Abstract
Imidazolium ionic liquids (IILs) have a range of biotechnological applications, including as pretreatment solvents that extract cellulose from plant biomass for microbial fermentation into sustainable bioenergy. However, residual levels of IILs, such as 1-ethyl-3-methylimidazolium chloride ([C2C1im]Cl), are toxic to biofuel-producing microbes, including the yeast Saccharomyces cerevisiae. S. cerevisiae strains isolated from diverse ecological niches differ in genomic sequence and in phenotypes potentially beneficial for industrial applications, including tolerance to inhibitory compounds present in hydrolyzed plant feedstocks. We evaluated >100 genome-sequenced S. cerevisiae strains for tolerance to [C2C1im]Cl and identified one strain with exceptional tolerance. By screening a library of genomic DNA fragments from the [C2C1im]Cl-tolerant strain for improved IIL tolerance, we identified SGE1, which encodes a plasma membrane multidrug efflux pump, and a previously uncharacterized gene that we named ionic liquid tolerance 1 (ILT1), which encodes a predicted membrane protein. Analyses of SGE1 sequences from our panel of S. cerevisiae strains together with growth phenotypes implicated two single nucleotide polymorphisms (SNPs) that associated with IIL tolerance and sensitivity. We confirmed these phenotypic effects by transferring the SGE1 SNPs into a [C2C1im]Cl-sensitive yeast strain using CRISPR/Cas9 genome editing. Further studies indicated that these SNPs affect Sge1 protein stability and cell surface localization, influencing the amount of toxic IILs that cells can pump out of the cytoplasm. Our results highlight the general potential for discovering useful biotechnological functions from untapped natural sequence variation and provide functional insight into emergent SGE1 alleles with reduced capacities to protect against IIL toxicity.
Keywords: Saccharomyces cerevisiae, yeast, ionic liquid, natural variation, major facilitator superfamily, biofuels, toxin tolerance
IONIC liquids are neutral salts that attain a liquid state at temperatures mostly <100° (reviewed in Welton 1999) and have a broad range of biological applications from biomedicine (reviewed in Dias et al. 2017; Egorova et al. 2017) to production of biochemicals and bioenergy. In renewable bioenergy applications, ionic liquids, particularly imidazolium ionic liquids (IILs) such as 1-ethyl-3-methylimidazolium chloride ([C2C1im]Cl), 1-ethyl-3-methylimidazolium acetate ([C2C1im][OAc]), and 1-butyl-3-methylimidazolium chloride ([C4C1im]Cl) are effective in solubilizing plant biomass for purification of cellulose through a process called pretreatment (Binder and Raines 2010; Li et al. 2010; Elgharbawy et al. 2016). After pretreatment, cellulose is highly accessible to cellulase enzymes that hydrolyze it into monomeric glucose, which is then fermented into bioethanol or other biofuels by industrial microbes. However, two disadvantages of these solvents are their high cost (Blanch et al. 2011; Konda et al. 2014) and toxicity to biofuel-producing microbes, both of which impose a demand for IIL recovery after pretreatment. Escherichia coli strains used in the industrial production of biofuels and biochemicals are growth impaired in laboratory media containing 200–270 mM [C2C1im]Cl (Khudyakov et al. 2012; Ruegg et al. 2014). The dominant biofuel-producing microbe, Saccharomyces cerevisiae, is even more sensitive; 30–60 mM [C2C1im]Cl can inhibit growth (Ouellet et al. 2011; Dickinson et al. 2016). After biomass pretreatment and hydrolysis, up to 270 mM IIL may persist during fermentation (Datta et al. 2010); IILs at these concentrations severely impair both yeast growth and biofuel production. Thus genetically modified yeasts that better tolerate inhibitory IIL concentrations are highly desirable to improve the production of lignocellulosic biofuels and bioproducts.
To circumvent IIL toxicity, gene sequences from IIL-tolerant microbes can be inserted into biofuel-producing microbes to improve tolerance to IILs. For example, two genes, eilA and eilR, are determined to be primarily responsible for IIL tolerance in Enterobacter lignolyticus (Ruegg et al. 2014), an IIL-tolerant rain forest bacterium (Khudyakov et al. 2012). A member of the major facilitator superfamily (MFS) of proteins, the inner membrane transporter EilA exports quaternary ammonium cations and is transcriptionally regulated by EilR, which is induced by the [C2C1im]+ cation. Because of this, when expressed in E. coli, the eilAR gene cassette increases both cell growth and biofuel production in media containing IILs (Ruegg et al. 2014).
In yeast, chemical genomics screening of S. cerevisiae mutants determined that deletion of PTK2 increases cell fitness and sugar metabolism in the presence of inhibitory IIL concentrations (Dickinson et al. 2016). PTK2 encodes a putative serine/threonine protein kinase that activates the plasma membrane H+-ATPase Pma1 (Eraso et al. 2006), and it was suggested that deletion of PTK2 blocks Pma1 proton-coupled import of IILs into the cytoplasm (Dickinson et al. 2016), where IILs appear to affect mitochondrial function (Mehmood et al. 2015; Dickinson et al. 2016). Although deletion of PTK2 improves IIL tolerance, the resulting reduction in Pma1 activity and altered ion homeostasis also causes decreased strain fitness in other conditions (Giaever et al. 2002; Qian et al. 2012).
Functional screening of homologous DNA libraries has been an effective means to identify overexpressed genes in yeast that confer tolerance to industrially relevant inhibitors, such as ethanol (Anderson et al. 2012) and toxins in molasses fermentations (Ness and Aigle 1995). As an alternative approach, we explored the genetic variation in natural S. cerevisiae isolates to identify additional genes or sequence variants that enable IIL tolerance. The growth and fermentation phenotypes of numerous wild and domesticated S. cerevisiae strains have been examined across a wide range of media conditions (Fay and Benavides 2005; Liti et al. 2009; Schacherer et al. 2009; Strope et al. 2015), including media that contained various inhibitory compounds generated from biomass pretreatment (Parreiras et al. 2014; Sato et al. 2014; Wohlbach et al. 2014; Kong et al. 2018). Individual strains exhibited a wide range of growth tolerances, indicating that some natural isolates contain genetic differences that are protective against toxins present in hydrolyzed plant biomass. Here, we combined phenotypic and genotypic analyses with functional screening to identify the MFS transporter SGE1 and an uncharacterized open reading frame (ORF) YDR090C with important roles in IIL tolerance. Our results uncovered the impact of natural genetic variation in IIL tolerance and identified an SGE1 allele that offers a clear technological application for biofuel production.
Materials and Methods
Media
Standard yeast laboratory media were prepared as described elsewhere (Sherman 2002), with modifications. YPD (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose) and synthetic complete (SC) media were adjusted to pH 5.0 with HCl. For experiments described in Supplemental Material, Figure S4, the pH was adjusted to the indicated values with HCl or NaOH accordingly. Cationic compounds were purchased from the following vendors: [C2C1im]Cl (catalog #272841, Sigma Aldrich; or catalog #AC354080250, Fisher Scientific, Pittsburgh, PA), [C4C1im]Cl (catalog #94128; Sigma Aldrich), [C2C1im][OAc] (catalog #689483; Sigma Aldrich), and Crystal Violet (CV) (catalog #NC9002731; Fisher Scientific). Cationic compounds were added directly to YPD or SC media and sterilized by passing through 2 μm filters. The following concentrations were used to select for plasmid and PCR product transformations in yeast and E. coli: 200 μg/ml Geneticin (catalog #10131027; Life Technologies), 100 μg/ml nourseothricin sulfate (catalog #RC-187; G-Biosciences), 200 μg/ml hygromycin B (catalog #10687010; GIBCO, Grand Island, NY), 200 μg/ml Zeocin (catalog #R25001; GIBCO), and 100 μg/ml carbenicillin (catalog #00049; Chem-Impex).
Yeast strain construction
Genotypes and sources of S. cerevisiae strains used in this study are described in File S1 and Table S2. Deletion mutant strains from the Yeast Knockout (YKO) Collection (Winzeler et al. 1999) were obtained from Open Biosystems/Dharmacon. Deletion of SGE1 and ionic liquid tolerance 1 (ILT1) were performed by integration of PCR products generated from LoxP-KanMX-LoxP and LoxP-hphMX-LoxP templates (Guldener et al. 1996; Gueldener et al. 2002), primers containing 50–60 bp of homology flanking the SGE1 or ILT1 ORF, and Phusion DNA polymerase (New England Biolabs, Beverly, MA). For deletion of ILT1 with KanMX4, strain 4025 from the YKO Collection was used as the PCR template. PCR products were purified (PCR Purification Kit, QIAGEN, Valencia, CA) and transformed into the appropriate strains (Gietz and Schiestl 2007). Antibiotic selection markers flanked by LoxP sequences were excised by Cre recombinase as published (Guldener et al. 1996). Both GLBRCY412 and GLBRCY490 were generated in the homozygous diploid 378604X (hereafter named 378) strain background, thus requiring two gene deletions to create the complete null mutants. To generate GLBRCY412, one copy of ILT1 was deleted by replacement with the bleMX4 selection marker, followed by a subsequent replacement of the second ILT1 copy with KanMX. Construction of GLBRCY490 was conducted by deleting the first copy of SGE1 with LoxP-KanMX-LoxP and the second copy with LoxP-hphMX-LoxP, followed by Cre-mediated excision of both KanMX and hphMX selection markers. SGE1 and ILT1 deletions were confirmed by Sanger sequencing (University of Wisconsin-Madison Biotechnology Center DNA Sequencing Facility) of purified PCR products generated from purified genomic DNA (gDNA) (Epicentre MasterPure Yeast DNA Purification kit) and primers that annealed outside of the homologous sequences were used for gene deletion. In-frame, genomic insertion of MYC or green fluorescence protein (GFP) (S65T) at the 3′ ends of SGE1 or ILT1 were performed by transformation of a PCR product generated from the pFA6a-13Myc-KanMX6 or pFA6a-GFP(S65T)-KanMX (Bähler et al. 1998) plasmid templates and the following primer pairs:
For SGE1 fusions: SGE1MycFOR, CTTTGGAATATTCACTTCGAGTAAGAAAACAACAATATCAGCCAAAAAGCAACAA cggatccccgggttaattaa;
SGE1MycREV, GTACTGTCTAGTTTTATCGAACTACGATAAGTTAATTTATACGTTGGAAAATTGT gaattcgagctcgtttaaac.
For ILT1 fusions: YDR090MYCfor, TGTCCATGGAGTTGTGGTTAGAACAGATCCTGATCGTTATTCGAGGCTAAGTGTG cggatccccgggttaattaa;
YDR090MYCrev,
AAGCGTGCTATCAAAAAGAGATGAAAACGTGCTAACTAAAAAGGACTCAGATTCG gaattcgagctcgtttaaac.
The uppercase nucleotides correspond to the sequences used for homologous recombination at 3′ ends of SGE1 and ILT1. The lowercase nucleotides correspond to the annealing sequences for the pFA6a plasmids. Sanger sequencing of PCR products confirmed proper construction of all MYC- and GFP-tagged strains. Strains are available upon request.
Plasmid construction
E. coli strains E. cloni (Lucigen), DH10B (New England Biolabs), and EPI300 (Epicentre) were used for bacterial transformation, plasmid amplification, and assembly. Yeast genes were amplified by PCR of gDNA or fosmid DNA and primer pairs that annealed 1137 bp 5′ and 180 bp 3′ of the SGE1 ORF or 885 bp 5′ and 96 bp 3′ of the ILT1 ORF. SGE1 and ILT1 PCR products were cloned into pRS416 and pRS415 plasmids (Christianson et al. 1992), respectively, by sequence- and ligation-independent cloning (SLIC) (Li and Elledge 2007). Gene splicing by overlap extension (SOE) (Horton 1995) and SLIC cloning were used to generate mutant SGE1PLL plasmids. For experiments in YPD medium, SGE1 and ILT1 plasmids containing KanMX or hphMX antibiotic selection markers in place of URA3 or TRP1 auxotrophic markers, respectively, were used. Additionally, empty vector controls (lacking SGE1 or ILT1) with KanMX and hphMX selection markers were also generated. To generate doxycycline-inducible SGE1 expression plasmids, the SGE1 ORF was amplified by PCR with primers containing 60 bp of flanking sequence that were homologous to the CYC1 minimal promoter and 6× glycine linker/16× MYC tag in pBM5155 (Alexander et al. 2016). Purified PCR product was then cotransformed with NotI-digested pBM5155 into BY4741 (BY) yeast for gapped plasmid repair (Muhlrad et al. 1992). Plasmids were rescued from nourseothricin-resistant yeast colonies as described elsewhere (Mülleder et al. 2016) with modifications: cells were resuspended in 200 μl 1 M sorbitol with 20 units of zymolyase (Zymo Research) and incubated at 37° for 1 hr. Zymolyase-treated cells were then centrifuged at 3000 relative centrifugal force for 3 min, supernatant was aspirated, and then they were resuspended in P1 buffer for glass bead lysis. Rescued plasmids were then transformed into E. coli, miniprepped (QIAGEN), and fully sequenced to confirm proper in-frame insertion of SGE1 into the pBM5155 plasmid. Plasmids are available upon request.
CRISPR/Cas9-mediated genome editing
CRISPR/Cas9 editing was performed by modification of plasmids published elsewhere (Kuang et al. 2018). In brief, a protospacer adjacent motif single guide RNA (sgRNA) sequence (TTTCATTTTCTGTCATTATC) that targeted adjacent to the SGE1 SLS site along with an HDV ribozyme were cloned between the SNR52 promoter and terminator. This sgRNA expression cassette was then amplified by PCR and cloned into the NotI site of the pXIPHOS vector (accession MG897154; GenBank), which contains a codon-optimized Cas9 gene driven by the constitutive RNR2 promoter and the NatMX selection marker by gapped plasmid repair. SGE1 repair templates were generated by PCR amplification of 378 sge1SLS, BY SGE1PLL plasmid, or BY SGE1PLS and BY SGE1SLL sequences generated by gene SOE. Purified repair templates were cotransformed in 20-fold molar excess with the pXIPHOS-SGE1 sgRNA plasmid into the BY or 378 yeast strain. Single nourseothricin-resistant colonies were restreaked two times on YPD agar plates. SGE1 was amplified by PCR of gDNA from single colonies, purified, and sequenced to confirm the SGE1 allele swap. For the 378 sge1SLS/sge1SLS homozygous mutant strain, Sanger sequencing only identified the presence of the SLS allele. Confirmed strains were also tested for loss of nourseothricin resistance, which indicates that the strains also lost the pXIPHOS-SGE1 sgRNA plasmid.
Fosmid library construction and screening
A yeast fosmid library vector was prepared by adding the yeast replicative origin and URA3 gene from the pRS416 yeast shuttle vector into the pCC1FOS fosmid vector (Epicentre Biotechnologies). Yeast maintenance regions were amplified from pRS416 by PCR using the following primer pair: GACGGGCGGCCACCTGGGTCCTTTTCATCA and GACGGGCGGCTCTGTGCGGTATTTCACACC. The resulting ∼1.9-kb fragment and the pCC1FOS fosmid vector were digested with KasI restriction enzyme and ligated together using T4 ligase (Thermo Fisher). Plasmids was transformed into E. coli and sequence verified. The resulting plasmid, pDH219, was further digested with PmlI to release a small unnecessary fragment bordered by PmlI sites. The backbone was religated together, transformed into E. coli EPI300 cells, and verified by Sanger sequencing. The resulting plasmid (pDH241) was digested with PmlI to yield a blunt linear vector for fosmid library construction.
S. cerevisiae 378 gDNA was isolated as described elsewhere (Hoffman and Winston 1987). The metagenomic library was constructed following the manufacturer’s protocol for the pCC1FOS fosmid vector (Epicentre), with the modification that PmlI-linearized pDH241 was substituted for the pCC1FOS vector. E. coli transductions were plated on LB supplemented with 12.5 µg/ml chloramphenicol (henceforth cm 12.5), resulting in ∼3600 E. coli colonies. E. coli cells were swabbed up, diluted into LB cm 12.5 with CopyControl Fosmid Autoinduction Solution (Epicentre), grown, and purified by plasmid miniprepping (QIAGEN).
BY yeast-competent cells were prepared and transformed (Gietz and Schiestl 2007) with the fosmid library. Yeast transformants were selected on SC (pH 5) agar plates lacking uracil and supplemented with 0 or 125 mM [C2C1im]Cl. A total of 19 colonies that grew to large size on 125 mM [C2C1im]Cl were restreaked to confirm IIL tolerance. Fosmids were harvested from confirmed transformant yeast cells and transformed into electrocompetent E. coli EPI300 cells. Fosmid preparations were recovered from E. coli and sequenced from the fosmid backbone into the chromosome inserts, which were subsequently mapped to the S288c yeast genome sequence (Saccharomyces Genome Database).
Yeast growth assays
To assess IIL tolerance of wild and domesticated yeast isolates, total cell growth for each strain was determined as previously described (Parreiras et al. 2014) with modifications. Individual strains were cultured aerobically at 30° in 96-well plates containing YPD (pH 5), YPD (pH 5) with 250 mM [C2C1im]Cl and 250 mM [C2C1im][OAc]. Total cell growth for each strain was determined by subtracting the OD600 measurement after 24 hr of growth from the initial OD600 value. Relative total cell growth was calculated by dividing the total cell growth in YPD (pH 5) with 250 mM [C2C1im]Cl or [C2C1im][OAc] by the total cell growth in YPD (pH 5) alone. For BY strains transformed with pRS415-ILT1 or pRS416-SGE1 plasmids (Figure 1, B and C), triplicate cultures of S. cerevisiae were inoculated and grown to stationary phase in SC − leucine or SC − uracil medium, respectively. Cells were diluted to an OD600 of 0.05 in the appropriate SC medium containing 0–250 mM [C2C1im]Cl or 125 mM [C2C1im][OAc]. Cell growth was monitored by OD600 measurements every 20 min at 30° with shaking using an Infinite F-200, F-200 PRO, or Safire multimode reader (Tecan).
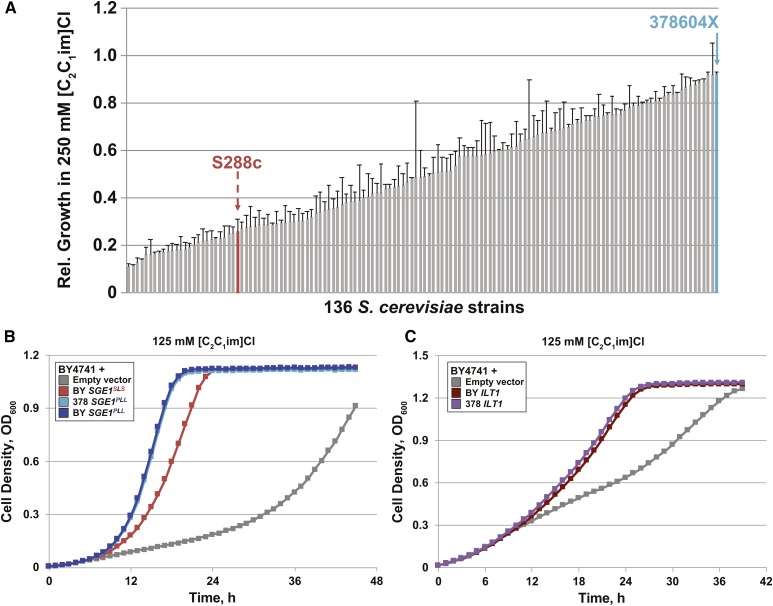
Figure 1.
SGE1 and YDR090C/ILT1 function in [C2C1im]+ tolerance. (A) Relative total cell densities for 136 wild or domesticated S. cerevisiae strains cultured aerobically in 96-well plates containing YPD (pH 5) + 250 mM [C2C1im]Cl medium, relative to total cell growth in YPD (pH 5) alone. Red and blue ↓’s indicate the locations of the S288c laboratory and 378 wild strains, respectively. Average values and SEM were determined from independent biological triplicates. (B and C) Representative aerobic growth of BY cells transformed with the indicated low-copy plasmids and cultured in SC (pH 5) medium + 125 mM [C2C1im]Cl for 40–46 hr. Vector-specific growth differences were observed in (B and C). Rel., relative.
Growth assays in 24-well plates were similarly carried out as described for 96-well plates with modifications. Yeast strains were cultured overnight at 30° in tubes containing 5 ml of YPD medium and any appropriate antibiotic for plasmid selection. The following morning, cultures were diluted in fresh medium and regrown to log phase (OD600 = 0.8, as measured in a 1-cm path length cuvette with a Beckman Coulter spectrophotometer). Cells were centrifuged, washed with sterile water, and inoculated into 24-well plates at an OD600 of 0.1 in 1.5 ml YPD (pH 5) media containing serial dilutions of [C2C1im]Cl, [C2C1im][OAc], [C4C1im]Cl, or CV and any appropriate antibiotics. Cell densities were determined from OD600 measurements taken every 10–40 min for 24–70 hr with a Tecan Infinite M200Pro multimode reader. Relative total cell growth was determined by dividing the total cell growth after 18–24 hr for a strain in media containing specified concentrations of CV, [C4C1im]Cl, or [C2C1im]Cl by the cell growth for the same strain in media lacking those compounds in the same time frame. For pH-dependency experiments, relative total cell growth was determined by dividing the total cell growth after 24 hr for each strain in 125 mM [C2C1im]Cl by the total cell growth for the same strain in media lacking [C2C1im]Cl at a specific pH. For experiments using tetracycline-inducible SGE1-MYC, BY sge1Δ yeast transformed with empty pBM5155, pBM5155-SGE1SLS, or pBM5155-SGE1PLL were cultured in 1.5 ml YPD (pH 5), 0 or 125 mM [C2C1im]Cl, 100 μg/ml nourseothricin, and 0–625 ng/ml doxycycline hydrochloride (BP26535; Fisher Scientific). Relative total cell growth was determined by dividing the total cell growth after 24 hr for each strain by the total cell growth for yeast with empty pBM515 in 0 ng/ml doxycycline hydrochloride.
Aerobic and anaerobic growth experiments were performed with tubes and flasks, respectively, as previously described (Parreiras et al. 2014) with some modifications. Cells grown to log phase in 10–30 ml YPD (pH 5) medium containing the appropriate antibiotic for plasmid selection were washed with sterile water and inoculated to a concentration of OD600 = 0.1 in YPD (pH 5) medium containing 0–250 mM [C2C1im]Cl and the appropriate antibiotic for plasmid selection. Cell densities were determined by OD600 spectrophotometer (Beckman Coulter) measurements as described above. Extracellular glucose and ethanol concentrations were determined by high performance liquid chromatography and refractive index detection (Keating et al. 2014).
SGE1 sequence analysis
Single nucleotide polymorphisms (SNPs) for the SGE1 gene were extracted from a whole genome variant data set for the strains phenotyped in this study (Sardi et al. 2018). Briefly, whole genome Illumina sequences for S. cerevisiae strains from publicly available sequencing projects (Skelly et al. 2013; Bergström et al. 2014; Hose et al. 2015; Strope et al. 2015) were mapped to reference genome S288c [NC_001133 version 64 (Engel et al. 2014)] using bwa-mem. Variants were identified using the GATK pipeline for Unified Genotyper (McKenna et al. 2010) using default parameters with a -mbq of 25 to reduce false positives. Annotation of variants was performed with SNPEff (Cingolani et al. 2012).
Additional genome sequences from S. cerevisiae and non-S. cerevisiae yeast strains were obtained as described in previous publications (Scannell et al. 2011; Liti et al. 2013; Almeida et al. 2014; Bing et al. 2014; Gayevskiy and Goddard 2016; Gonçalves et al. 2016; Peris et al. 2016; Yue et al. 2017). SGE1 sequences from additional S. cerevisiae and non-S. cerevisiae strains (File S3) were retrieved by using two approaches: (1) BLASTing the S288c SGE1 gene sequence to a local database (Altschul et al. 1990), and/or (2) downloading the Illumina reads and mapping them to the reference SGE1 nucleotide sequence to extract and assemble the SGE1 alleles using the HybPiper wrapper (Johnson et al. 2016). Sequence alignment and amino acid comparisons were performed in Geneious version 6.1.6 (Kearse et al. 2012).
Western blotting
Protein from whole cell lysates were prepared as previously described (Zhang et al. 2011) with modifications. Yeast cells were grown in 24-well plates as described above. After 24 hr of growth, 1.5 ml of cell culture was transferred to tubes, centrifuged, and the supernatant was aspirated. Cell pellets were washed with sterile water and then resuspended in 67 μl of 2 M lithium acetate per OD600 of cells. LiAc-treated cells were then centrifuged, the supernatant was aspirated, and the cells were resuspended in 67 μl/OD600 of cells in 0.4 M NaOH and placed on ice for 5 min. Treated cells were then centrifuged, the supernatant was aspirated, and protein was extracted by lysing cells in 1× Laemmli sample buffer with β-mercaptoethanol at 100° for 5 min.
Protein electrophoresis and Western blotting were performed using the Mini-Protean system according to the manufacturer’s protocol (Bio-Rad, Hercules, CA). Total protein (30 mg) from each cell sample was loaded on the 4–15% acrylamide gels, along with 5 μl chemiluminescent protein standards (Precision Plus Protein WesternC Standards; Bio-Rad). Blots were cut into two separate pieces along the 50-kD protein standard, with the top half used to for detection of Sge1-Myc, and the bottom half used for actin. Sge1-Myc and actin proteins were detected by incubation with anti-Myc (9E10; Sigma Aldrich) or anti-actin (mAbGEa; Thermo Fisher Scientific) mouse monoclonal primary antibodies at dilutions of 1:1000 in TBST (Tris buffered saline + 0.2% Tween-20; Bio-Rad) and 5% nonfat dried milk (TBSTM) or 1:5000 in TBST with 3% bovine serum albumin (TBSTB), respectively. After primary incubation, washed blots were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (Bio-Rad) and StrepTactin-HRP conjugate (Bio-Rad) in corresponding TBSTM or TBSTB buffer. Sge1-Myc and actin protein bands were visualized by enhanced chemiluminescence (Clarity Western ECL; Bio-Rad) and quantified by densitometry (Quantity One Software; Bio-Rad).
Quantification of chemiluminescent signals for Sge1-Myc, actin, and an ∼130-kD nonspecific (NS) band was performed by densitometry (Quantity One Software; Bio-Rad). Volume intensities for each were measured using identical areas for each protein across replicate experiments. Actin and Sge1-Myc/NS band signals were captured from 5 or 30–60 sec exposures, respectively. Each Sge1-Myc chemiluminescent signal was normalized relative to the NS or actin bands from the same sample for three to five biological replicates. For correlations, Sge1-Myc protein levels normalized to NS band intensities were paired to relative total cell growth (see above) from the same sample. Paired values were analyzed by Spearman’s rank correlation in Spotfire (TIBCO).
Fluorescence microscopy
Yeast strains were cultured to exponential growth phase in YPD at 30°. Cells were harvested by centrifugation and resuspended at a ratio of 1:5 in fresh YPD media. A total of 3 μl of cells were then spotted onto a poly-l-lysine-coated glass slide and covered with an 8 × 8 mm coverslip. GFP fusions were visualized at 100× magnification using fluorescence (with an EVOS GFP LED cube) or transmitted light sources, an EVOS FL Auto 2 microscope (Invitrogen, Carlsbad, CA), and EVOS FL Auto 2 Imaging System (Invitrogen). Contrast and brightness for whole images were adjusted uniformly using Adobe Photoshop in accordance with the journal’s image-manipulation policy.
Data availability
All yeast strains and plasmids used in this study are available upon request. Table S2 contains a list of the strains described in this study. Data necessary to confirm findings of this article are present within the article text and figures as well as supplemental files, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6731201.
Results
Environmental isolates of S. cerevisiae display a range of growth abilities in IIL media
To identify strains with innate tolerance to IILs, we measured the growth of 136 wild and domesticated S. cerevisiae strains cultured in media containing 250 mM [C2C1im]Cl (Figure 1A and File S1), [C2C1im][OAc] (Figure S1 and File S1), or lacking IILs. Strains displayed a wide range of maximum cell growth in IIL-containing media relative to the control medium lacking IILs, with more strains growing to lower cell densities in [C2C1im][OAc] than [C2C1im]Cl (File S1). Weak correlation (R2 = 0.27) in relative cell growth between the different IILs indicated that the Cl− and [OAc]− anions synergize differently with [C2C1im]+ to inhibit yeast growth, consistent with an earlier report (Ouellet et al. 2011). The strain with highest relative growth in both [C2C1im]Cl and [C2C1im][OAc] was 378, a clinical isolate from Newcastle, United Kingdom. In contrast, the common laboratory strain, S288c, achieved significantly lower relative cell growth in [C2C1im]Cl and [C2C1im][OAc], suggesting that the 378 strain contains different genetic sequences that permit greater cell growth in the presence of [C2C1im]+.
SGE1 and YDR090C/ILT1 function in ionic liquid tolerance
Reasoning that genetic factors in the 378 strain are responsible for its IIL tolerance, we used a selection scheme adapted from our previous work with bacteria (Ruegg et al. 2014). Specifically, we generated a fosmid library containing large (∼30–40 kb) fragments of gDNA from this IIL-tolerant strain and transformed the library into an IIL-sensitive laboratory derivative of S288c, BY (Figure S2). We then selected transformants that grew normally on solid medium containing 125 mM [C2C1im]Cl. Fosmid inserts from selected transformants were recovered and partially sequenced onto the S. cerevisiae genome. We identified nine distinct inserts that clustered in two genome regions. Five of these nine DNA segments coincided in a core 14-kb region of chromosome XVI that contained four genes: HPA2, encoding a histone acetyl transferase (Angus-Hill et al. 1999); OPT2, an oligopeptide transporter (Wiles et al. 2006) that may also function in drug detoxification (Aouida et al. 2009); SKI3, involved in exosome-mediated messenger RNA decay (Anderson and Parker 1998); and SGE1, an MFS multidrug efflux pump that exports toxic cationic dyes out of the cytoplasm (Amakasu et al. 1993; Ehrenhofer-Murray et al. 1994, 1998; Jacquot et al. 1997). Because we previously found that a bacterial MFS efflux pump, EilA, functions in [C2C1im]+ tolerance (Ruegg et al. 2014), we were immediately drawn to investigate SGE1.
The remaining four fosmid inserts coincided in a core 18-kb region of chromosome IV containing nine genes: STN1, RRP8, TVP23, AFR1, SSS1, RRP1, SLU7, YDR089W, and YDR090C (Figure S2). Unlike SGE1, none of these genes had immediately obvious functions predicted to be relevant to IIL tolerance. Considering that deletions of IIL-tolerance genes would sensitize yeast to [C2C1im]Cl, we examined five available strains from the BY deletion library (Winzeler et al. 1999) containing individual deletions in genes within this region of chromosome IV; only ydr090cΔ displayed reduced growth in medium with a subtoxic 31 mM [C2C1im]Cl concentration compared to medium without IIL (Table S2). Based on these results, we propose ILT1 as the standard name for the YDR090C ORF. Protein structure and homology analyses (Altschul et al. 1990; Claros and von Heijne 1994) indicated that ILT1 encodes a membrane protein with seven transmembrane helices and has a putative PQ-loop motif found in Ypq1, Ypq2, and Rtc2/Ypq3, which are putative vacuolar membrane transporters of cationic amino acids (Jézégou et al. 2012).
Next, we investigated whether specific expression of SGE1 or ILT1 could explain the tolerance phenotypes of the fosmid-carrying strains. The individual promoters, ORFs, and terminators for SGE1 and ILT1 from the tolerant 378 and sensitive BY strains were cloned into a low-copy plasmid and expressed in the BY strain (Figure 1, B and C, and Figure S3). In media containing 125 mM [C2C1im]Cl, BY transformants expressing the SGE1 and ILT1 ORFs from the tolerant 378 strain grew faster than BY containing the same plasmid but lacking SGE1 (empty vector), providing evidence that these specific genes contributed to IIL-tolerance effects from the fosmid constructs. Expression of BY SGE1 and ILT1 alleles also increased growth in 125 mM [C2C1im]Cl over the empty vector control, indicating that additional copies of the identical BY SGE1 and ILT1 alleles granted IIL protection. However, expression of the 378 SGE1 gene conferred faster growth than BY SGE1 in [C2C1im]Cl-containing media, suggesting that differences in the BY and 378 SGE1 sequences affect IIL tolerance. In contrast, there were no growth differences between cells expressing ILT1 from BY or 378.
The SGE1 sequences from the sensitive BY and tolerant 378 strains were examined for coding differences that could explain the IIL-tolerance phenotypes. Two nonsynonymous SNPs corresponding to amino acid positions Ser 282 and Ser 284 (hereafter denoted as the “SLS” sequence), which flank Leu 283, in BY were found to encode Pro 282 and Leu 284 (the “PLL” sequence) in 378. Transmembrane prediction models place these amino acid residues in the fifth cytoplasmic loop between the eighth and ninth transmembrane helices of Sge1p. To exclude any effects from promoter and terminator sequence differences or silent mutations between the BY and 378 strains, we generated site-directed S282P and S284L mutations in our low copy plasmid-borne SGE1 from the sensitive BY strain (BY SGE1PLL). Expression of the BY SGE1PLL mutant in the BY strain enabled an equivalent growth rate as 378 SGE1PLL (Figure 1B), indicating that the Pro 282 and Leu 284 amino acid differences were specifically responsible for the increased [C2C1im]Cl tolerance. There were no differences in the ILT1 coding sequence between the BY and 378 strains, explaining why no growth differences were seen for strains expressing BY or 378 ILT1 (Figure 1C).
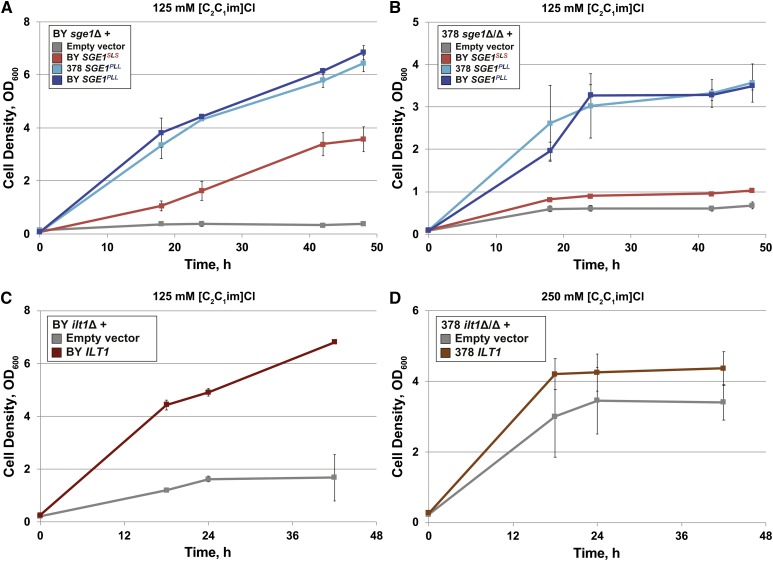
We next examined the requirements for SGE1 and ILT1 in IIL tolerance by deleting SGE1 or ILT1 genes in the haploid BY and homozygous diploid 378 strains. Both sge1Δ and ilt1Δ deletion mutants were transformed with an empty vector or plasmids containing the various SGE1 and ILT1 sequences and examined for growth in media containing [C2C1im]Cl (Figure 2). The BY sge1Δ and 378 sge1Δ/sge1Δ null mutant strains grew to significantly lower cell densities in 125 mM [C2C1im]Cl than strains expressing their corresponding wild-type SGE1 sequences (Figure 2, A and B). Expression of the BY SGE1PLL or 378 SGE1PLL alleles resulted in significantly faster growth and higher cell densities than the BY SGE1SLS allele in both BY sge1Δ and 378 sge1Δ/sge1Δ mutant strains. The BY ilt1Δ strain transformed with the empty control vector grew significantly slower than the deletion strain complemented with the ILT1 sequence (Figure 2C). In contrast, no significant differences in cell growth were seen between 378 ilt1Δ/ilt1Δ mutants transformed with empty or ILT1 plasmids (Figure 2D), suggesting that ILT1 has strain-specific functions. Together, these results indicate that both SGE1 and ILT1 function in [C2C1im]+ tolerance, and that natural sequence differences in SGE1 alleles can influence tolerance levels.
Figure 2.
SGE1 alleles from BY and 378 determine [C2C1im]+-tolerance phenotypes. Haploid BY or diploid 378 strains harboring (A and B) sge1Δ or (C and D) ilt1Δ null mutations were transformed with plasmids containing the indicated SGE1 or ILT1 sequences. In (A and B), a plasmid containing site-directed mutations in the BY SGE1 gene sequence (BY SGE1PLL) was included. Transformed strains were then cultured aerobically in tubes containing 10 ml YPD (pH 5), 125 mM [C2C1im]Cl, and the appropriate antibiotic. Average cell densities (OD600) ± SD are reported from three independent biological replicates.
Since some biofuels, such as ethanol, are industrially produced under anaerobic, fermentative conditions, we assessed whether SGE1 and ILT1 were important for ionic liquid tolerance in the absence of oxygen. Consistent with the aerobic results, both BY sge1Δ and ilt1Δ strains transformed with empty plasmids grew more slowly and fermented less glucose to ethanol anaerobically than strains expressing their native SGE1 or ILT1 sequences in media containing 250 mM [C2C1im]Cl (Figure S4). BY sge1Δ mutants expressing the PLL allele grew to higher cell densities and fermented more glucose into ethanol in the presence of 250 mM [C2C1im]Cl than the BY strain expressing the native SGE1SLS allele (Figure S4, A–C). Thus, swapping the SGE1PLL allele into the BY genome enabled greater anaerobic bioethanol production in the presence of [C2C1im]Cl.
SGE1 and ILT1 confer resistance to multiple cationic compounds
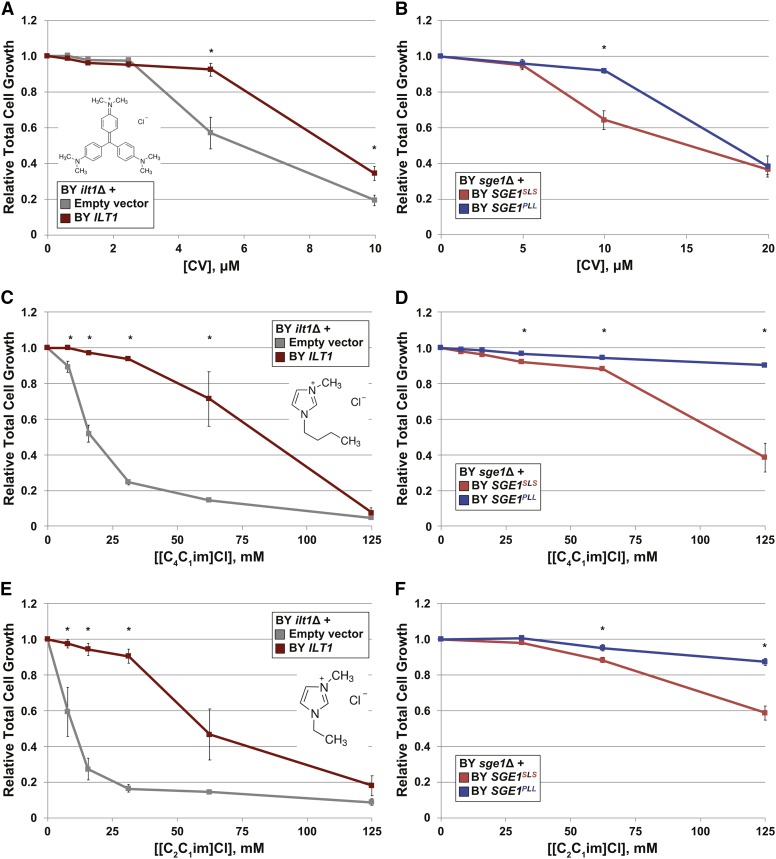
Previously, SGE1 was identified for its role in resistance to the toxic cationic dyes CV, 10-N-nonyl acridine orange, and ethidium bromide in other S. cerevisiae strain backgrounds (Ehrenhofer-Murray et al. 1994; Jacquot et al. 1997). We wanted to determine if ILT1 functioned similarly in cationic dye resistance and whether the SGE1PLL allele conferred greater resistance to other cationic dyes than the SGE1SLS allele. Compared to the BY parent strain, we found that BY ilt1Δ mutants grew to lower cell densities in 10 and 20 μM CV (Figure 3A). In contrast, deletions of both ILT1 copies in the 378 strain background, which retained two copies of the SGE1PLL allele, had insignificant effects on tolerance to CV (Figure S5A) and [C2C1im]Cl (Figure S5B), consistent with results seen with plasmid-transformed strains (Figure 2D). Additionally, BY and 378 strains containing SGE1PLL grew to relatively higher cell densities than strains containing the SGE1SLS alleles in 10 or 20 μM CV (Figure 3B and Figure S5B) and, as expected, in 125 or 250 mM [C2C1im]Cl (Figure S5D). As a further test of specificity, we also compared wild-type and mutant cell growth in media containing [C4C1im]Cl (Figure 3, C and D), another IIL used in pretreatment of plant feedstocks (Binder and Raines 2010), to growth in media containing [C2C1im]Cl (Figure 3, E and F). Similar to [C2C1im]Cl, deletion of ILT1 significantly reduced the growth tolerance to [C4C1im]Cl, and expression of the BY SGE1PLL allele conferred greater resistance to [C4C1im]Cl than the SGE1SLS allele. These results indicate that ILT1 functions in tolerance to a range of cationic toxins in the BY strain background, while the SGE1PLL allele enables greater tolerance than the SGE1SLS allele across multiple cationic toxins in both BY and 378 strains.
Figure 3.
SGE1 and ILT1 function in resistance to multiple cationic inhibitors. The sge1Δ or ilt1Δ mutants were transformed with empty vector or a plasmid containing unmodified or mutant SGE1 or ILT1 from BY. Transformed strains were then cultured aerobically in YPD (pH 5) and the indicated concentrations of (A and B) CV, (C and D) [C4C1im]Cl, or (E and F) [C2C1im]Cl. Relative growth was determined by measuring the total cell growth after 18 hr of culturing in YPD (pH 5) containing the cationic compound normalized to total growth in YPD (pH 5) alone. Average relative total growth ± SD was plotted from three independent biological triplicates. Statistical significance determined by paired Student’s t-test. * P < 0.05.
The Sge1PLL H+ antiporter maintains IIL tolerance across a wider extracellular pH range than Sge1SLS protein
MFS protein member Sge1 is a Dha2-like, 14-span transmembrane H+ antiporter that couples the import of protons from the extracellular medium with the export of toxins out of the cytoplasm (Sá-Correia et al. 2009; Dos Santos et al. 2014). To test whether the efflux of IILs by the Sge1 variants is coupled with proton influx, we examined the interaction between extracellular pH and IIL tolerance for both the SGE1SLS and SGE1PLL alleles. Between pH 8 and 9, both the SGE1SLS and SGE1PLL strains failed to grow significantly in the presence of 125 mM [C2C1im]Cl (Figure S6). From pH 5 to 7, the SGE1PLL strain grew to relatively higher cell densities than the SGE1SLS strain, whereas both strains grew to similar cell densities at pH 4. These observations indicated that the tolerant SGE1PLL allele enables greater IIL tolerance in a wider pH range than the sensitive SGE1SLS allele.
The SGE1PLL allele is the ancestral sequence associated with IIL tolerance across natural S. cerevisiae strains
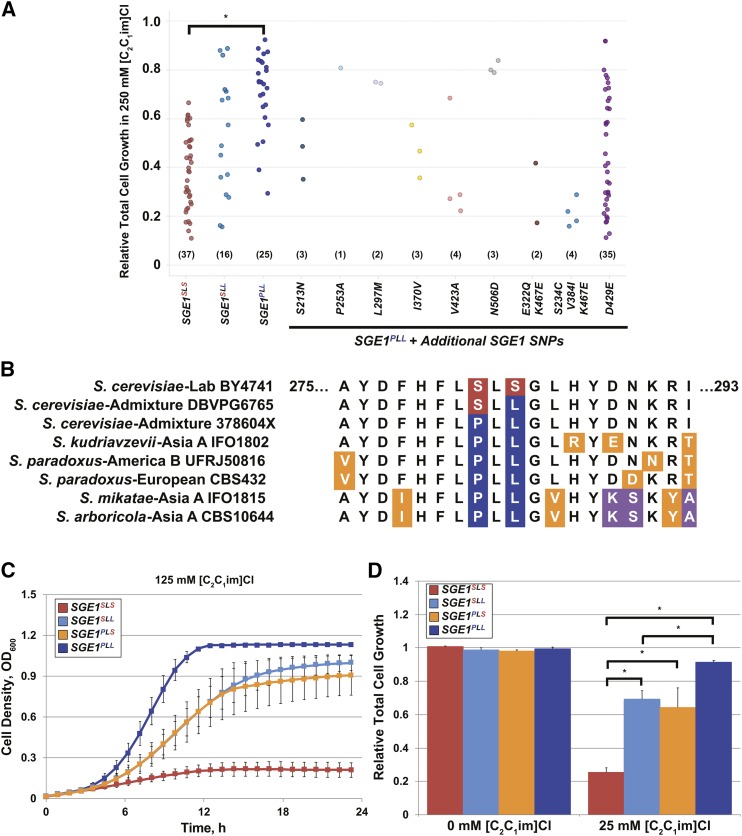
The discovery that two different alleles of SGE1 conferred differential tolerance to [C2C1im]+ prompted us to investigate whether the tolerance phenotypes of 136 haploid or homozygous diploid S. cerevisiae strains examined in Figure 1 correlated with the SLS and PLL genotypes. Using published genome sequences (Sardi et al. 2018) and targeted Sanger sequencing, we identified 37 strains containing the SGE1SLS allele, 16 with an SGE1SLL allele, 25 with SGE1PLL, and 58 strains containing the SGE1PLL allele along with one or more additional polymorphisms (see “SGE1PLL + Additional SGE1 SNPs” in Figure 4A, File S1, and File S2). No sequenced strains from our collection contained an SGE1PLS allele. In media containing 250 mM [C2C1im]Cl (Figure 4A), the relative growth for strains containing only the SGE1PLL allele (average relative growth = 0.709 ± 0.159) was significantly higher (P < 0.05) than strains containing the SGE1SLS allele (average relative growth = 0.386 ± 0.155). We did not observe any statistically significant correlations between IIL tolerance and strains containing both the SGE1PLL sequence and additional SGE1 SNPs, although the small number of representatives limited our statistical power. These results indicate that yeast strains containing SGE1SLS, SGE1SLL, and SGE1PLL alleles persist in the natural environment and influence tolerance to cationic toxins.
Figure 4.
The SGE1PLL allele confers greater tolerance to [C2C1im]Cl than the SGE1SLS variant. (A) The individual S. cerevisiae strains were grouped based on their SGE1 genotype and plotted according to their relative growth in 250 mM [C2C1im]Cl. For strains containing SGE1PLL and additional SGE1 SNPs, the amino acid changes in each strain are relative to S288c reference sequence. The numbers of strains for each genotype are listed in parentheses. Statistical significance of growth differences between strains containing the SGE1PLL alone and SGE1SLS alleles was determined by unpaired Student’s t-test. * P < 2e−10. (B) Sge1 protein sequences encompassing the region surrounding the Sge1SLS/Sge1PLL variants from different strains and species from the genus Saccharomyces were aligned to display the amino acid sequences at the corresponding SLS/PLL residues. For each species, their locations of isolation as well as common strain identifiers are listed. (C) Average ± SEM aerobic cell densities of BY SGE1SLS cells or the indicated SGE1 mutations introduced into the genome by CRISPR/Cas9 from three biological replicates are shown. (D) Average total cell growth + SD was normalized relative to growth for BY SGE1SLS strain after 10 hr of culturing and plotted with SD from three independent biological replicates. Statistical significance determined by paired Student’s t-tests. * P < 0.05.
The identification of the SGE1SLL allele along with the inability to detect the SGE1PLS allele suggested that the SGE1SLS or SGE1PLL allele was derived from an ancestral sequence through two successive nucleotide changes. To ascertain the order in which the derived alleles emerged, we identified the ancestral sequence by aligning analogous Sge1 protein sequences from other species within the genus Saccharomyces (Scannell et al. 2011; Bing et al. 2014; Yue et al. 2017), which diverged from S. cerevisiae up to 20 MYA (Figure 4B). Interestingly, SGE1 from five different species of Saccharomyces contained the SGE1PLL sequence, whereas the SGE1SLS allele was not identified in any other sequenced non-S. cerevisiae species. We also found that the genome sequences of multiple S. eubayanus, S. uvarum, S. paradoxus, and even S. cerevisiae strains lacked the SGE1 gene, and two strains of S. cerevisiae also appeared to contain premature stop codons (File S3). This suggests that SGE1PLL was the tolerant ancestral allele and that the sensitive SGE1SLS allele was derived from an initial P282S mutation followed by the secondary L284S mutation.
To understand the emergence of the derived SGE1SLS-sensitive allele, we determined the functional effects that each individual sequence variant contributed to the IIL-tolerance phenotypes. First, CRISPR/Cas9 and homologous recombination were used to precisely edit the BY genome to generate strains containing specific SGE1SLL, SGE1PLS, or SGE1PLL mutations. When cultured in 125 mM [C2C1im]Cl, BY strains harboring the single SGE1SLL or SGE1PLS mutations displayed growth rates and total cell growth phenotypes that were intermediate from the tolerant SGE1PLL and sensitive SGE1SLS strains (Figure 4, C and D). Furthermore, the phenotypes of the SGE1SLL and SGE1PLS strains were statistically indistinguishable, indicating that both P282S and L284S changes were needed to derive the SGE1SLS phenotype. These results further suggest the order of the P282S and the L284S mutations were interchangeable in the context of cationic toxin tolerance.
Sge1PLL protein is present at higher abundance than Sge1SLS in cells
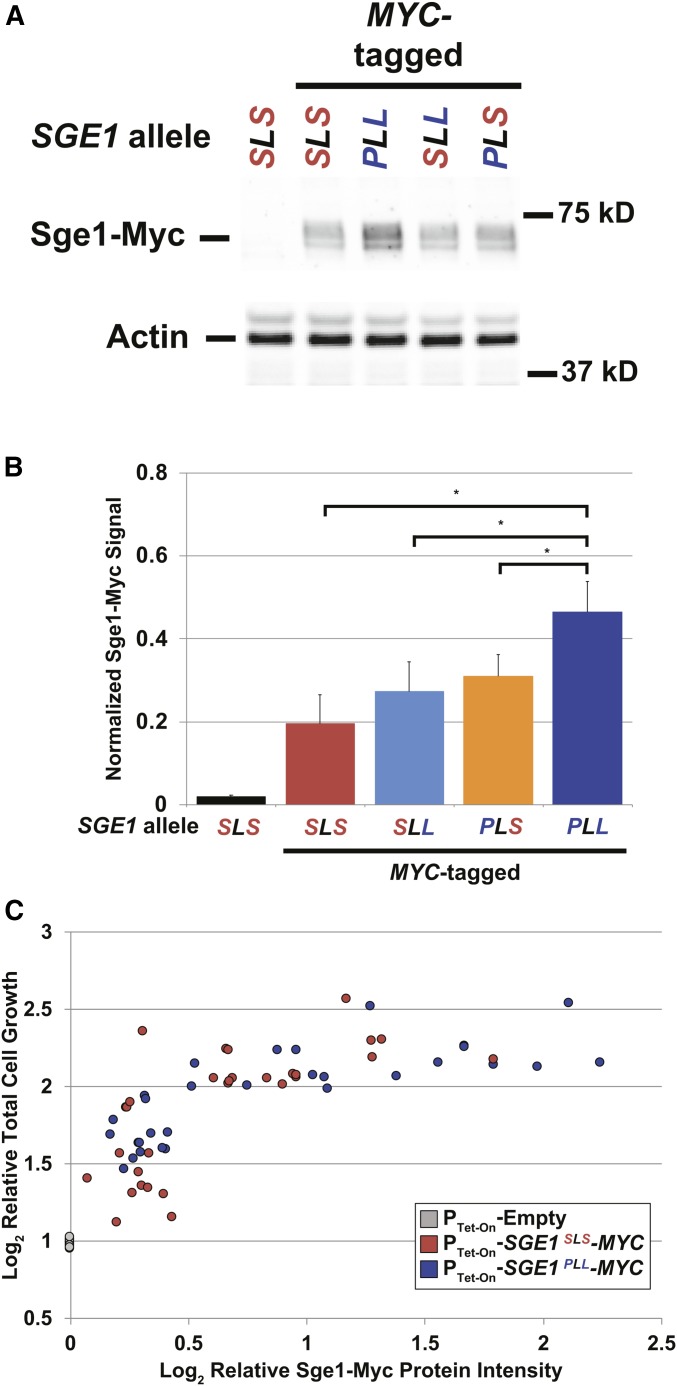
One potential explanation for the increased tolerance to ionic liquids is that the Sge1PLL protein is more abundant in cells than Sge1SLS protein, resulting in greater export of toxic IILs from the cytoplasm. We investigated this possibility by comparing Myc-tagged Sge1SLS and Sge1PLL protein levels in the BY strain background by Western blotting. A Myc epitope tag was inserted in frame at the 3′ end of wild-type SGE1SLS, mutant SGE1SLL, SGE1PLS, and SGE1PLL sequences generated in the BY genome by CRISPR/Cas9. Strains containing the various Sge1-Myc fusions grew similarly to the untagged strains in the presence of 125 mM [C2C1im]Cl (Figure 4C and Figure S7), confirming that the fusion of Myc tags did not significantly alter Sge1p function. Equal amounts of total protein from strains expressing the variant Sge1-Myc protein fusions were electrophoresed and blotted with anti-Myc and anti-actin antibodies. The normalized Sge1PLL-Myc protein signal was significantly greater than that of Sge1SLS-Myc (Figure 5, A and B). Moreover, both Sge1PLL-Myc and Sge1SLS-Myc proteins appeared to separate into more than one band, suggesting that Sge1 protein may be post-translationally modified. These results indicate that the SGE1PLL allele may confer greater tolerance to IILs and cationic dyes due to greater protein abundance and stability than the SGE1SLS variant.
Figure 5.
Increased Sge1 protein abundance correlates with increased tolerance to IILs. (A) A representative Western blot of different alleles of Myc-tagged Sge1 or actin protein from total cell lysates harvested from the indicated BY strains. Chemiluminescence signal for Sge1-Myc was normalized to actin signal from the same sample. (B) Average normalized Sge1-Myc signals + SEM were plotted from five independent biological replicates. Statistical significance was determined by paired Student’s tests. * P < 0.05. (C) sge1Δ mutant cells containing a plasmid with SGE1-MYC alleles driven by a tetracycline-inducible promoter were cultured in YPD (pH 5) medium containing 0–625 ng/ml doxycycline and 125 mM [C2C1im]Cl for 24 hr. Total cell growth was recorded and cells were harvested for total cellular protein lysates after 24 hr. Sge1-Myc protein was quantified with anti-Myc antibodies and normalized for protein loading (see Figure S7 and Materials and Methods). Normalized Sge1-Myc signal from each strain condition was plotted against the total cell growth relative to sge1Δ cells transformed with PTet-On-Empty plasmid and grown in 0 ng/ml doxycycline and 125 mM [C2C1im]Cl.
If SGE1 SNPs determine IIL tolerance primarily by affecting Sge1p abundance, rather than by affecting transporter activities, we hypothesized that yeast strains expressing equivalent levels of Sge1SLS and Sge1PLL protein would display similar IIL tolerance. To test this model, we cultured the sge1Δ mutant strain transformed with a plasmid containing SGE1-MYC under the control of a doxycycline-inducible promoter in a range of doxycycline concentrations. Increasing doxycycline concentrations did not affect the growth of cells containing the empty control vector in the presence of 125 mM [C2C1im]Cl (Figure S8A) or the growth of cells containing SGE1-MYC in the absence of [C2C1im]Cl. In contrast, strains harboring the Tet-inducible SGE1-MYC plasmid displayed doxycycline-dependent increases in cell growth and Sge1SLS-Myc (Figure S8B) and Sge1PLL-Myc (Figure S8C) protein levels in the presence of [C2C1im]Cl. We then compared the normalized Sge1-Myc protein signal coupled to the relative total cell growth for each strain across multiple doses of doxycycline (Figure 5C). By Spearman rank analysis (not including data from PTet-On-Empty samples), we found that Sge1-Myc protein levels significantly correlated with relative total cell growth (R2 = 0.6, P < 3.2e−13), regardless of the Sge1 protein sequence. Together, these results support a model in which the natural SGE1SLS and SGE1PLL alleles primarily affect Sge1 protein abundance, which in turn determines cellular IIL tolerance.
Sge1PLL and Ilt1 proteins function independently at the plasma membrane
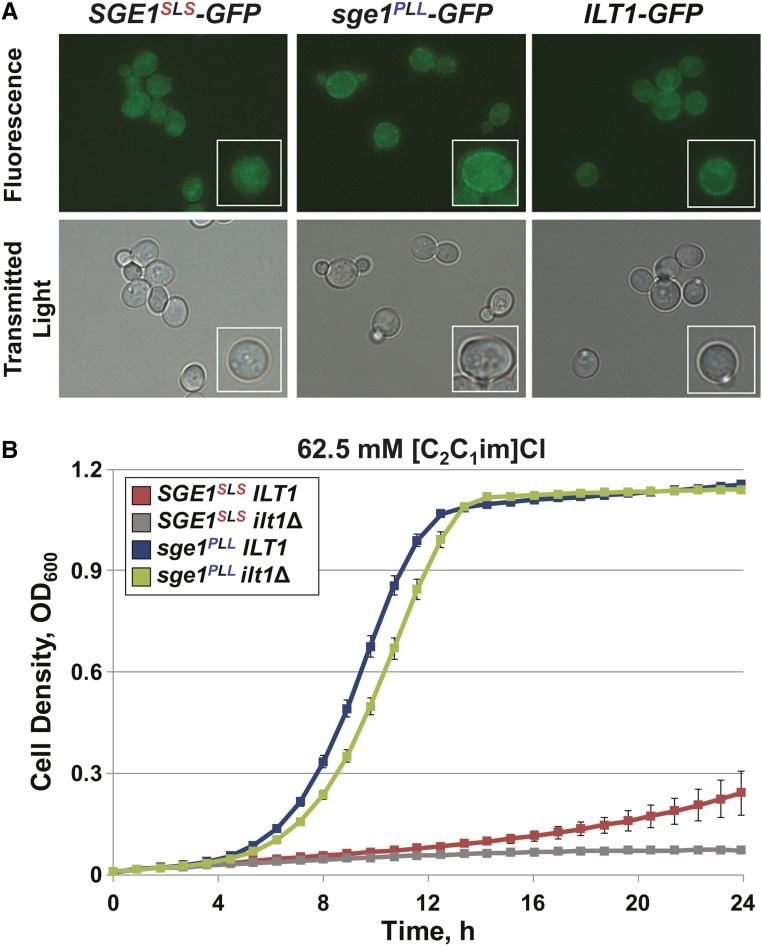
Sge1 protein has been proposed to function at the plasma membrane to extrude cationic toxins from the cytoplasm (Ehrenhofer-Murray et al. 1998). Given this, we predicted that increased abundance of Sge1PLL protein could also result in greater localization to the plasma membrane than the Sge1SLS variant protein. We tested this prediction by inserting GFP in frame at the carboxyl termini of chromosomal SGE1 and ILT1. Strains containing these gene fusions did not grow significantly differently from strains containing the untagged genes (Figure S8). By fluorescence microscopy, both Sge1PLL-GFP and Ilt1-GFP fusion proteins localized to the plasma membrane, whereas the Sge1SLS-GFP fusion appeared to localize weakly to the plasma membrane and to internal organelles (Figure 6A). This suggested that, in the BY strain background, the PLL sequence promotes greater Sge1 abundance at the plasma membrane than the IIL-sensitive SLS sequence.
Figure 6.
Plasma membrane-localized Sge1PLL protein functions independently of ILT1. BY strains containing GFP fused to the indicated genes were cultured in YPD medium. (A) GFP fluorescence from representative cells was visualized with 100× magnification. Insets in the bottom right corners display a single representative cell with an additional 50% higher magnification. (B) ILT1 was deleted from BY strains containing SGE1SLS or sge1PLL alleles. Resulting strains were cultured in YPD (pH 5) medium containing 62.5 mM [C2C1im]Cl. Cell growth is reported as average cell densities ± SEM from three independent biological replicates.
The localization of both Sge1PLL and Ilt1 protein to the plasma membrane suggested the possibility that Ilt1 functionally interacts with Sge1 in IIL tolerance. To examine this possibility, we compared the growth of BY strains containing deletion mutations in ILT1 in medium containing [C2C1im]Cl (Figure 6B). The BY sge1PLLilt1Δ double mutant strain was similarly tolerant to [C2C1im]Cl as the sge1PLL strain containing wild-type ILT1. This indicated that sge1PLL function does not require ILT1 for IIL tolerance in the BY strain background.
Discussion
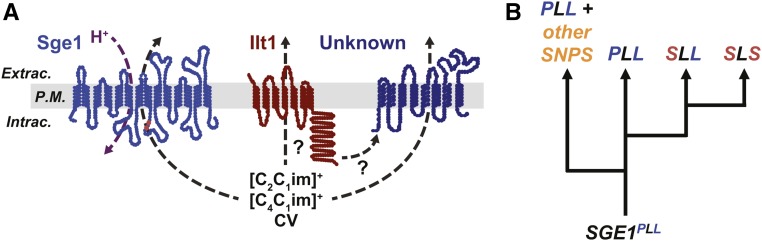
In this investigation of natural variants of S. cerevisiae, we discovered novel functions for YDR090C/ILT1 and two alleles of the SGE1 efflux pump in tolerance to IIL solvents and cationic toxins. Our genetic and biochemical analyses support a model in which Sge1 and Ilt1 function in resistance to cationic toxins (Figure 7A). The H+ antiporter Sge1 presumably exports toxic IIL cations from the cytoplasm through the plasma membrane in a similar manner as has been shown for cationic dyes (Ehrenhofer-Murray et al. 1994). The greater abundance of Sge1PLL protein at the plasma membrane likely enables greater extrusion of IILs and cationic toxins from the cell over the less abundant Sge1SLS protein. Ilt1 may also function at the plasma membrane and through a mechanism distinct from Sge1, but whether it directly exports ionic liquids and cationic compounds out of the cell remains unclear. Ilt1p shares homology to the paralogs Ypq1p, Ypq2p, and Rtc2p/Ypq3p, solely through a PQ-loop domain (Ponting et al. 2001) which is present on vacuolar/lysosomal transporters of cationic amino acids (Cherqui et al. 2001; Jézégou et al. 2012), suggesting the possibility that Ilt1 may also function in transport. Additional studies are needed to better understand the role of Ilt1p function in IIL tolerance.
Figure 7.
A model for the role of yeast transmembrane proteins in resistance to cationic toxins and the emergence of SGE1 alleles. The model in (A) proposes the functions of ILT1 and SGE1 determined from this study. Sge1 functions in exporting cationic toxins, including CV and IILs, out of the cell through proton exchange. Alleles of SGE1 determine its protein abundance and the ability to tolerate high concentrations of cationic inhibitors. ILT1 may also directly or indirectly export cationic toxins out of the cytoplasm through the plasma membrane (P.M.). The dendogram in (B) proposes the evolutionary path for the emergence of the SGE1SLS sequence from the ancestral SGE1PLL allele. Extrac., Extracellular; Intrac., Intracellular.
The existence of multiple SGE1 alleles encoding proteins of differing stabilities suggests that these sequence variations are tolerated in specific genetic backgrounds or environmental contexts. Since the tolerant SGE1PLL allele is ancestral (Figure 7B) and has been conserved across millions of years of evolution, man-made cationic toxins, such as IILs and CV, cannot be invoked as recent selective agents for adaptation at this locus. The SGE1SLS allele likely increased in frequency due to a bottleneck or reduced selective pressure to maintain higher levels of Sge1p expression. For example, there may have been reduced exposure to natural cationic toxins in the environment, or some genetic backgrounds may have conferred partly redundant mechanisms for coping with these toxins. Interestingly, several strains of multiple different Saccharomyces species lack the SGE1 gene entirely (File S3), further suggesting the importance of its function may be conditional. Although the derived alleles are most likely conditionally neutral or slightly deleterious, the observation that many S. cerevisiae strains contain the sensitive SGE1SLS and intermediate SGE1SLL sequences could suggest their importance in some natural environments. The Sge1SLS or SgeSLL proteins may have altered specificity for unknown natural toxins, causing fitness trade-offs in reduced resistance to man-made IILs and cationic dyes. Alternatively, the differences in protein abundances between natural variants suggest that Sge1 protein stability is regulated. Sge1 is ubiquitinated in vivo (Swaney et al. 2013), which could affect Sge1 stability by targeting Sge1 protein for endocytosis and delivery to the vacuole for degradation (Piper et al. 2014). Further molecular and genetic studies are needed to better understand the roles of each allele in Sge1 function across a broad range of ecologically and industrially relevant conditions.
To make cost-effective and sustainable biofuels and bioproducts, microbial catalysts will need to metabolize lignocellulosic sugars efficiently in the presence of inhibitory compounds such as ionic liquids, which are necessary for deconstruction of lignocellulosic feedstocks into fermentable sugars. Our results suggest that CRISPR/Cas9-based gene editing or engineered overexpression of SGE1 sequences in industrial yeast strains may enable wider use of ionic liquid-pretreated biomass for biofuel production. Genetic modifications of other MFS transporters have been shown to improve biofuel production (Farwick et al. 2014; Li et al. 2016), further adding to the idea that exploring natural or experimental variation in MFS sequences may enable phenotypes for industrial applications. Our approach to screen and identify allelic sequences is not limited to the ionic liquid tolerance trait, but can be applied to phenotypes gathered from a large number of wild and domesticated S. cerevisiae strains cultured across a variety of media conditions (Fay and Benavides 2005; Liti et al. 2009; Schacherer et al. 2009; Parreiras et al. 2014; Sato et al. 2014; Wohlbach et al. 2014; Strope et al. 2015). Through this approach, distinct phenotypes can be assigned to specific genetic variants within the same species, thus providing a better understanding of how sequence differences among strains can be used to improve production of industrial biofuels and products.
Acknowledgments
We thank Amanda Reider Apel, Sarah Rodriguez, Shu Shen, Tom Ruegg, Charles Denby, Rago Avanasi Narasimhan, Li Hinchman, Lucas Parreiras, Austin Pier, Rebecca Breuer, Maika Vang-Smith, Michael Graham, Mike Place, Mick McGee, Bill Alexander, Jan-Fang Cheng, Jeff Piotrowski, Dave Katzmann, Yaoping Zhang, and Donna Bates for advice and technical assistance; Justin Fay, Clete Kurtzman, and John McCusker for yeast strains; and James Runde and Matthew Wisniewski for assistance in generating figures. This material is partly based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under award number DE-SC0018409, and work funded by the Department of Energy Great Lakes Bioenergy Research Center (Department of Energy Office of Science BER DE-FC02-07ER64494). This work was also part of the Department of Energy Joint BioEnergy Institute (http://www.jbei.org), supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. DP is a Marie Sklodowska-Curie fellow of the European Union’s Horizon 2020 research and innovation programme, grant agreement No. 747775. D.A.H., M.P.T., and Lawrence Livermore National Security have filed a provisional patent application entitled “Engineered Microorganisms Having Resistance to Ionic Liquids,” based on some findings of this manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6731201.
Communicating editor: J. Akey
Literature Cited
- Alexander W. G., Peris D., Pfannenstiel B. T., Opulente D. A., Kuang M., et al. , 2016. Efficient engineering of marker-free synthetic allotetraploids of Saccharomyces. Fungal Genet. Biol. 89: 10–17. 10.1016/j.fgb.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P., Goncalves C., Teixeira S., Libkind D., Bontrager M., et al. , 2014. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 5: 4044 10.1038/ncomms5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amakasu H., Suzuki Y., Nishizawa M., Fukasawa T., 1993. Isolation and characterization of SGE1: a yeast gene that partially suppresses the gal11 mutation in multiple copies. Genetics 134: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. S., Parker R. P., 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. 10.1093/emboj/17.5.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. J., Barker S. L., Boone C., Measday V., 2012. Identification of RCN1 and RSA3 as ethanol-tolerant genes in Saccharomyces cerevisiae using a high copy barcoded library. FEMS Yeast Res. 12: 48–60. 10.1111/j.1567-1364.2011.00762.x [DOI] [PubMed] [Google Scholar]
- Angus-Hill M. L., Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V., 1999. Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol. 294: 1311–1325. 10.1006/jmbi.1999.3338 [DOI] [PubMed] [Google Scholar]
- Aouida M., Khodami-Pour A., Ramotar D., 2009. Novel role for the Saccharomyces cerevisiae oligopeptide transporter Opt2 in drug detoxification. Biochem. Cell Biol. 87: 653–661. 10.1139/O09-045 [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., III, et al. , 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Bergström A., Simpson J. T., Salinas F., Barre B., Parts L., et al. , 2014. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 31: 872–888. 10.1093/molbev/msu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder J. B., Raines R. T., 2010. Fermentable sugars by chemical hydrolysis of biomass. Proc. Natl. Acad. Sci. USA 107: 4516–4521. 10.1073/pnas.0912073107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing J., Han P. J., Liu W. Q., Wang Q. M., Bai F. Y., 2014. Evidence for a far East Asian origin of lager beer yeast. Curr. Biol. 24: R380–R381. 10.1016/j.cub.2014.04.031 [DOI] [PubMed] [Google Scholar]
- Blanch H. W., Simmons B. A., Klein-Marcuschamer D., 2011. Biomass deconstruction to sugars. Biotechnol. J. 6: 1086–1102. 10.1002/biot.201000180 [DOI] [PubMed] [Google Scholar]
- Cherqui S., Kalatzis V., Trugnan G., Antignac C., 2001. The targeting of cystinosin to the lysosomal membrane requires a tyrosine-based signal and a novel sorting motif. J. Biol. Chem. 276: 13314–13321. 10.1074/jbc.M010562200 [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M. G., von Heijne G., 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10: 685–686. [DOI] [PubMed] [Google Scholar]
- Datta S., Holmes B., Park J. I., Chen Z., Dibble D. C., et al. , 2010. Ionic liquid tolerant hyperthermophilic cellulases for biomass pretreatment and hydrolysis. Green Chem. 12: 338–345. 10.1039/b916564a [DOI] [Google Scholar]
- Dias A. R., Costa-Rodrigues J., Fernandes M. H., Ferraz R., Prudencio C., 2017. The anticancer potential of ionic liquids. ChemMedChem 12: 11–18. 10.1002/cmdc.201600480 [DOI] [PubMed] [Google Scholar]
- Dickinson Q., Bottoms S., Hinchman L., McIlwain S., Li S., et al. , 2016. Mechanism of imidazolium ionic liquids toxicity in Saccharomyces cerevisiae and rational engineering of a tolerant, xylose-fermenting strain. Microb. Cell Fact. 15: 17 10.1186/s12934-016-0417-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos S. C., Teixeira M. C., Dias P. J., Sá-Correia I., 2014. MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Front. Physiol. 5: 180 10.3389/fphys.2014.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova K. S., Gordeev E. G., Ananikov V. P., 2017. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem. Rev. 117: 7132–7189. 10.1021/acs.chemrev.6b00562 [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Würgler F. E., Sengstag C., 1994. The Saccharomyces cerevisiae SGE1 gene product: a novel drug-resistance protein within the major facilitator superfamily. Mol. Gen. Genet. 244: 287–294. 10.1007/BF00285456 [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Seitz M. U., Sengstag C., 1998. The Sge1 protein of Saccharomyces cerevisiae is a membrane-associated multidrug transporter. Yeast 14: 49–65. [DOI] [PubMed] [Google Scholar]
- Elgharbawy A. A., Alam M. Z., Moniruzzaman M., Goto M., 2016. Ionic liquid pretreatment as emerging approaches for enhanced enzymatic hydrolysis of lignocellulosic biomass. Biochem. Eng. J. 109: 252–267. 10.1016/j.bej.2016.01.021 [DOI] [Google Scholar]
- Engel S. R., Dietrich F. S., Fisk D. G., Binkley G., Balakrishnan R., et al. , 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 4: 389–398. 10.1534/g3.113.008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso P., Mazon M. J., Portillo F., 2006. Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim. Biophys. Acta 1758: 164–170. 10.1016/j.bbamem.2006.01.010 [DOI] [PubMed] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M., Boles E., 2014. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc. Natl. Acad. Sci. USA 111: 5159–5164. 10.1073/pnas.1323464111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Benavides J. A., 2005. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1: e5 10.1371/journal.pgen.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayevskiy V., Goddard M. R., 2016. Saccharomyces eubayanus and Saccharomyces arboricola reside in North Island native New Zealand forests. Environ. Microbiol. 18: 1137–1147. 10.1111/1462-2920.13107 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31–34. 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- Gonçalves M., Pontes A., Almeida P., Barbosa R., Serra M., et al. , 2016. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26: 2750–2761. 10.1016/j.cub.2016.08.040 [DOI] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H., 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30: e23 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H., 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. 10.1093/nar/24.13.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F., 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. 10.1016/0378-1119(87)90131-4 [DOI] [PubMed] [Google Scholar]
- Horton R. M., 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3: 93–99. 10.1007/BF02789105 [DOI] [PubMed] [Google Scholar]
- Hose J., Yong C. M., Sardi M., Wang Z., Newton M. A., et al. , 2015. Dosage compensation can buffer copy-number variation in wild yeast. eLife 4: e05462 [corrigenda: Elife 5: e15743 (2016)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquot C., Julien R., Guilloton M., 1997. The Saccharomyces cerevisiae MFS superfamily SGE1 gene confers resistance to cationic dyes. Yeast 13: 891–902. [DOI] [PubMed] [Google Scholar]
- Jézégou A., Llinares E., Anne C., Kieffer-Jaquinod S., O’Regan S., et al. , 2012. Heptahelical protein PQLC2 is a lysosomal cationic amino acid exporter underlying the action of cysteamine in cystinosis therapy. Proc. Natl. Acad. Sci. USA 109: E3434–E3443. [corrigenda: Proc. Natl. Acad. Sci. USA 110: 3197 (2013)]. 10.1073/pnas.1211198109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. G., Gardner E. M., Liu Y., Medina R., Goffinet B., et al. , 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4: apps.1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., et al. , 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating D. H., Zhang Y., Ong I. M., McIlwain S., Morales E. H., et al. , 2014. Aromatic inhibitors derived from ammonia-pretreated lignocellulose hinder bacterial ethanologenesis by activating regulatory circuits controlling inhibitor efflux and detoxification. Front. Microbiol. 5: 402 10.3389/fmicb.2014.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov J. I., D’Haeseleer P., Borglin S. E., Deangelis K. M., Woo H., et al. , 2012. Global transcriptome response to ionic liquid by a tropical rain forest soil bacterium, Enterobacter lignolyticus. Proc. Natl. Acad. Sci. USA 109: E2173–E2182. 10.1073/pnas.1112750109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konda N. M., Shi J., Singh S., Blanch H. W., Simmons B. A., et al. , 2014. Understanding cost drivers and economic potential of two variants of ionic liquid pretreatment for cellulosic biofuel production. Biotechnol. Biofuels 7: 86 10.1186/1754-6834-7-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong I. I., Turner T. L., Kim H., Kim S. R., Jin Y. S., 2018. Phenotypic evaluation and characterization of 21 industrial Saccharomyces cerevisiae yeast strains. FEMS Yeast Res. 18 10.1093/femsyr/foy001 [DOI] [PubMed] [Google Scholar]
- Kuang M. C., Kominek J., Alexander W. G., Cheng J. F., Wrobel R. L., et al. , 2018. Repeated cis-regulatory tuning of a metabolic bottleneck gene during evolution. Mol. Biol. Evol. 35: 1968–1981. 10.1093/molbev/msy102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Knierim B., Manisseri C., Arora R., Scheller H. V., et al. , 2010. Comparison of dilute acid and ionic liquid pretreatment of switchgrass: biomass recalcitrance, delignification and enzymatic saccharification. Bioresour. Technol. 101: 4900–4906. 10.1016/j.biortech.2009.10.066 [DOI] [PubMed] [Google Scholar]
- Li H., Schmitz O., Alper H. S., 2016. Enabling glucose/xylose co-transport in yeast through the directed evolution of a sugar transporter. Appl. Microbiol. Biotechnol. 100: 10215–10223. 10.1007/s00253-016-7879-8 [DOI] [PubMed] [Google Scholar]
- Li M. Z., Elledge S. J., 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4: 251–256. 10.1038/nmeth1010 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Nguyen Ba A. N., Blythe M., Muller C. A., Bergstrom A., et al. , 2013. High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome. BMC Genomics 14: 69 10.1186/1471-2164-14-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood N., Husson E., Jacquard C., Wewetzer S., Buchs J., et al. , 2015. Impact of two ionic liquids, 1-ethyl-3-methylimidazolium acetate and 1-ethyl-3-methylimidazolium methylphosphonate, on Saccharomyces cerevisiae: metabolic, physiologic, and morphological investigations. Biotechnol. Biofuels 8: 17 10.1186/s13068-015-0206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Hunter R., Parker R., 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8: 79–82. 10.1002/yea.320080202 [DOI] [PubMed] [Google Scholar]
- Mülleder M., Campbell K., Matsarskaia O., Eckerstorfer F., Ralser M., 2016. Saccharomyces cerevisiae single-copy plasmids for auxotrophy compensation, multiple marker selection, and for designing metabolically cooperating communities. F1000Res. 5: 2351 10.12688/f1000research.9606.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F., Aigle M., 1995. RTM1: a member of a new family of telomeric repeated genes in yeast. Genetics 140: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet M., Datta S., Dibble D. C., Tamrakar P. R., Benke P. I., et al. , 2011. Impact of ionic liquid pretreated plant biomass on Saccharomyces cerevisiae growth and biofuel production. Green Chem. 13: 2743–2749. 10.1039/c1gc15327g [DOI] [Google Scholar]
- Parreiras L. S., Breuer R. J., Avanasi Narasimhan R., Higbee A. J., La Reau A., et al. , 2014. Engineering and two-stage evolution of a lignocellulosic hydrolysate-tolerant Saccharomyces cerevisiae strain for anaerobic fermentation of xylose from AFEX pretreated corn stover. PLoS One 9: e107499 10.1371/journal.pone.0107499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris D., Langdon Q. K., Moriarty R. V., Sylvester K., Bontrager M., et al. , 2016. Complex ancestries of lager-brewing hybrids were shaped by standing variation in the wild yeast Saccharomyces eubayanus. PLoS Genet. 12: e1006155 10.1371/journal.pgen.1006155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Dikic I., Lukacs G. L., 2014. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb. Perspect. Biol. 6 a016808 10.1101/cshperspect.a016808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Mott R., Bork P., Copley R. R., 2001. Novel protein domains and repeats in Drosophila melanogaster: insights into structure, function, and evolution. Genome Res. 11: 1996–2008. 10.1101/gr.198701 [DOI] [PubMed] [Google Scholar]
- Qian W., Ma D., Xiao C., Wang Z., Zhang J., 2012. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2: 1399–1410. 10.1016/j.celrep.2012.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg T. L., Kim E. M., Simmons B. A., Keasling J. D., Singer S. W., et al. , 2014. An auto-inducible mechanism for ionic liquid resistance in microbial biofuel production. Nat. Commun. 5: 3490 10.1038/ncomms4490 [DOI] [PubMed] [Google Scholar]
- Sá-Correia I., dos Santos S. C., Teixeira M. C., Cabrito T. R., Mira N. P., 2009. Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 17: 22–31. 10.1016/j.tim.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Sardi M., Paithane V., Place M., Robinson D. E., Hose J., et al. , 2018. Genome-wide association across Saccharomyces cerevisiae strains reveals substantial variation in underlying gene requirements for toxin tolerance. PLoS Genet. 14: e1007217 10.1371/journal.pgen.1007217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. K., Liu T., Parreiras L. S., Williams D. L., Wohlbach D. J., et al. , 2014. Harnessing genetic diversity in Saccharomyces cerevisiae for fermentation of xylose in hydrolysates of alkaline hydrogen peroxide-pretreated biomass. Appl. Environ. Microbiol. 80: 540–554. 10.1128/AEM.01885-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D. R., Zill O. A., Rokas A., Payen C., Dunham M. J., et al. , 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 1: 11–25. 10.1534/g3.111.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345. 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 2002. Getting started with yeast. Methods Enzymol. 350: 3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Skelly D. A., Merrihew G. E., Riffle M., Connelly C. F., Kerr E. O., et al. , 2013. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res. 23: 1496–1504. 10.1101/gr.155762.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope P. K., Skelly D. A., Kozmin S. G., Mahadevan G., Stone E. A., et al. , 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25: 762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., et al. , 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10: 676–682. 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton T., 1999. Room-temperature ionic liquids. Solvents for synthesis and catalysis. Chem. Rev. 99: 2071–2084. 10.1021/cr980032t [DOI] [PubMed] [Google Scholar]
- Wiles A. M., Cai H., Naider F., Becker J. M., 2006. Nutrient regulation of oligopeptide transport in Saccharomyces cerevisiae. Microbiology 152: 3133–3145. 10.1099/mic.0.29055-0 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Wohlbach D. J., Rovinskiy N., Lewis J. A., Sardi M., Schackwitz W. S., et al. , 2014. Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production. Genome Biol. Evol. 6: 2557–2566. 10.1093/gbe/evu199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J. X., Li J., Aigrain L., Hallin J., Persson K., et al. , 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 49: 913–924. 10.1038/ng.3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Lei J., Yang H., Xu K., Wang R., et al. , 2011. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28: 795–798. 10.1002/yea.1905 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All yeast strains and plasmids used in this study are available upon request. Table S2 contains a list of the strains described in this study. Data necessary to confirm findings of this article are present within the article text and figures as well as supplemental files, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6731201.