Organisms respond to environmental conditions by altering gene expression; however, it is unclear if organisms retain epigenetic memory of their ancestors’ environmental conditions. Webster et al. assessed the descendants of two genetically identical...
Keywords: dauer, diapause, starvation, transgenerational, plasticity, epigenetic
Abstract
Phenotypic plasticity is facilitated by epigenetic regulation, and remnants of such regulation may persist after plasticity-inducing cues are gone. However, the relationship between plasticity and transgenerational epigenetic memory is not understood. Dauer diapause in Caenorhabditis elegans provides an opportunity to determine how a plastic response to the early-life environment affects traits later in life and in subsequent generations. We report that, after extended diapause, postdauer worms initially exhibit reduced reproductive success and greater interindividual variation. In contrast, F3 progeny of postdauers display increased starvation resistance and lifespan, revealing potentially adaptive transgenerational effects. Transgenerational effects are dependent on the duration of diapause, indicating an effect of extended starvation. In agreement, RNA-seq demonstrates a transgenerational effect on nutrient-responsive genes. Further, postdauer F3 progeny exhibit reduced gene expression plasticity, suggesting a trade-off between plasticity and epigenetic memory. This work reveals complex effects of nutrient stress over different time scales in an animal that evolved to thrive in feast and famine.
EPIGENETIC regulation mediates phenotypic plasticity, such that a single genotype can produce different phenotypes in response to environmental conditions (Kelly et al. 2012). Most epigenetic modifications are reset at the beginning of each generation, but those that persist have the potential to impact environmental adaptation and evolution independent of DNA sequence (Rapp and Wendel 2005; Jablonka and Raz 2009; Heard and Martienssen 2014). Consequently, there is substantial interest in identifying environmental perturbations that elicit transgenerational epigenetic effects. Epigenetic inheritance has been reported in the roundworm Caenorhabditis elegans, but these studies typically focus on an aphysiological stimulus, such as a mutation or exogenous RNAi trigger (Greer et al. 2011; Buckley et al. 2012), or they assay a molecular rather than organismal phenotype, such as gene expression (Schott et al. 2014; Ni et al. 2016; Klosin et al. 2017; Minkina and Hunter 2017). Consequently, the importance of epigenetic inheritance to environmental adaptation is unclear. Identification and characterization of plasticity-induced transgenerational effects manifest at the organismal level can address important questions. Do epigenetic changes in gene regulation affect organismal traits? Does epigenetic memory provide a predictive adaptive response that improves fitness? Alternatively, does epigenetic memory arise due to a failure to reset the epigenetic program between generations, potentially posing a fitness cost?
C. elegans has a variety of developmental responses to nutrient availability (Angelo and Van Gilst 2009; Baugh 2013; Schindler et al. 2014; Schulenburg and Felix 2017). In particular, larvae respond to specific environmental cues (high population density, low food availability, and high temperature) and undergo an alternative developmental program resulting in dauer arrest (Hu 2007). Dauer arrest is a classic example of diapause since dauer larvae develop in response to cues perceived in advance of arrest (Kostal 2006). Dramatic morphological and gene regulatory changes occur during dauer development (Wang and Kim 2003; McElwee et al. 2004; Baugh et al. 2011) under the regulation of highly conserved signaling pathways (Ren et al. 1996; Schackwitz et al. 1996; Li et al. 2003; Bargmann 2006; Fielenbach and Antebi 2008). Dauers have a thicker cuticle and plugged pharynx, supporting increased stress resistance but preventing feeding. Dauers also have an altered nervous system, gut, and muscles compared to nondauers (Albert and Riddle 1983; Keane and Avery 2003; Dixon et al. 2008). They are provisioned with fat stores that aid survival during prolonged starvation (Cunningham and Ashrafi 2009; Narbonne and Roy 2009). If environmental conditions improve, larvae exit dauer arrest and ultimately become reproductive adults.
C. elegans collected from the wild are mostly in the dauer stage, providing evidence that starvation stress during dauer diapause is a common feature of their life history (Barriere and Felix 2005). Time spent in dauer does not alter the length of the adult lifespan, suggesting an “ageless” state (Klass and Hirsh 1976). Nonetheless, dauer larvae do eventually die, with about one-third failing to recover at 60 days (Klass and Hirsh 1976). Further, germline defects increase in frequency as the duration of dauer diapause increases (Kim and Paik 2008), suggesting that at least some consequences of starvation incurred during diapause persist in postdauers. It is currently unknown if effects of long-term dauer diapause persist upon recovery to affect important life-history traits, including progeny quality and starvation resistance. The consequences of long-term dauer diapause beyond one generation are, to our knowledge, completely unexplored.
Here, we report the consequences of long-term dauer diapause over multiple generations. We found that long-term dauers recovered to produce fewer, smaller, and starvation-sensitive F1 progeny that exhibited greater interindividual variation, suggesting proximal fitness costs. In contrast, great-grandprogeny (F3 generation) of long-term dauers exhibited increased starvation resistance and lifespan, consistent with potentially adaptive transgenerational effects. Increased starvation resistance was dependent on the time spent in the ancestral dauer diapause, suggesting that it is starvation during dauer, rather than dauer development itself, that leads to these transgenerational effects. Consistent with this, we identified epigenetic differences in nutrient-responsive gene expression in the F3 generation including a dampened response to nutrient availability. These results indicate that ancestral environment can influence how worms respond to their current environment, and they suggest a trade-off between epigenetic memory and plasticity.
Materials and Methods
Dauer and control cultures
The wild-type strain N2 was used for all experiments. N2 was obtained from the Sternberg collection at the California Institute of Technology, originally from the CGC in 1987. Worms were maintained for at least three generations in standard laboratory conditions without starving prior to beginning experiments. Approximately 10 adults were picked onto each of four to five 10-cm NGM plates seeded with Escherichia coli OP50 and maintained at 20°. Embryos were obtained by standard hypochlorite treatment after 4 days in culture. For dauer-forming conditions, embryos were suspended in S-complete at a density of five/μl with 1 mg/ml E. coli HB101 (Baugh et al. 2011). HB101 was prepared as described previously (Hibshman et al. 2016). For control conditions, embryos were suspended at one/μl with 38 mg/ml HB101. Worms were cultured at 180 rpm and 20° in 25 ml glass Erlenmeyer flasks in a volume of 5 ml (25,000 worms for dauer conditions and 5000 for control conditions). For experiments that required >5000 control worms, a 20 ml volume in a 250 ml Erlenmeyer flask with the same density and food concentration was used. Dauer formation occurs in ∼4 days with nearly 100% penetrance. Short-term dauers remained in culture for 10 days, being arrested as dauers for 6 days. Long-term dauers remained in culture for 40–49 days, being arrested as dauers for 36–45 days. Survival was 90–100% in long-term dauer cultures. Control worms were in culture for 40–44 hr and were plated as L4 larvae. The L4 stage was chosen because dauers recover to become L4 larvae.
Dauer recovery and maintenance
Dauer and control conditions were paired such that control cultures were set up to be recovered at the same time as the dauer culture. Thus, recovery, maintenance, sampling, and assaying were done in parallel. Worms were taken from liquid cultures and plated on 10 cm NGM plates seeded with OP50 and incubated at 20°. To obtain F1 progeny, ∼1000 P0 worms were plated per seeded plate, and these were hypochlorite treated 2 days later to obtain F1 progeny for analysis. To obtain F3 progeny, 10 P0 worms were plated per seeded plate. These worms laid F1 embryos, which grew to become gravid adults on the same plates (∼1000 F1 worms per plate). Five days after plating, the F1 worms on these plates were hypochlorite treated to obtain F2 embryos. Approximately 1000 F2 embryos were plated per new seeded plate, and these worms were hypochlorite treated 3 days later to obtain F3 progeny. In all cases, hypochlorite treatment was performed prior to worms starving on plates.
L1 starvation survival
Embryos were suspended following hypochlorite treatment in virgin S-basal (no cholesterol or ethanol) at a density of one embryo/μl in a volume of 5 ml in a 16-mm glass test tube and placed on a tissue culture roller drum at ∼25 rpm and 21–22°. Beginning at day 1 (24 hr after hypochlorite treatment) and continuing every other day, a 100 μl sample was taken and plated on a 6 cm NGM plate with a spot of OP50 in the center. The sample was plated to the side of the OP50. The number of worms plated was scored (total plated = TP). Two days later, the number of worms that were alive on the plate were scored (total alive = TA). The proportion alive at each time point was calculated as TA/TP.
Brood size
Worms were singled onto 6 cm NGM plates with OP50 as L4 larvae. Worms were transferred to new plates each day until they stopped producing progeny. The number of progeny per plate was scored 2 days after removal of the mother. The total brood size per worm was determined by summing the progeny per plate across all plates for a single worm. Worms were censored if they died during egg laying. This affected 11 worms total in P0 brood size (five postdauers, six controls). In the F3 generation following long-term dauer, for 0 days of arrest, number of worms that died during egg laying included zero controls and one postdauer. For 8 days of arrest, number of worms that died during egg laying include 15 controls and 13 postdauers; two controls and two postdauers were sterile. In the F3 generation following short-term dauer, 10 control worms and 9 postdauer worms died during egg laying; one postdauer worm was sterile.
Embryo length
Embryos were plated onto unseeded 10 cm NGM plates. Embryos were imaged with a Zeiss Discovery. V20 stereomicroscope with a 10× objective (KSC 190–975). The images were analyzed with FIJI as described previously (Hibshman et al. 2016). Lengths of embryos were measured by thresholding embryos and calculating the long axis from ellipse fitting. The background was subtracted, images were thresholded, converted to binary, holes were filled, and particles were analyzed. Analysis was done in batch and the results were manually curated to ensure only quality embryo images were included.
Worm length
L1 larvae that had been starved for 1 or 8 days were recovered by plating on 10 cm NGM plates with OP50 as previously described (Hibshman et al. 2016). After 48 hr of recovery, worms were washed off the plates with virgin S-basal and plated on unseeded 10 cm NGM plates for imaging. Images were taken on a ZeissDiscovery. V20 stereomicroscope with automated zoom. Images were analyzed with the WormSizer plugin for FIJI to determine worm length and manually passed or failed (Moore et al. 2013).
Lifespan
For each condition and replicate, 150 L1 larvae arrested for 1 day in virgin S-basal were plated on 6 cm NGM plates seeded with OP50. After 2 days, 72 worms were randomly picked onto new seeded plates in groups of 12. Adults were picked away from their progeny onto fresh plates every day until egg laying ceased. Worms that responded to gentle prodding with a platinum wire were transferred to fresh plates every 2–3 days. Worms were considered dead when they failed to respond to prodding. Worms that crawled off the agar were considered lost and subtracted from the total n. No other animals were censored. Lifespan curves were analyzed to determine mean survival using OASIS (Yang et al. 2011).
Statistical analysis
Statistics were calculated in R or Microsoft Excel. To test for differences in means across groups, linear mixed effect models were fit to the data using the “nlme” package in R. The summary function was used to calculate P-values for the models, which implements the Wald test. Fixed effects included condition (postdauer vs. control) and, where applicable, length of starvation (0 or 1 day vs. 8 days). An interaction term was included for experiments with both types of fixed effects. A random effect of biological replicate was included for all models. To test for differences in interindividual variation across conditions, data were mean normalized within each biological replicate and condition, individual worms across all replicates were pooled, and Levene’s test was used to assess homogeneity of variance across conditions. For starvation survival (Figure 2, Figure 4, and Figure 5), logistic curves were fit to survival data and median survival times were calculated (Kaplan et al. 2015). Paired t-tests were performed on median survival times. Statistical tests and significance are indicated in figure legends. Plots were generated using ggplot2 in R.
Figure 2.
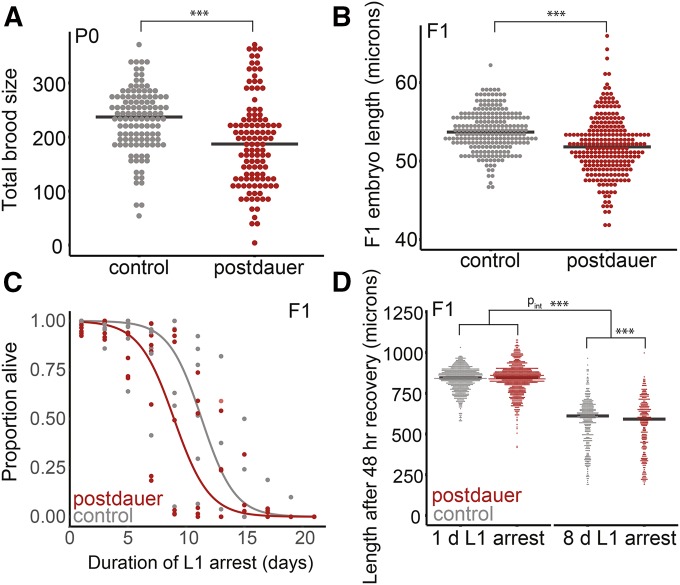
Long-term dauers recover to produce fewer, smaller F1 progeny that are starvation sensitive. (A) Brood sizes following long-term dauer diapause or control conditions. Seven biological replicates were scored with 13–18 individuals per biological replicate per condition. Effect of condition, P = 4.46 × 10−7. (B) Embryo length in the F1 progeny of postdauers and controls. Six biological replicates were scored with 23–70 individual embryos scored per biological replicate. Effect of condition, P = 9.0 × 10−13. (C) L1 starvation survival for seven biological replicates of F1 progeny of postdauers and controls. Logistic regression curves were fit, and median survival was determined for each replicate. Paired t-test on median survival, P = 0.10. (D) Worm length after 48 hr of recovery from 1 to 8 days of L1 arrest was scored in the F1 progeny of postdauers and controls. Eight biological replicates were scored. For 1 day, 50–335 worms were scored per biological replicate; following 8 days, 7–118 worms scored per replicate. Effect of condition for 1 day, P = 0.95; effect of condition for 8 days, P < 2.0 × 10−16. A linear mixed-effect model was fit with condition and length of starvation as fixed effects and biological replicate as a random effect. An interaction term was included for fixed effects. Effect of the interaction of condition and length of starvation, P = 5.1 × 10−14; effect of starvation, P < 2.0 × 10−16; effect of condition, P = 0.20. (A, B, and D). Linear mixed-effect models were fit with condition (postdauer vs. control) as a fixed effect and biological replicate as a random effect. P-values were calculated using the Wald test. Horizontal black lines represent medians. For means of biological replicates and grand mean, see Figure S1. *** P < 0.001
Figure 4.
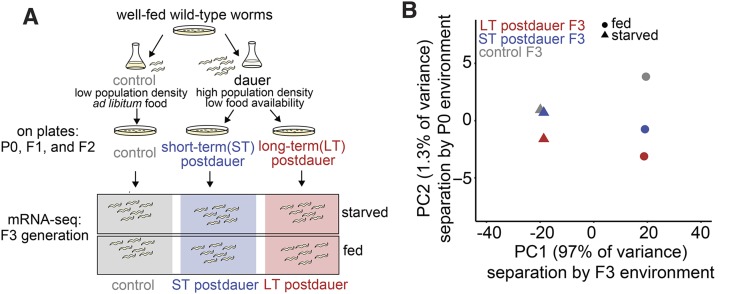
mRNA-seq reveals relative contributions of current environment and ancestral environment in shaping gene expression variation. (A) Schematic of experimental set-up for collection of RNA-seq samples. (B) Principal component analysis (PCA) of six conditions including all 8649 reliably detected genes; 97% of variance explained by whether worms were fed or starved at collection (PC1), and 1.3% of variance explained by whether ancestors experienced control, short-term dauer, or long-term dauer conditions (PC2). Mean CPM of biological replicates were used for each condition for PCA. Number of biological replicates: control starved (12), ST dauer starved (9), LT dauer starved (6), control fed (4), ST dauer fed (4), and LT dauer fed (3).
Figure 5.
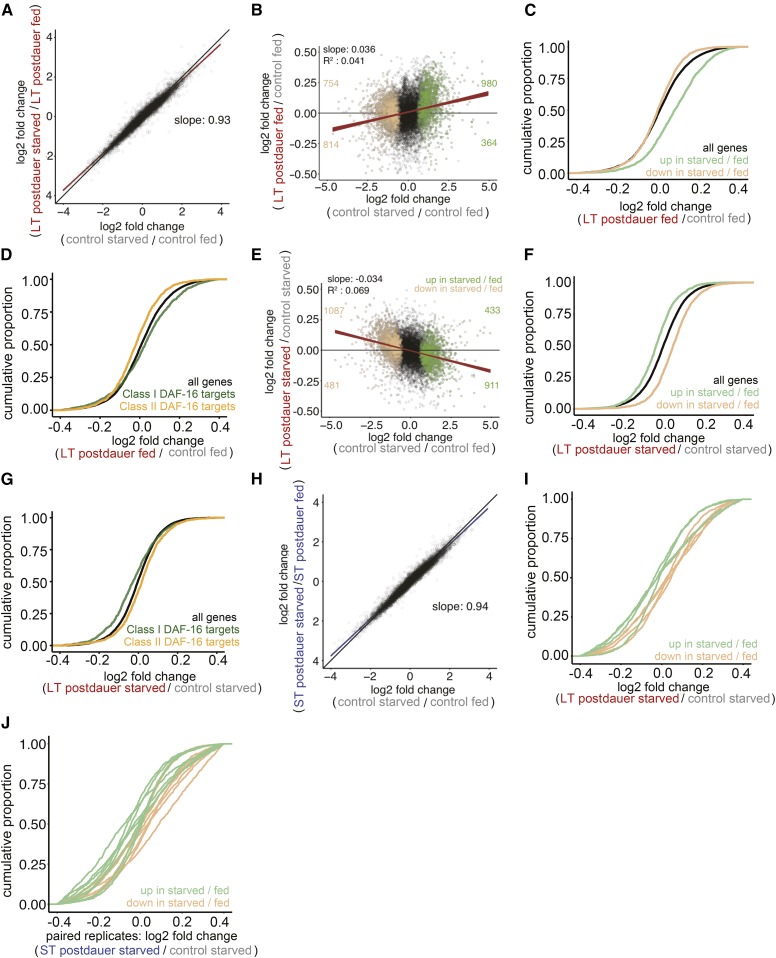
F3 progeny of dauers exhibit reduced gene expression plasticity driven by differences in both fed and starved worms. (A) Log2 fold changes of all genes in control starved/control fed plotted against LT postdauer starved/LT postdauer fed. Red line is linear regression through all points and width indicates 95% confidence interval (CI). Black line is the line y = x. Slope is 0.93, with CI from 0.926 to 0.934. Slope different from 1: P < 2.2 × 10−16. (B) Log2 fold changes of all genes in control starved/control fed plotted against log2 fold changes of LT postdauer fed F3/control fed plotted in black. Slope is significantly different from 0, P < 2.2 × 10−16. (C) All genes compared to genes up in starved/fed: P = 0; to genes down in starved/fed: P = 3.3 × 10−8. (D) Class I and Class II targets defined in Tepper et al. (2013). All genes compared to Class I targets: P = 1.6 × 10−5; to Class II targets: P = 2.3 × 10−11. (E) Log2 fold changes of all genes in control starved/control fed plotted against log2 fold changes of LT postdauer starved F3/control starved plotted in black. Slope is significantly different from 0, P < 2.2 × 10−16. (F) All genes compared to genes up in starved/fed: P = 0; to genes down in starved/fed: P = 0. (G) All genes compared to Class I targets: P = 1.0 × 10−12; to Class II targets: P = 4.0 × 10−7. (H) Log2 fold change of all genes in control starved/control fed compared to log2 fold change in ST postdauer starved/ST postdauer fed. Blue line indicates simple linear regression through all points, and thickness of line indicates 95% CI. Slope is 0.94, with CI from 0.936 to 0.943. Slope different from 1: P < 2.2 × 10−16 (I) LT postdauer starved/control starved comparison, P = 0.0083. (J) ST postdauer starved/control starved comparison, P = 0.088. (A and H) The summary.lm() function was used in R to generate slope and SE estimates, and a t-test was used to assess if the slope differed from 1. (B and E) Genes up in control starved/control fed using FDR 1 × 10−10 are plotted in green. Genes down in control starved/control fed are plotted in tan. Red line is a simple linear regression through all points, and thickness of line indicates 95% confidence interval. Number of genes differentially expressed in control starved/control fed in each quadrant is indicated. The summary.lm() function was used in R to generate t-values and their corresponding P-values. (C, D, F, and G) Within the indicated comparison, cumulative distribution functions (CDFs) of indicated gene lists are plotted. Kolmogorov-Smirnov test used to assess significance of gene list distribution compared to all genes. (I and J) Cumulative distribution function plots for individual paired replicates using the same groups of genes defined as upregulated and downregulated in control starved/control fed comparison. Paired t-test on median fold changes for these two gene groups.
Sample collection for RNA-seq
F3 embryos were suspended at one embryo/μl in S-complete either with or without 25 mg/ml HB101 to obtain fed or starved L1s, respectively. At least 10,000 embryos were used per condition per replicate. Fed larvae were collected 18 hr after hypochlorite treatment as early-stage developing L1 larvae. Starved larvae were collected 24 hr after hypochlorite treatment as arrested L1 larvae that had hatched ∼12 hr earlier. To collect starved samples, cultures were transferred to 15 ml conical tubes and spun for 1 min at 3000 rpm. Liquid was aspirated to <100 μl containing the worm pellet. Worms were washed 0–1× with S-basal. The worm pellet was transferred to a 1.5 ml microcentrifuge tube, flash frozen in liquid nitrogen, and stored at −80° until RNA isolation. To collect fed samples, cultures were transferred to 15 ml conical tubes and spun at 3000 rpm for 10 sec. Liquid was aspirated to <100 μl containing the worm pellet. The pellet was quickly washed 3–4× with 10 ml S-basal, visually inspected to ensure removal of the vast majority of bacteria, transferred to a 1.5 ml microcentrifuge tube, flash frozen, and stored at −80° until RNA isolation.
RNA isolation and library preparation
RNA was isolated using TRIzol (Invitrogen) using the manufacturer’s protocol with minor modifications; 1 ml of TRIzol was used per sample along with 100 μl of acid-washed sand. mRNA-seq libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530) in two batches, utilizing either 500 or 100 ng of starting RNA per library and 12 or 15 PCR cycles, respectively. Libraries were sequenced using Illumina HiSeq 4000 to obtain single-end 50 bp reads.
RNA-seq analysis
Bowtie was used to map reads to the WS210 genome (Langmead et al. 2009). We also included transcripts annotated in WS220 mapped back to the WS210 genome coordinates, as described previously (Maxwell et al. 2012). Mapping efficiencies ranged from 81 to 86% for all libraries. HTSeq was used to generate count tables for each library (Anders et al. 2015). Count tables were analyzed for differential expression using the edgeR package in R, which utilizes a negative binomial model to estimate dispersions (Robinson et al. 2010). Detected genes were considered those expressed at a level of at least four counts per million (CPM) across all libraries for all conditions, reducing the number of genes included in the analysis to 8649. The “calcNormFactors” function was used to normalize for RNA composition and the tagwise dispersion estimate was used for differential expression. The exact test was used for pairwise comparisons. Log2 fold change estimates from differential expression analysis in edgeR were used for generating plots in Figure 5 and Supplemental Material, Figure S5. Kolmogorov-Smirnov tests were used to determine differences in cumulative distributions of log2 fold changes.
Data availability
Raw and processed RNA-seq data are available through the GEO NCBI database with accession number GSE113500. Raw data for Figure 2 and Figure 3 are in Files S2–S4. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6510158.
Figure 3.
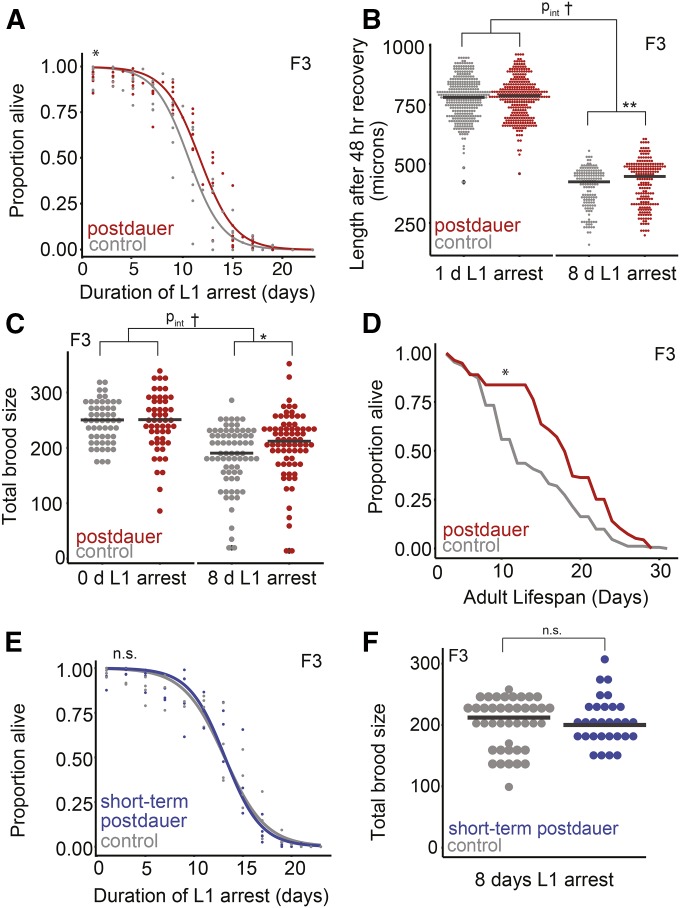
F3 progeny of long-term dauers exhibit increased starvation resistance and lifespan. (A) L1 starvation survival was scored in the F3 progeny of postdauers and controls. Eight biological replicates were scored, logistic curves were fit, and median survival times were determined. Paired t-test on median survival, P = 0.01. (B) Worm body length following 48 hr of recovery from either 1 or 8 days of L1 arrest. Four biological replicates were scored, consisting of 342 controls starved 1 day, 323 postdauers starved 1 day, 148 controls starved 8 days, and 204 postdauers starved 8 days. Effect of condition for 1 day, P = 0.25; effect of condition for 8 days, P = 0.0048. Effect of the interaction between condition and length of starvation, P = 0.066; effect of the length of starvation, P < 2.0 × 10−16; effect of condition, P = 0.93. (C) Brood size was scored for F3 progeny of controls and postdauers that experienced 0 or 8 days of L1 arrest. Three biological replicates for 0 days; five biological replicates for 8 days. For 0 day L1 arrest, 54 controls and 53 postdauers; for 8 days of L1 arrest, 72 controls and 74 postdauers. Effect of condition for 0 days, P = 0.82; effect of condition for 8 days, P = 0.03. Effect of the interaction between condition and length of starvation, P = 0.087; effect of the length of starvation, P < 1.0 × 10−4; effect of condition, P = 0.87. (D) Lifespans of at least 135 F3 progeny of postdauers and controls from three biological replicates (See Figure S3, C–E for individual replicates). Paired t-test on the means of biological replicates, P = 0.026. (E) Starvation survival in the F3 progeny of controls and short-term dauers (6 days as a dauer). Four biological replicates were scored, logistic curves were fit to data, and median survival times were determined. Paired t-test on median survival, P = 0.73. (F) Brood size of F3 progeny of controls and short-term dauers was scored after worms experienced 8 days of L1 arrest. Three biological replicates of 5–18 individual worms per condition were scored. Grand mean for control: 200; for postdauers: 201. A linear mixed-effect model was fit to brood size data with condition (postdauer vs. control) as a fixed effect and biological replicate as a random effect. P-values were calculated using the Wald test. Effect of condition, P = 0.75. (B and C) Linear mixed-effect models were fit with condition (postdauer vs. control) as a fixed effect and biological replicate as a random effect. Next, a linear mixed-effect model was fit with condition (postdauer vs. control) and length of starvation (0 or 1 day vs. 8 days) as fixed effects and biological replicate as a random effect. An interaction term was included for fixed effects. P-values were calculated using the Wald test. * P < 0.05, ** P < 0.01, *** P < 0.001; † interaction P < 0.1; n.s. not significant. Horizontal black lines represent medians. For means of biological replicates, see Figure S2.
Results
We carefully controlled worm population density and bacterial food concentration in a liquid culture system to cause wild-type worms to either enter dauer diapause or to bypass it (Baugh et al. 2011) (Figure 1). Ancestors of dauer and control worms were well fed for multiple generations (>3) to control for multigenerational effects of starvation or dietary restriction. Long-term dauers remained dauers for 36–45 days, and then were put on plates with food to recover. In parallel, control worms that did not experience dauer were plated from liquid culture so that postdauer and control worms were paired for experiments.
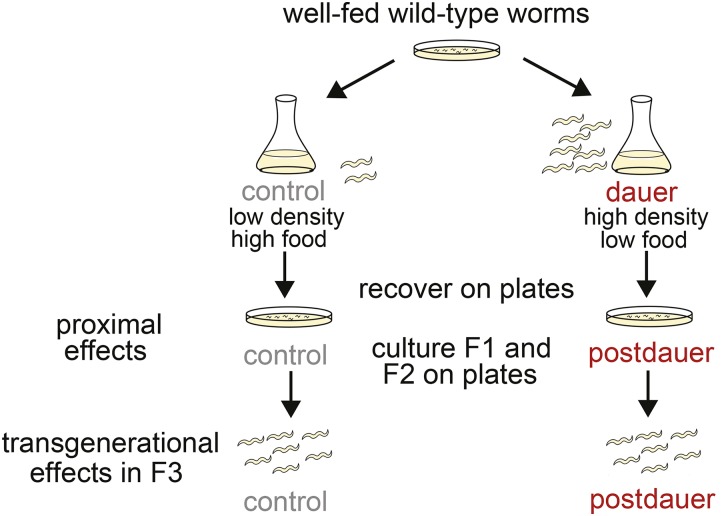
Figure 1.
Schematic of experimental design. Well-fed, wild-type worms were hypochlorite treated as gravid adults to obtain embryos, which were placed in dauer or control conditions. For long-term dauer conditions, worms remained in culture for 40–49 days and were dauers for 36–45 days. Worms from dauer and control cultures were plated on food in parallel. These lay F1 progeny, some of which were used for experiments of proximal effects of long-term dauer diapause. Other F1 adults were hypochlorite treated to obtain F2 embryos. F2 adults were hypochlorite treated to obtain F3 embryos. F3 postdauer and control worms were used for experiments.
Long-term dauer diapause reduces reproductive success in the P0 generation
We determined the proximal consequences of long-term dauer diapause on reproductive success. Following diapause, postdauer worms recovered to produce fewer F1 progeny (Figure 2A and Figure S1A). Postdauer adults also produced smaller embryos (Figure 2B and Figure S1B). C . elegans embryos hatch as L1 larvae. In the absence of food, L1 larvae survive starvation by remaining arrested as L1s, and this is reversible upon feeding. Survival during L1 arrest is a measure of starvation resistance (Baugh 2013). We found that postdauers produced F1 progeny that were relatively sensitive to starvation by two metrics: L1 starvation survival (Figure 2C) and growth rate during recovery from L1 starvation (Figure 2D and Figure S1C). Thus, long-term dauer arrest incurs costs to average fecundity as well as to progeny quality and starvation resistance, indicating reduced reproductive success.
Because we identified differences in mean trait values for postdauer and control worms upon recovery and in the F1 generation, we also considered the possibility that experiencing long-term dauer diapause could alter the interindividual variation of key traits within a worm population. We found that postdauer worms exhibited greater variation in brood size than controls, and that F1 embryos were more variable in length (Figure S2, A and B). Consistent with these observations, once these embryos hatched as L1s, they appeared to exhibit greater variation in body length after recovery from 8 days of starvation, but not 1 day of starvation (Figure S2C). This suggests that increased interindividual variation is uncovered by the stress of extended L1 starvation. Together our results suggest that the proximal consequences of long-term diapause include decreased reproductive success and increased phenotypic variation.
Long-term dauer diapause increases starvation resistance and lifespan transgenerationally
We asked whether effects of long-term dauer diapause persisted beyond the F1 generation to the F3 generation. Persistence to the F3 generation suggests transgenerational epigenetic inheritance, while effects in the F1 and F2 generations could be due to maternal effects (Skinner 2008). We measured starvation resistance in three ways: L1 starvation survival, body length following 48 hr of recovery from L1 starvation, and brood size following recovery from L1 starvation. In contrast to F1 L1 larvae, we found that F3 progeny of long-term dauers exhibited increased L1 starvation survival (Figure 3A). Likewise, F3 progeny of long-term dauers allowed to recover for 48 hr from 8 days of L1 starvation were larger than controls recovering from 8 days of L1 starvation, demonstrating increased growth rate (Figure 3B and Figure S3A). In addition, the broods of F3 progeny of long-term dauers following 8 days of L1 starvation were larger than the broods of F3 controls following 8 days of L1 starvation (Figure 3C and Figure S3B). We also assayed growth rate and brood size in the F3 postdauers with just 1 day of L1 starvation, and found no significant differences in these traits (Figure 3, B and C and Figure S3, A and B). In addition, we did not find evidence of differences in interindividual variation in the F3 generation (Figure S3, F and G), suggesting that the differences in mean trait values in the F3 generation were not due to a difference in the shape of the underlying trait distribution. Together these data support the conclusion that long-term dauer diapause leads to epigenetic inheritance of increased starvation resistance in F3 progeny.
We found transgenerational effects of long-term dauer arrest that were present beyond the L1 stage and into adulthood in F3 worms that were never starved. F3 progeny of long-term dauers lived longer as adults than controls (Figure 3D and Figure S3, C–E), demonstrating that there are transgenerational consequences of long-term dauer diapause outside the context of starvation.
We next wanted to determine whether the transgenerational increase in starvation resistance depends on the duration of dauer diapause. When worms were dauers for just 6 days, F3 progeny of these short-term dauers did not exhibit differences in starvation survival or brood size following starvation (Figure 3, E and F). We had the statistical power to detect differences in starvation survival of less than a day (Table S1; power analysis not shown). This suggests that the transgenerational effects of experiencing long-term dauer arrest are dependent on the length of starvation during arrest as opposed to the dauer-inducing culture conditions, dauer formation, or dauer recovery.
Epigenetic memory of dauer diapause affects gene expression globally
Transgenerational effects of long-term dauer diapause on important organismal traits suggest epigenetic regulation of gene expression. We used mRNA-seq to determine if gene expression patterns in both fed and starved F3 progeny of controls, short-term dauers, and long-term dauers differed (Figure 4A). This two-factor design allowed us to analyze both transgenerational and instantaneous effects of nutrient availability on gene expression. We performed between 3 and 12 biological replicates per condition (Table S2). Replicates within the same condition were highly correlated (Table S3), with an average Pearson correlation coefficient of 0.99 across all fed libraries and also across all starved libraries. We performed principal component analysis (PCA) on the normalized mean CPM values for six conditions as well as on individual biological replicates (Figure 4B and Figure S4). PC1 separated condition means depending on whether the worms were fed or starved as F3 L1s, while PC2 separated them according to length of time the initial generation spent in dauer diapause (Figure 4B). Notably, instantaneous effects of nutrient availability (PC1) appeared to explain substantially greater variance in gene expression than epigenetic effects (PC2). Nonetheless, these results suggest that ancestral environmental conditions play a small, but detectable, role in shaping gene expression in the F3 generation.
We compared gene expression plasticity in F3 long-term postdauers to controls. That is, we assessed gene expression differences between starved and fed F3 worms with long-term dauer ancestors to starved and fed F3 worms with control ancestors (Figure 5A). Regardless of ancestral background, starved and fed worms exhibited dramatic gene expression differences (Figure 5A and File S1), consistent with PC1. Gene expression responses to nutrient availability were also highly correlated. Notably, no genes had large differences in one comparison and not the other. Consistent with this, virtually no genes were individually significantly differentially expressed when only ancestral conditions differed (File S1). However, the slope of the linear regression was significantly <1 (Figure 5A), suggesting that gene expression plasticity in response to nutrient availability in the F3 generation following long-term dauer diapause is dampened relative to controls.
Dampened plasticity could occur if fed F3 progeny of long-term dauers had a transcriptional profile that appeared “less fed” compared to fed controls, or if starved F3 progeny of long-term dauers appeared transcriptionally “less starved” compared to starved controls. First, we found fed F3 progeny of long-term dauers appeared less fed (or, equivalently, more starved) relative to fed controls by comparing the effect of plasticity in controls to the epigenetic effect in fed larvae (Figure 5, B and C). We further tested this effect using the most differentially expressed nutrient-responsive genes. These genes, highlighted in Figure 5B, showed that genes that were significantly upregulated or downregulated in starved worms compared to fed worms tend to also be upregulated or downregulated, respectively, in fed F3 progeny of long-term dauers relative to fed controls (Figure 5, B and C). We also found that the regulatory targets of the transcription factor daf-16 (Tepper et al. 2013), which promotes starvation survival (Baugh 2013), overlap with the nutrient-responsive genes and display a similar epigenetic effect of long-term dauer diapause (Figure 5D and Figure S5A). This result corroborates the epigenetic effect on nutrient-responsive genes using gene sets defined outside the context of this study. These results provide evidence that dampened gene expression plasticity is driven, at least in part, by differences in nutrient-responsive gene expression in fed F3 progeny of long-term dauers.
We wanted to determine whether there are differences in nutrient-responsive gene expression in the starved F3 progeny of long-term dauers compared to controls. In contrast to the fed F3 progeny, the starved F3 progeny of long-term dauers exhibited a negative correlation with the control nutrient response (Figure 5, E and F). This was corroborated by the fact that daf-16 target genes displayed similar behavior (Figure 5G). Together, these results suggest that there are transgenerational effects on gene expression in both fed and starved F3 larvae that collectively contribute to reduced gene expression plasticity.
F3 progeny of short-term dauers did not display detectable epigenetic effects on starvation resistance, but we did detect effects on gene expression. We found that F3 progeny of short-term dauers showed similar transcriptional changes as identified in the F3 progeny of long-term dauers, including dampening of nutrient-responsive gene expression (Figure 5H and Figure S5, B–D). However, transgenerational changes in gene expression following short-term dauer appeared to be less consistent and possibly of a smaller magnitude than those elicited by long-term dauer. PC2 aligned means of short-term dauer samples between control and long-term dauer samples (Figure 4A). The slope of the dampening response of F3 progeny of short-term dauers compared to controls was closer to 1 than F3 progeny of long-term dauers, and F3 progeny of long-term dauers exhibited dampening relative to short-term dauers (Figure 5H, Figure S4B, and Figure S5B). In addition, we examined gene expression shifts within individual paired replicates (replicates in which an F3 dauer sample was collected in parallel with a control). All paired replicates for starved F3 progeny of long-term dauers compared to starved controls showed shifts in nutrient-responsive genes in the expected direction (Figure 5I). Paired replicates for F3 progeny of short-term dauers exhibited shifts in gene expression for the majority, but not all, pairs (Figure 5J). Collectively, these results suggest that transgenerational epigenetic effects of short-term dauer are detectable and qualitatively similar to those of long-term dauer, but that they are less consistent, likely accounting for lack of detectable effects on starvation resistance.
Discussion
We utilized a well-studied model of phenotypic plasticity, dauer diapause, to interrogate the proximal and transgenerational consequences of early-life environment on organismal traits and gene expression. Postdauer worms displayed reduced reproductive success after long-term diapause, but their F3 progeny exhibited increased starvation resistance and lifespan. F3 progeny of both short- and long-term dauers exhibited changes in gene expression in nutrient-responsive genes, consistent with a reduction in gene expression plasticity in response to nutrient availability. This work has broad implications for understanding the consequences of starvation, including the potential for epigenetic inheritance in response to nutrient availability and potentially adaptive responses across generations.
Reduced reproductive success and increased phenotypic variation after long-term dauer diapause
Our results suggest that, while it is adaptive to be able to enter dauer diapause to survive adverse conditions, there is a cost to extended dauer diapause. That is, long-term dauers recovered to produce fewer, smaller F1 progeny that are starvation sensitive compared to worms that do not enter dauer. As a caveat, postdauer adults may be developmentally delayed compared to controls by several hours. Maternal age differences of 1 day affect L1 length (Hibshman et al. 2016; Perez et al. 2017). Though embryo length was not assessed in these previous studies, differences in maternal age could affect F1 embryo size in our results. However, the overall assessment of postdauers exhibiting reduced reproductive success is still valid, as total brood size is not affected by developmental delay. The proximal costs of long-term dauer diapause are consistent with previous work in our laboratory identifying costs to 8 days of L1 starvation, as worms recovered from L1 starvation are more sensitive to subsequent starvation and produce smaller broods with decreased progeny quality (Jobson et al. 2015). Both extended L1 starvation and dauer diapause appear costly, but this is likely due to the costs of starvation experienced during these states, rather than simply arresting development. Postdauer worms that experienced dauer for just 1 day live longer and produce larger broods (Hall et al. 2010). Together these observations suggest that hormesis, in which a mild stress can increase fitness (Calabrese and Baldwin 2002), may occur when starvation is brief, but that extended starvation is costly as the buffering capacity of the organism is overwhelmed.
In addition to reduced reproductive success, we found that long-term postdauers exhibit increased interindividual variation, consistent with developmental decanalization (Chen et al. 2015) stemming from deleterious effects of extended starvation. Greater interindividual variability could occur as a result of (1) decanalization or (2) an evolutionary bet-hedging strategy (Waddington 1957). Decanalization occurs when phenotypic robustness is compromised, as a consequence of, for example, conditions that result in pathology. We believe the observed decreases in average P0 brood size and F1 progeny size and starvation resistance following long-term dauer arrest reflect pathological consequences of extended starvation exceeding the buffering capacity of the organism. Consistent with this interpretation, appreciable lethality and developmental abnormalities affecting the reproductive system have been reported after 60 and 25 days of dauer arrest, respectively (Klass and Hirsh 1976; Kim and Paik 2008). Alternatively, bet hedging is thought to be evolutionarily adaptive in unpredictable environments (Starrfelt and Kokko 2012). In a bet-hedging scenario, a population exhibits interindividual variability such that it is suited for variable environmental conditions. In a particular condition, the bet-hedging population may be less fit than a highly specialized population. However, the bet-hedging population reduces variation in fitness across variable environments. If postdauer worms display a bet-hedging strategy, then a subpopulation of postdauers would be expected to perform better than controls in at least some environmental condition. Our data do not provide compelling evidence for a subpopulation of postdauers or their progeny that performs better than controls for any of the proximal traits assayed, despite increased variability. However, we cannot formally exclude the possibility that there are other environmental conditions that would reveal an advantage to increased phenotypic variation as a consequence of extended dauer diapause. We also emphasize that this consideration of bet hedging is in the context of proximal traits. Since we found that the great-grandprogeny of long-term dauers survive starvation better, it is possible that any advantage to a bet-hedging strategy is only apparent on the scale of multiple generations.
Transgenerational effects of long-term dauer diapause on starvation survival
We found that the F3 progeny of long-term dauers exhibit increased starvation resistance as larvae and live longer as fed adults. These results indicate that transgenerational effects manifest in both fed and starved animals and that they persist through the life cycle. We considered whether these transgenerational effects could be due to factors other than epigenetic inheritance, including mutation and selection, confounding stressors, or developmental timing differences, and we conclude that the transgenerational effects observed are most consistent with epigenetic inheritance. First, control and dauer worms for each replicate originated from the same isogenic population. Dauer worms, despite being starved for an extended period, were not subject to genetic selection, as the vast majority survived long-term dauer diapause. Additionally, a de novo mutation would likely not have sufficient generational time to affect population-level phenotypes by the F3 generation, since 10 P0 worms were plated for each experiment. Further, because we performed multiple biological replicates, it is unlikely that de novo mutations could repeatedly affect population-level phenotypes in the F3 generation. Worms were also cultured for the F1 and F2 generations with ample food and frequent passage such that any inadvertent selection would be for rapid growth and early fecundity, traits that theoretically trade off with starvation resistance. Embryos were prepared with hypochlorite treatment, a standard protocol that may nonetheless provide an additional stress. Since both control and postdauer worms receive this treatment, however, we do not consider this an explanation of transgenerational effects. An initial delay exiting dauer diapause also does not lead to effects in the F3 generation, as postdauer and control populations are highly synchronized beyond the P0 generation. The lack of transgenerational effects in the F3 generation following short-term dauer provides additional support that entering, experiencing, or exiting dauer arrest do not confer transgenerational effects. Rather, the extended nature of long-term dauer diapause is uniquely associated with these effects.
Multigenerational consequences of L1 starvation have been previously examined and exhibit some similarities to those of long-term dauer diapause. Following 8 days of L1 starvation in buffer with no carbon source, we previously observed increased starvation resistance in the F2 but not F3 generation (Jobson et al. 2015). Notably, transmission of increased starvation resistance to F1 and F2 progeny of worms that experienced L1 starvation required sorting larvae by size after 2 days of recovery from starvation (Jobson et al. 2015), and in the present study such sorting was not required. Increased lifespan in the F3 generation following 6 days of L1 starvation on plates has been reported in another study (Rechavi et al. 2014). We were not able to reproduce this lifespan extension following L1 starvation, suggesting critical differences in experimental conditions. Still, both L1 and dauer starvation paradigms reveal apparent proximal fitness costs of extended starvation followed by increases in starvation resistance in subsequent generations. The switch from proximal starvation sensitivity to heritable starvation resistance in both paradigms is of particular interest.
Transgenerational effects of long-term dauer diapause on gene expression plasticity
In addition to life-history trait differences, we identified transgenerational effects of dauer diapause on gene expression plasticity in response to nutrient availability. Specifically, we found that F3 progeny of both short-term and long-term dauers exhibited reduced gene expression plasticity, with starved F3 worms appearing less starved and fed F3 worms appearing less fed. Theory suggests that there should be costs and limits to plasticity, though there are relatively few empirical examples of such costs (Dewitt et al. 1998; Murren et al. 2015). While we emphasize that the transgenerational effect on mRNA expression plasticity does not necessarily indicate an effect at other levels of regulation, one interpretation of this result is that maintaining an epigenetic memory of ancestral environmental conditions limits or canalizes plastic responses to current environmental conditions. In other words, there is hypothetically a fixed capacity to respond to environmental conditions, but this capacity can be exhausted by instantaneous and epigenetic regulation.
Evolutionary significance of epigenetic inheritance
It is possible that subtle changes in starvation resistance and gene expression reflect physiological fine-tuning in anticipation of future conditions based on ancestral history. In contrast, it is possible that epigenetic memory of ancestral conditions occurs but is not necessarily adaptive, as a potentially neutral or even costly byproduct of some of other selected trait. Epigenetic stability from parent to offspring is favorable given that four conditions are met: (1) the environment is variable; (2) parental environment has some predictive power over offspring environment; (3) transgenerational effects increase fitness of parents and/or offspring; and (4) costs associated with the transgenerational response are low (Herman et al. 2014). Though it is clear that C. elegans experience variable nutrient conditions in the wild (Felix and Braendle 2010), it is currently unknown how predictive parental environment is of offspring environment over multiple generations. Specifically, it is unknown whether conditions leading to extended dauer diapause are predictive of environmental conditions three generations later, particularly when dauer-inducing conditions are not experienced in the intervening generations. Here, we find that F3 progeny of long-term dauers are more starvation resistant, and this is not accompanied by detectable costs in growth or fecundity, consistent with the possibility that selection for an epigenetic memory of extended dauer diapause could occur. Recognizing that it is ultimately difficult to determine if such a complex trait as epigenetic inheritance is evolutionarily adaptive, we hope that future studies in the context of the natural ecology of C. elegans will shed light on this question.
Acknowledgments
We thank the Duke University School of Medicine and the Center for Genomic and Computational Biology for use of the Sequencing and Genomic Technologies core resource, which provided RNA sequencing service. This work was funded by the National Institutes of Health (R01GM117408). A.K.W. is supported by the National Science Foundation Graduate Research Fellowship.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6510158.
Present address: Department of Biology, University of North Carolina, Chapel Hill, NC 27599.
Communicating editor: B. Grant
Literature Cited
- Albert P. S., Riddle D. L., 1983. Developmental alterations in sensory neuroanatomy of the Caenorhabditis elegans dauer larva. J. Comp. Neurol. 219: 461–481. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo G., Van Gilst M. R., 2009. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science 326: 954–958. 10.1126/science.1178343 [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., 2006. Chemosensation in C. elegans (October 25, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.123.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere A., Felix M. A., 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15: 1176–1184. [DOI] [PubMed] [Google Scholar]
- Baugh L. R., 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Kurhanewicz N., Sternberg P. W., 2011. Sensitive and precise quantification of insulin-like mRNA expression in Caenorhabditis elegans. PLoS One 6: e18086 10.1371/journal.pone.0018086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., et al. , 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E. J., Baldwin L. A., 2002. Defining hormesis. Hum. Exp. Toxicol. 21: 91–97. [DOI] [PubMed] [Google Scholar]
- Chen J., Nolte V., Schlotterer C., 2015. Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster. PLoS Genet. 11: e1004883 10.1371/journal.pgen.1004883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. A., Ashrafi K., 2009. Fat rationing in dauer times. Cell Metab. 9: 113–114. 10.1016/j.cmet.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Dewitt T. J., Sih A., Wilson D. S., 1998. Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 13: 77–81. [DOI] [PubMed] [Google Scholar]
- Dixon S. J., Alexander M., Chan K. K., Roy P. J., 2008. Insulin-like signaling negatively regulates muscle arm extension through DAF-12 in Caenorhabditis elegans. Dev. Biol. 318: 153–161. 10.1016/j.ydbio.2008.03.019 [DOI] [PubMed] [Google Scholar]
- Felix M. A., Braendle C., 2010. The natural history of Caenorhabditis elegans. Curr. Biol. 20: R965–R969. 10.1016/j.cub.2010.09.050 [DOI] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Maures T. J., Ucar D., Hauswirth A. G., Mancini E., et al. , 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479: 365–371. 10.1038/nature10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. E., Beverly M., Russ C., Nusbaum C., Sengupta P., 2010. A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr. Biol. 20: 149–155. 10.1016/j.cub.2009.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Martienssen R. A., 2014. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157: 95–109. 10.1016/j.cell.2014.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. J., Spencer H. G., Donohue K., Sultan S. E., 2014. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68: 632–643. 10.1111/evo.12324 [DOI] [PubMed] [Google Scholar]
- Hibshman J. D., Hung A., Baugh L. R., 2016. Maternal diet and insulin-like signaling control intergenerational plasticity of progeny size and starvation resistance. PLoS Genet. 12: e1006396 10.1371/journal.pgen.1006396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P. J., 2007 Dauer (August 08, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.144.1, http://www.wormbook.org.
- Jablonka E., Raz G., 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84: 131–176. [DOI] [PubMed] [Google Scholar]
- Jobson M. A., Jordan J. M., Sandrof M. A., Hibshman J. D., Lennox A. L., et al. , 2015. Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics 201: 201–212. 10.1534/genetics.115.178699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R. E., Chen Y., Moore B. T., Jordan J. M., Maxwell C. S., et al. , 2015. dbl-1/TGF-beta and daf-12/NHR signaling mediate cell-nonautonomous effects of daf-16/FOXO on starvation-Induced developmental arrest. PLoS Genet. 11: e1005731 10.1371/journal.pgen.1005731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane J., Avery L., 2003. Mechanosensory inputs influence Caenorhabditis elegans pharyngeal activity via ivermectin sensitivity genes. Genetics 164: 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. A., Panhuis T. M., Stoehr A. M., 2012. Phenotypic plasticity: molecular mechanisms and adaptive significance. Compr. Physiol. 2: 1417–1439. 10.1002/cphy.c110008 [DOI] [PubMed] [Google Scholar]
- Kim S., Paik Y. K., 2008. Developmental and reproductive consequences of prolonged non-aging dauer in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 368: 588–592. 10.1016/j.bbrc.2008.01.131 [DOI] [PubMed] [Google Scholar]
- Klass M., Hirsh D., 1976. Non-ageing developmental variant of Caenorhabditis elegans. Nature 260: 523–525. [DOI] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B., 2017. Transgenerational transmission of environmental information in C. elegans. Science 356: 320–323. 10.1126/science.aah6412 [DOI] [PubMed] [Google Scholar]
- Kostal V., 2006. Eco-physiological phases of insect diapause. J. Insect Physiol. 52: 113–127. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kennedy S. G., Ruvkun G., 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17: 844–858. 10.1101/gad.1066503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C. S., Antoshechkin I., Kurhanewicz N., Belsky J. A., Baugh L. R., 2012. Nutritional control of mRNA isoform expression during developmental arrest and recovery in C. elegans. Genome Res. 22: 1920–1929. 10.1101/gr.133587.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J. J., Schuster E., Blanc E., Thomas J. H., Gems D., 2004. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279: 44533–44543. [DOI] [PubMed] [Google Scholar]
- Minkina O., Hunter C. P., 2017. Stable heritable germline silencing directs somatic silencing at an endogenous locus. Mol. Cell 65: 659–670.e5. 10.1016/j.molcel.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. T., Jordan J. M., Baugh L. R., 2013. WormSizer: high-throughput analysis of nematode size and shape. PLoS One 8: e57142 10.1371/journal.pone.0057142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murren C. J., Auld J. R., Callahan H., Ghalambor C. K., Handelsman C. A., et al. , 2015. Constraints on the evolution of phenotypic plasticity: limits and costs of phenotype and plasticity. Heredity (Edinb) 115: 293–301. 10.1038/hdy.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., Roy R., 2009. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457: 210–214. 10.1038/nature07536 [DOI] [PubMed] [Google Scholar]
- Ni J. Z., Kalinava N., Chen E., Huang A., Trinh T., et al. , 2016. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenet. Chromatin 9: 3 10.1186/s13072-016-0052-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M. F., Francesconi M., Hidalgo-Carcedo C., Lehner B., 2017. Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature 552: 106–109. 10.1038/nature25012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp R. A., Wendel J. F., 2005. Epigenetics and plant evolution. New Phytol. 168: 81–91. [DOI] [PubMed] [Google Scholar]
- Rechavi O., Houri-Ze’evi L., Anava S., Goh W. S., Kerk S. Y., et al. , 2014. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158: 277–287. 10.1016/j.cell.2014.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P., Lim C. S., Johnsen R., Albert P. S., Pilgrim D., et al. , 1996. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274: 1389–1391. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., Thomas J. H., 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17: 719–728. [DOI] [PubMed] [Google Scholar]
- Schindler A. J., Baugh L. R., Sherwood D. R., 2014. Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 10: e1004426 10.1371/journal.pgen.1004426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D., Yanai I., Hunter C. P., 2014. Natural RNA interference directs a heritable response to the environment. Sci. Rep. 4: 7387 10.1038/srep07387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H., Felix M. A., 2017. The natural biotic environment of Caenorhabditis elegans. Genetics 206: 55–86. 10.1534/genetics.116.195511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. K., 2008. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 25: 2–6. 10.1016/j.reprotox.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrfelt J., Kokko H., 2012. Bet-hedging–a triple trade-off between means, variances and correlations. Biol. Rev. Camb. Philos. Soc. 87: 742–755. 10.1111/j.1469-185X.2012.00225.x [DOI] [PubMed] [Google Scholar]
- Tepper R. G., Ashraf J., Kaletsky R., Kleemann G., Murphy C. T., et al. , 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154: 676–690. 10.1016/j.cell.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H., 1957. The Strategy of the Genes; A Discussion of Some Aspects of Theoretical Biology. Allen & Unwin, London. [Google Scholar]
- Wang J., Kim S. K., 2003. Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130: 1621–1634. [DOI] [PubMed] [Google Scholar]
- Yang J. S., Nam H. J., Seo M., Han S. K., Choi Y., et al. , 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6: e23525 10.1371/journal.pone.0023525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw and processed RNA-seq data are available through the GEO NCBI database with accession number GSE113500. Raw data for Figure 2 and Figure 3 are in Files S2–S4. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6510158.
Figure 3.
F3 progeny of long-term dauers exhibit increased starvation resistance and lifespan. (A) L1 starvation survival was scored in the F3 progeny of postdauers and controls. Eight biological replicates were scored, logistic curves were fit, and median survival times were determined. Paired t-test on median survival, P = 0.01. (B) Worm body length following 48 hr of recovery from either 1 or 8 days of L1 arrest. Four biological replicates were scored, consisting of 342 controls starved 1 day, 323 postdauers starved 1 day, 148 controls starved 8 days, and 204 postdauers starved 8 days. Effect of condition for 1 day, P = 0.25; effect of condition for 8 days, P = 0.0048. Effect of the interaction between condition and length of starvation, P = 0.066; effect of the length of starvation, P < 2.0 × 10−16; effect of condition, P = 0.93. (C) Brood size was scored for F3 progeny of controls and postdauers that experienced 0 or 8 days of L1 arrest. Three biological replicates for 0 days; five biological replicates for 8 days. For 0 day L1 arrest, 54 controls and 53 postdauers; for 8 days of L1 arrest, 72 controls and 74 postdauers. Effect of condition for 0 days, P = 0.82; effect of condition for 8 days, P = 0.03. Effect of the interaction between condition and length of starvation, P = 0.087; effect of the length of starvation, P < 1.0 × 10−4; effect of condition, P = 0.87. (D) Lifespans of at least 135 F3 progeny of postdauers and controls from three biological replicates (See Figure S3, C–E for individual replicates). Paired t-test on the means of biological replicates, P = 0.026. (E) Starvation survival in the F3 progeny of controls and short-term dauers (6 days as a dauer). Four biological replicates were scored, logistic curves were fit to data, and median survival times were determined. Paired t-test on median survival, P = 0.73. (F) Brood size of F3 progeny of controls and short-term dauers was scored after worms experienced 8 days of L1 arrest. Three biological replicates of 5–18 individual worms per condition were scored. Grand mean for control: 200; for postdauers: 201. A linear mixed-effect model was fit to brood size data with condition (postdauer vs. control) as a fixed effect and biological replicate as a random effect. P-values were calculated using the Wald test. Effect of condition, P = 0.75. (B and C) Linear mixed-effect models were fit with condition (postdauer vs. control) as a fixed effect and biological replicate as a random effect. Next, a linear mixed-effect model was fit with condition (postdauer vs. control) and length of starvation (0 or 1 day vs. 8 days) as fixed effects and biological replicate as a random effect. An interaction term was included for fixed effects. P-values were calculated using the Wald test. * P < 0.05, ** P < 0.01, *** P < 0.001; † interaction P < 0.1; n.s. not significant. Horizontal black lines represent medians. For means of biological replicates, see Figure S2.