001). The influence of mutational bias has previously been observed for beneficial mutations to single amino acids in the laboratory (Stoltzfus and McCandlish 2015, 2017; Sackman et al. 2017) and in the wild (Stoltzfus and McCandlish 2017; Zhu et al. 2018). Here, we demonstrate it for more radical mutations, namely the de novo birth of entire protein-coding genes.
Acknowledgments
We thank Rafik Neme and Matt Cordes for comments on an earlier draft of this manuscript, Scott Foy for sharing his IUPred interface script, David Karlin for alerting us to issues annotating the relative ages of TGBp2 and TGBp3, and Arlin Stolzfus for pointers to the mutation-driven evolution literature. This work was supported by the John Templeton Foundation (39667, 60814), and the National Institutes of Health (GM104040).
Footnotes
Communicating editor: D. Begun
Literature Cited
- Ángyán A. F., Perczel A., Gáspári Z., 2012. Estimating intrinsic structural preferences of de novo emerging random-sequence proteins: is aggregation the main bottleneck? FEBS Lett. 586: 2468–2472. 10.1016/j.febslet.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Belshaw R., Pybus O. G., Rambaut A., 2007. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 17: 1496–1504. 10.1101/gr.6305707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E., Alba M. M., 2013. Dynamics and adaptive benefits of modular protein evolution. Curr. Opin. Struct. Biol. 23: 459–466. 10.1016/j.sbi.2013.02.012 [DOI] [PubMed] [Google Scholar]
- Brown C. J., Takayama S., Campen A. M., Vise P., Marshall T. W., et al. , 2002. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 55: 104–110. 10.1007/s00239-001-2309-6 [DOI] [PubMed] [Google Scholar]
- Buljan M., Frankish A., Bateman A., 2010. Quantifying the mechanisms of domain gain in animal proteins. Genome Biol. 11: R74 10.1186/gb-2010-11-7-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillo-Balderas J. A., Lazcano A., Becerra A., 2015. Viral genome size distribution does not correlate with the antiquity of the host lineages. Front. Ecol. Evol. 3: 143 10.3389/fevo.2015.00143 [DOI] [Google Scholar]
- Carter J. J., Daugherty M. D., Qi X., Bheda-Malge A., Wipf G. C., et al. , 2013. Identification of an overprinting gene in merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc. Natl. Acad. Sci. USA 110: 12744–12749. 10.1073/pnas.1303526110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A.-R., Rolland T., Wapinski I., Calderwood M. A., Yildirim M. A., et al. , 2012. Proto-genes and de novo gene birth. Nature 487: 370–374. 10.1038/nature11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico N., Vianelli A., Belshaw R., 2010. Why genes overlap in viruses. Proc. R. Soc. Lond. B Biol. Sci. 277: 3809–3817. 10.1098/rspb.2010.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.-Y., Wadhawan S., Szklarczyk R., Pond S. K., Nekrutenko A., 2007. A first look at ARFome: dual-coding genes in mammalian genomes. PLOS Comput. Biol. 3: e91 10.1371/journal.pcbi.0030091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z., Csizmók V., Tompa P., Simon I., 2005. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347: 827–839. 10.1016/j.jmb.2005.01.071 [DOI] [PubMed] [Google Scholar]
- Echave J., Spielman S. J., Wilke C. O., 2016. Causes of evolutionary rate variation among protein sites. Nat. Rev. Genet. 17: 109–121. 10.1038/nrg.2015.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman D., Elofsson A., 2010. Identifying and quantifying orphan protein sequences in fungi. J. Mol. Biol. 396: 396–405. 10.1016/j.jmb.2009.11.053 [DOI] [PubMed] [Google Scholar]
- Fernandes J. D., Faust T. B., Strauli N. B., Smith C., Crosby D. C., et al. , 2016. Functional segregation of overlapping genes in HIV. Cell 167: 1762–1773.e12. 10.1016/j.cell.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Longhi S., Canard B., Karlin D., 2006. A practical overview of protein disorder prediction methods. Proteins 65: 1–14. 10.1002/prot.21075 [DOI] [PubMed] [Google Scholar]
- Foy S. G., Wilson B. A., Cordes M. H., Masel J., 2017. Progressively more subtle aggregation avoidance strategies mark a long-term direction to protein evolution. bioRxiv 176867. https//.org/10.1101/176867. [Google Scholar]
- Graur D., 2016. Molecular and Genome Evolution, pp. 140–141, Ed. 1 Sinauer Associates Inc., Sunderland, MA. [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., et al. , 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese P. K., Gibbs A., 1992. Origins of genes: “big bang” or continuous creation? Proc. Natl. Acad. Sci. USA 89: 9489–9493. 10.1073/pnas.89.20.9489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-S., Cho C.-Y., Huh J.-W., Kim H.-S., Cho H.-G., 2008. Evog: a database for evolutionary analysis of overlapping genes. Nucleic Acids Res. 37: D698–D702. 10.1093/nar/gkn813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E., Tompa P., Liliom K., Kalmar L., 2010. Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc. Natl. Acad. Sci. USA 107: 5429–5434. 10.1073/pnas.0907841107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Huang Y., 2014. Advantages of proteins being disordered. Protein Sci. 23: 539–550. 10.1002/pro.2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandlish D. M., Stoltzfus A., 2014. Modeling evolution using the probability of fixation: history and implications. Q. Rev. Biol. 89: 225–252. 10.1086/677571 [DOI] [PubMed] [Google Scholar]
- McLysaght A., Hurst L. D., 2016. Open questions in the study of de novo genes: what, how and why. Nat. Rev. Genet. 17: 567–578. 10.1038/nrg.2016.78 [DOI] [PubMed] [Google Scholar]
- Miller A. D., 1997. Principles of retroviral vector design in Retroviruses, edited by Coffin J., Hughes S., Varmus H. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Mizokami M., Orito E., Ohba K.-i., Ikeo K., Lau J. Y., et al. , 1997. Constrained evolution with respect to gene overlap of hepatitis B virus. J. Mol. Evol. 44: S83–S90. 10.1007/PL00000061 [DOI] [PubMed] [Google Scholar]
- Moore A. D., Bornberg-Bauer E., 2011. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 29: 787–796. 10.1093/molbev/msr250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov S. Y., Solovyev A. G., 2003. Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84: 1351–1366. 10.1099/vir.0.18922-0 [DOI] [PubMed] [Google Scholar]
- Moyers B. A., Zhang J., 2017. Further simulations and analyses demonstrate open problems of phylostratigraphy. Genome Biol. Evol. 9: 1519–1527. 10.1093/gbe/evx109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Gojobori T., Ikemura T., 2000. Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28: 292 10.1093/nar/28.1.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Asai S., Takahashi Y., Maekawa O., Kasama Y., 2007. Overlapping of genes in the human genome. Int. J. Biomed. Sci. 3: 14–19. [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A., Wadhawan S., Goetting-Minesky P., Makova K. D., 2005. Oscillating evolution of a mammalian locus with overlapping reading frames: an XLαs/ALEX relay. PLoS Genet. 1: e18 10.1371/journal.pgen.0010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R., Tautz D., 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14: 117 10.1186/1471-2164-14-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielly-Thibault L., Landry C. R., 2018. Differences between the de novo proteome and its non-functional precursor can result from neutral constraints on its birth process, not necessarily from natural selection alone. bioRxiv 289330. https//.org/10.1101/289330. [Google Scholar]
- Oberg A. L., Mahoney D. W., 2007. Linear mixed effects models, pp. 213–234 in Topics in Biostatistics. Springer, Totowa, NJ: 10.1007/978-1-59745-530-5_11 [DOI] [PubMed] [Google Scholar]
- Pavesi A., Magiorkinis G., Karlin D. G., 2013. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of deltaretroviruses. PLOS Comput. Biol. 9: e1003162 10.1371/journal.pcbi.1003162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancurel C., Khosravi M., Dunker A. K., Romero P. R., Karlin D., 2009. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 83: 10719–10736. 10.1128/JVI.00595-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribrioux S., Brüngger A., Baumgarten B., Seuwen K., John M. R., 2008. Bioinformatics prediction of overlapping frameshifted translation products in mammalian transcripts. BMC Genomics 9: 122 10.1186/1471-2164-9-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N., Graur D., Landan G., 2008a Same-strand overlapping genes in bacteria: compositional determinants of phase bias. Biol. Direct 3: 36 10.1186/1745-6150-3-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N., Landan G., Graur D., 2008b A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS One 3: e3996 10.1371/journal.pone.0003996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N., Wagner A., Karlin D., 2012. Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 29: 3767–3780. 10.1093/molbev/mss179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackman A. M., McGee L. W., Morrison A. J., Pierce J., Anisman J., et al. , 2017. Mutation-driven parallel evolution during viral adaptation. Mol. Biol. Evol. 34: 3243–3253. 10.1093/molbev/msx257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Hilgenfeld R., 2015. Acquisition of new protein domains by coronaviruses: analysis of overlapping genes coding for proteins N and 9b in SARS coronavirus. Virus Genes 50: 29–38. 10.1007/s11262-014-1139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E. C., Pagán I., 2013. The effect of gene overlapping on the rate of RNA virus evolution. Mol. Biol. Evol. 30: 1916–1928. 10.1093/molbev/mst094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R., Rohlf J., 1994. Biometry, Ed. 3 W. H. Freeman, New York. [Google Scholar]
- Stoltzfus A., McCandlish D. M., 2015. Mutation-biased adaptation in Andean house wrens. Proc. Natl. Acad. Sci. USA 112: 13753–13754. 10.1073/pnas.1518490112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A., McCandlish D. M., 2017. Mutational biases influence parallel adaptation. Mol. Biol. Evol. 34: 2163–2172. 10.1093/molbev/msx180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N., Oldfield C. J., Uversky V. N., Berezovsky I. N., Tawfik D. S., 2009. Do viral proteins possess unique biophysical features? Trends Biochem. Sci. 34: 53–59. 10.1016/j.tibs.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Tretyachenko V., Vymětal J., Bednárová L., Kopeckỳ V., Hofbauerová K., et al. , 2017. Random protein sequences can form defined secondary structures and are well-tolerated in vivo. Sci. Rep. 7: 15449 10.1038/s41598-017-15635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N., Dunker A. K., 2010. Understanding protein non-folding. Biochim. Biophys. Acta 1804: 1231–1264. 10.1016/j.bbapap.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni V., Makalowski W., Galdzicki M., Sood R., Makalowska I., 2004. Mammalian overlapping genes: the comparative perspective. Genome Res. 14: 280–286. 10.1101/gr.1590904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y., 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56: 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. A., Foy S. G., Neme R., Masel J., 2017. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat. Ecol. Evol. 1: 0146 10.1038/s41559-017-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Blocquel D., Habchi J., Uversky A. V., Kurgan L., et al. , 2014. Structural disorder in viral proteins. Chem. Rev. 114: 6880–6911. 10.1021/cr4005692 [DOI] [PubMed] [Google Scholar]
- Yampolsky L. Y., Stoltzfus A., 2001. Bias in the introduction of variation as an orienting factor in evolution. Evol. Dev. 3: 73–83. 10.1046/j.1525-142x.2001.003002073.x [DOI] [PubMed] [Google Scholar]
- Zhou T., Gu W., Ma J., Sun X., Lu Z., 2005. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems 81: 77–86. 10.1016/j.biosystems.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Zhu X., Guan Y., Signore A. V., Natarajan C., DuBay S. G., et al. , 2018. Divergent and parallel routes of biochemical adaptation in high-altitude passerine birds from the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. USA 115: 1865–1870. 10.1073/pnas.1720487115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Scripts and data tables used in this work may be accessed at: https://github.com/MaselLab/Willis_Masel_Overlapping_Genes_Structural_Disorder_Explained.