Here, Warecki and Sullivan investigate how late-segregating chromosome fragments pass through nuclear envelope channels to maintain euploidy in the nuclei of Drosophila neuroblasts during telophase. The authors find that Aurora B activity prevents the recruitment of HP1a to...
Keywords: acentric, nuclear envelope, micronuclei, genome integrity
Abstract
While it is known that micronuclei pose a serious risk to genomic integrity by undergoing chromothripsis, mechanisms preventing micronucleus formation remain poorly understood. Here, we investigate how late-segregating acentric chromosomes that would otherwise form micronuclei instead reintegrate into daughter nuclei by passing through Aurora B kinase-dependent channels in the nuclear envelope of Drosophila melanogaster neuroblasts. We find that localized concentrations of Aurora B preferentially phosphorylate H3(S10) on acentrics and their associated DNA tethers. This phosphorylation event prevents HP1a from associating with heterochromatin and results in localized inhibition of nuclear envelope reassembly on endonuclease- and X-irradiation-induced acentrics, promoting channel formation. Finally, we find that HP1a also specifies initiation sites of nuclear envelope reassembly on undamaged chromatin. Taken together, these results demonstrate that Aurora B-mediated regulation of HP1a-chromatin interaction plays a key role in maintaining genome integrity by locally preventing nuclear envelope assembly and facilitating the incorporation of late-segregating acentrics into daughter nuclei.
EUKARYOTIC cells have evolved sophisticated mechanisms that maintain genome integrity. Checkpoints halt cell cycle progression in response to damaged DNA to allow for the repair or elimination of compromised cells (Abbas et al. 2013). For example, the G1-S and the G2-M checkpoints prevent entry into S-phase and mitosis, respectively, when DNA is damaged (Elledge 1996). An additional checkpoint at the metaphase–anaphase transition delays progression into anaphase if DNA is damaged once a cell commits to mitosis (Mikhailov et al. 2002; Royou et al. 2005). Despite these checkpoints, cells sometimes enter anaphase with damaged DNA. Unrepaired double-stranded DNA breaks are particularly problematic, as they result in chromosome fragments lacking either a telomere or a centromere (Kaye et al. 2004). The latter, called acentrics, are unable to form traditional microtubule–kinetochore attachments, and are therefore expected to fail to segregate and to be excluded from the nascent daughter nuclei, leading to the formation of micronuclei (Kanda and Wahl 2000; LaFountain et al. 2001; Fenech et al. 2011). Historically, micronuclei have been a biomarker for cancer (Santos et al. 2010; Bonassi et al. 2011), and recent studies reveal that micronuclei drive genomic instability either through their loss during subsequent cell divisions or through chromothripsis, the dramatic shattering and rearrangement of micronuclear DNA that is then incorporated into the genome (Crasta et al. 2012; Zhang et al. 2015; Vázquez-Diez et al. 2016; Ly et al. 2017).
While the formation of micronuclei from lagging chromosomes has been widely documented, in some instances, lagging chromosomes avoid micronuclei formation by rejoining daughter nuclei before mitosis is completed. For example, in human colorectal cancer cells, a proportion of lagging whole chromosomes that would otherwise form micronuclei instead reincorporate into the daughter nuclei in late anaphase (Huang et al. 2012). In fission yeast, lagging chromatids that remain distinct from the main segregating chromosomes during anaphase eventually reunite with daughter nuclei in telophase (Pidoux et al. 2000; Sabatinos et al. 2015). In addition, in Drosophila neuroblast and papillar divisions, late-segregating acentric fragments induced by endonuclease activity or irradiation successfully rejoin daughter nuclei in late telophase (Royou et al. 2010; Bretscher and Fox 2016). Therefore, the fate of lagging acentric chromosomes is an important but underexplored area of cell biology. Here, we specifically examine the mechanisms that facilitate the incorporation of late-segregating acentric chromosomes into daughter nuclei, avoiding micronuclei formation.
In Drosophila, acentric behavior has been studied using transgenic flies containing a heat-shock inducible I-CreI endonuclease (Rong et al. 2002; Royou et al. 2010; Kotadia et al. 2012; Derive et al. 2015; Karg et al. 2015, 2017; Bretscher and Fox 2016), which targets rDNA near the base of the X chromosome (Rong et al. 2002; Maggert and Golic 2005; Paredes and Maggert 2009; Golic and Golic 2011). I-CreI-mediated double-stranded DNA breaks result in γH2Av foci that persist through mitosis, and chromosome fragments that do not recruit canonical centromere components and thus are considered acentrics (Royou et al. 2010). Even though I-CreI-induced acentrics initially lag on the metaphase plate while undamaged chromosomes segregate, acentrics ultimately undergo delayed but successful segregation (Royou et al. 2010). Acentric segregation is achieved through protein-coated DNA tethers connecting acentrics to their centric partners and microtubule bundles that encompass acentrics, enabling their poleward movement (Karg et al. 2017). The histone-based DNA tether is associated with Polo, BubR1, and the chromosome passenger proteins Aurora B and INCENP (Royou et al. 2010).
Because lagging and acentric chromosome segregation is significantly delayed, occurring late in anaphase, they often remain distinct from the main mass of chromosomes when nuclear envelope reassembly initiates (Fenech 2000; Cimini et al. 2002; Afonso et al. 2014; Karg et al. 2015). Despite the presence of the nascent nuclear envelope surrounding the main nuclear mass, in Drosophila neuroblasts, lagging acentrics are not “locked out” of daughter nuclei and do not form micronuclei. Rather, the late-segregating acentrics bypass the nuclear envelope barrier and enter telophase nuclei through channels in the nuclear envelope that are formed by highly localized delays in the completion of nuclear envelope reassembly (Karg et al. 2015). Nuclear envelope channel formation is dependent upon the Aurora B kinase activity associated with the acentric and DNA tether. When Aurora B activity is reduced, acentrics are unable to enter daughter nuclei and instead form lamin-coated micronuclei. The pool of Aurora B responsible for channel formation likely comes from Aurora B persisting on the DNA tethers and acentrics, as channel formation is not observed in divisions that lack both acentrics and their associated Aurora B-coated tethers (Karg et al. 2015).
The formation of nuclear envelope channels suggests localized inhibition of important steps in nuclear envelope reassembly. Key events in nuclear envelope reassembly include the reformation of nuclear pore complexes, reestablishment of connections between chromatin and inner nuclear membrane proteins that are disrupted in early mitosis, and fusion of nuclear envelope membrane microdomains (Baur et al. 2007; Anderson and Hetzer 2008; Dultz et al. 2008; Lu et al. 2011; Olmos et al. 2015; Vietri et al. 2015). Additionally, the nuclear lamina reassembles once nuclear pore complexes and inner nuclear membrane proteins are recruited to daughter nuclei (Newport et al. 1990; Chaudhary and Courvalin 1993; Daigle et al. 2001; Katsani et al. 2008).
Regulation of nuclear envelope reassembly is achieved through the global activity of mitotic kinases, among which Aurora B is a known negative regulator (Ramadan et al. 2007; Afonso et al. 2014; Karg et al. 2015). One mechanism by which Aurora B activity may inhibit nuclear envelope reassembly is through disrupting chromatin interactions with the heterochromatin component HP1α/HP1a (Schellhaus et al. 2015). In interphase, HP1α/HP1a binds both methylated histone H3 and nuclear envelope components (Ye and Worman 1996; Ye et al. 1997; Kourmouli et al. 2000; Polioudaki et al. 2001). As cells enter mitosis, Aurora B-mediated phosphorylation of H3(S10) acts as a switch to remove HP1α/HP1a from chromosomes (Fischle et al. 2005; Hirota et al. 2005; Dormann et al. 2006). During anaphase, when Aurora B relocalizes to the spindle midzone and H3(S10) phosphate groups are removed (Carmena et al. 2012), HP1α/HP1a is reloaded onto segregating chromosomes and subsequently reestablishes connections with nuclear envelope-associated components (Sugimoto et al. 2001; Poleshko et al. 2013), which is a possible early step in the reformation of the nuclear envelope (Kourmouli et al. 2000).
Understanding the mechanisms by which Aurora B kinase activity locally alters the events of nuclear envelope reassembly to mediate channel formation is of particular interest. Specifically, understanding the pathway through which Aurora B acts to allow the incorporation of late-segregating acentrics into daughter nuclei would reveal new mechanisms by which Aurora B prevents micronuclei formation and maintains genome integrity. In addition, studying the mechanisms by which nuclear envelope channel formation is regulated may provide a system for understanding how global nuclear envelope reassembly is regulated in wild-type divisions. Here, we explore these issues by generating acentrics using both the I-CreI endonuclease and X-irradiation, and find that Aurora B excludes HP1a from late-segregating acentrics and that HP1a exclusion allows late-segregating acentrics to reincorporate into daughter telophase nuclei.
Materials and Methods
Fly stocks
All stocks were raised on standard Drosophila food (Sullivan et al. 2000). Chromosome dynamics were monitored using H2Av-RFP [stock #23651; Bloomington Drosophila Stock Center (BDSC), Bloomington, IN]. The following Gal4 drivers were used: elav-Gal4 (Lin and Goodman 1994), Wor-Gal4 (Cabernard and Doe 2009), and Actin-Gal4 [#25708; BDSC; (Ito et al. 1997)]. To monitor nuclear envelope dynamics, we expressed upstream activating sequence (UAS)-lamin-GFP (#7376; BDSC) driven by elav-Gal4. HP1a localization was assessed through use of GFP-HP1a (#30561; BDSC). UAS-ial-shRNA (#28691; BDSC) driven by Wor-Gal4 was used to deplete Aurora B. UAS-Su(Var)205-shRNA (#33400) driven by either Actin-Gal4 or elav-Gal4, depending on the experiment, was used to deplete HP1a.
Fixed neuroblast cytology
Crawling female third-instar larvae bearing either hs-I-CreI and Wor-Gal4 or hs-I-CreI, Wor-Gal4, and UAS-ial-shRNA were heat shocked for 1 hr at 37°. Following 1 hr recovery at room temperature, brains were dissected in 0.7% NaCl then fixed in 3.7% formaldehyde for 30 min. Brains were washed in 45% acetic acid in PBS for 3 min then placed between siliconized coverslips and glass slides in 60% acetic acid in PBS. Brains were squashed by tracing over coverslips with watercolor paper. Slides were frozen in liquid nitrogen for 10 min and then washed in 20% ethanol for 10 min at −20°. After washing with PBST (10 min) and PBS (2 × 5 min), slides were blocked in a 5% dried milk solution in PBST for 1 hr. Samples were incubated with rabbit anti-phospho-H3(S10) antibody (#ab5176; Abcam) at a 1:500 dilution overnight at 4°. Samples were subsequently washed in PBST before incubation with goat anti-rabbit-alexa488 (#A-11008; ThermoFisher) at a 1:300 dilution for 1 hr at room temperature. Slides were washed in PBST, counterstained with DAPI in vectashield, and imaged the following day. Procedure adapted from Bonaccorsi et al. (2000), Cenci et al. (2003).
Quantitative immunofluorescence imaging
Fixed slides were imaged on a Leica DMI6000B wide-field inverted microscope equipped with a Hamamatsu Electron-multiplying charge-coupled device (EM-CCD) camera (ORCA C9100-02) with a binning of 1 and a 100× Plan-Apochromat objective with a numerical aperture (NA) of 1.4. For experiments determining the ratio of phospho-H3(S10)/DNA on the acentrics vs. on the main nuclei, phospho-H3(S10)/DNA pixel intensity was determined in ImageJ (National Institutes of Health, Bethesda, MD) by drawing individual region of interests (ROIs) around both main nuclei and the acentrics (as determined from DAPI staining) from sum projections of all z-slices in which the nuclei and acentrics were in focus. Corrected total fluorescence (CTF) was calculated for each ROI (acentrics and nuclei) for both DAPI and phospho-H3(S10) channels by subtracting the product of the ROI area and the mean pixel intensity of an arbitrarily-selected background region from the measured integrated density of the ROI. For each division set, CTFs were averaged for the two main nuclei and for the acentrics when more than one acentric ROI was drawn. Phospho-H3(S10)/DAPI ratios were calculated by dividing the CTF of phospho-H3(S10) by the CTF of DAPI for the averaged acentrics and the averaged nuclei. To determine the fold change for acentrics vs. main nuclei phospho-H3(S10)/DAPI ratios, the phospho-H3(S10)/DAPI ratio of acentrics was divided by that of the main nuclei for each imaged division.
To compare phospho-H3(S10) levels on acentrics in I-CreI vs. I-CreI; Aurora B RNA interference (RNAi) neuroblasts, control and Aurora B-depleted brains were imaged at the same laser settings. Quantification of phospho-H3(S10)/DAPI ratios were calculated as detailed above.
Live neuroblast cytology
For experiments involving acentrics, crawling female third-instar larvae bearing hs-I-CreI, elav-Gal4, H2Av-RFP, and a combination of GFP-HP1, UAS-Lamin-GFP, and/or UAS-Su(Var)205-dsRNA were heat shocked for 1 hr at 37°. Larvae were allowed to recover for at least 1 hr following heat shock. For experiments with no acentrics, female third-instar larvae bearing elav-Gal4, H2Av-RFP, and a combination of GFP-HP1, UAS-Lamin-GFP, and/or UAS-Su(Var)205-dsRNA were used. Brains were dissected in PBS and gently squashed between a slide and coverslip (Buffin et al. 2005). Neuroblasts along the periphery of the squashed brain provided the best imaging samples. Slides were imaged for up to 1 hr.
Data from time-lapse imaging experiments were acquired with both a Leica DMI6000B wide-field inverted microscope equipped with a Hamamatsu EM-CCD camera (ORCA C9100-02) with a binning of 1 and a 100× Plan-Apochromat objective with NA 1.4, and an inverted Nikon Eclipse TE2000-E spinning disk (CSLI-X1; Nikon, Garden City, NY) confocal microscope equipped with a Hamamatsu EM-CCD camera (ImageE MX2) with a 100 × 1.4 NA oil immersion objective. Successive time points were filmed at 20 sec for the wide-field microscope and 8 sec for the spinning disk microscope. Spinning disk images were acquired with MicroManager 1.4 software.
Small molecule inhibition of Aurora B kinase
For experiments involving the inhibition of Aurora B kinase, following dissection, brains were washed in a 25.5 μM solution containing Binucleine-2 (B1186; Sigma [Sigma Chemical], St. Louis, MO) for 5 min, after which brains were squashed in PBS between a slide and coverslip. For control experiments, dissected brains were washed in 0.15% DMSO (final concentration of DMSO in solution used to dissolve Binucleine-2) for 5 min and then squashed in PBS between a slide and coverslip. Neuroblasts entering anaphase were selected for imaging, and slides were imaged for only one division.
Quantitative GFP-HP1a analysis
Live neuroblasts expressing H2Av-RFP, GFP-HP1a, and I-CreI were imaged with a Nikon Eclipse TE2000-E spinning disk (CSLI-X1) confocal microscope equipped with a Hamamatsu EM-CCD camera (ImageE MX2) with a 100 × 1.4 NA oil immersion objective. Neuroblasts treated with DMSO and DMSO + Binucleine-2 were imaged with the same laser settings. GFP-HP1a pixel intensity was calculated by creating sum projections of movies and then measuring the background-subtracted integrated density of GFP signal on the acentric region. Measurements began at the initial point of acentric segregation and continued for 160 sec.
Temperature-regulated expression of RNAi and lethality studies
Flies bearing either Actin-Gal4 or Actin-Gal4 and UAS-Su(Var)205-dsRNA were grown at room temperature (measured as 22°) until they reached third-instar stage. At this point, larvae were collected into vials and either allowed to continue to grow at room temperature or were shifted to grow at 29°. Survivability was determined by counting the number of adult flies that eclosed in each vial.
Irradiation studies
Crawling third-instar female larvae were placed in an empty plastic vial and irradiated with 605 rad using a Faxitron CP160 X-ray machine. Larvae were allowed to recover for at least 1 hr before brains were dissected and mitotic neuroblasts were imaged as described above.
Statistical analyses
Statistical analyses were determined by χ2 tests (Figure 2, C, E, and F, Figure 3, E and F, Figure 5, C and E, and Supplemental Material, Figure S1C) and Student’s t-tests (Figure 5D, Figure S1C, and Figure S3B) performed in R (R Core Team 2014). For analyses involving χ2 tests, we assumed a null hypothesis that there should be no difference in values between control and experimental conditions. Therefore, in these cases, the values measured for the control conditions served as the theoretical predictions for what we should observe in the experimental conditions. We then compared the actual values measured in the experimental conditions to these theoretical predictions and determined the significance of difference with a χ2 test.
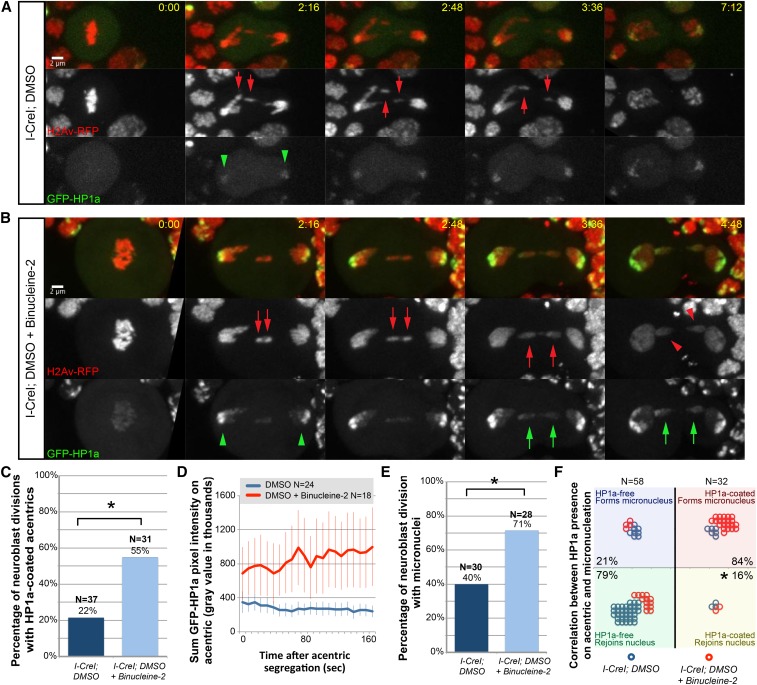
Figure 2.
Aurora B kinase activity blocks HP1a association with late-segregating acentrics. (A) Stills from a time-lapse movie of a mitotic neuroblast expressing I-CreI, H2Av-RFP (red fluorescent protein, red), and GFP-HP1a (green) treated with DMSO (control) (see Movie S1). GFP-HP1a detected on the centric heterochromatic region of segregating intact chromosomes is indicated by green arrowheads. Acentrics are indicated by red arrows. (B) Stills from a time-lapse movie of a mitotic neuroblast expressing I-CreI, H2Av-RFP (red), and GFP-HP1a (green) treated with the Aurora B inhibitor Binucleine-2 dissolved in DMSO (see Movie S2). GFP-HP1a observed on acentric chromosomes is indicated by green arrows. Micronuclei are indicated by red arrowheads. (C) Comparison of the percentage of divisions in which GFP-HP1a was detected on acentrics in DMSO-treated (left) and DMSO + Binucleine-2-treated (right) I-CreI-expressing neuroblasts. Asterisk indicates statistical significance by a χ2 test (P = 0.005). (D) Comparison of GFP-HP1a pixel intensity on acentrics from DMSO-treated neuroblasts (blue line) and DMSO + Binucleine-2-treated neuroblasts (red line). Error bars represent two SEs. (E) Comparison of the percentage of divisions in which acentrics formed micronuclei in DMSO-treated (left) and DMSO + Binucleine-2-treated (right) I-CreI-expressing neuroblasts. Asterisk indicates statistical significance by a χ2 test (P = 0.02). (F) Graph depicting a strong correlation between the lack of HP1a on the acentric and the ability of the acentric to rejoin the daughter nucleus. Each circle represents one acentric: blue and red circles depict single acentrics derived from DMSO- and DMSO + Binucleine-2-treated I-CreI-expressing neuroblasts, respectively. Asterisk indicates statistical significance by a χ2 test(P = 5.34 × 10−9). Time is written as min:sec after anaphase onset. Bar, 2 μm.
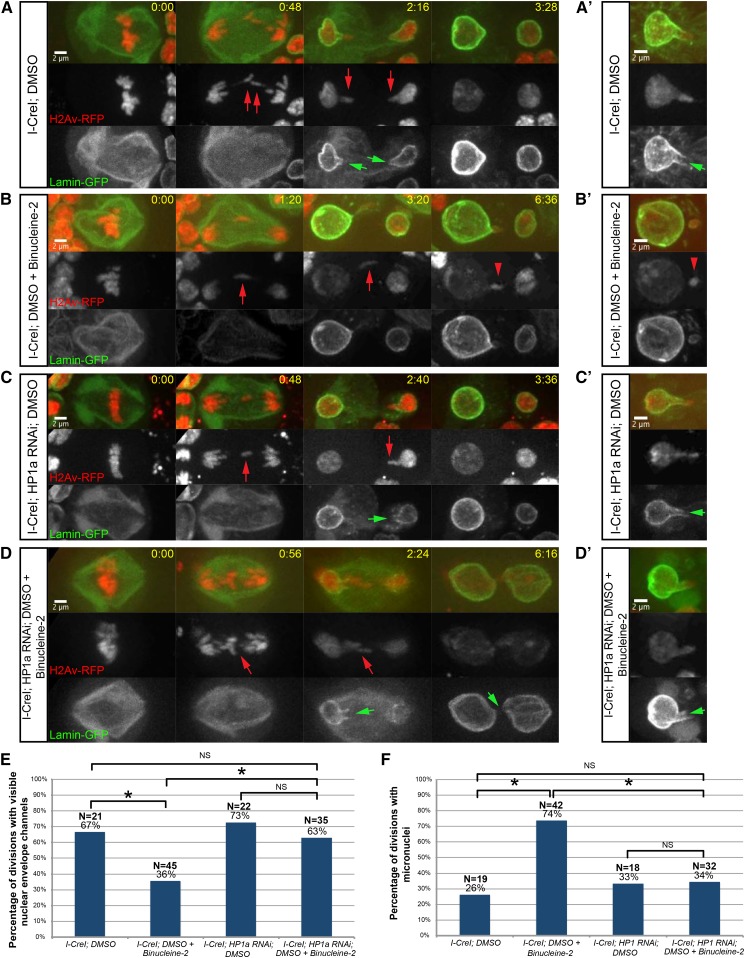
Figure 3.
Aurora B-mediated HP1a exclusion from acentrics/tethers results in nuclear envelope channel formation. (A and A’) Stills from time-lapse movies of two different mitotic neuroblasts expressing I-CreI, H2Av-RFP (red fluorescent protein, red), and Lamin-GFP (green) treated with DMSO (see Movie S3). Acentrics are indicated by red arrows. Channels are indicated by green arrows. (B and B’) Stills from time-lapse movies of two different mitotic neuroblasts expressing I-CreI, H2Av-RFP, and Lamin-GFP treated with DMSO + Binucleine-2 (see Movie S4). Micronuclei are indicated by red arrowheads. (C and C’) Stills from time-lapse movies of two different mitotic neuroblasts expressing I-CreI, H2Av-RFP, Lamin-GFP, and HP1a RNA interference (RNAi) treated with DMSO (see Movie S5). (D and D’) Stills from time-lapse movies of two different mitotic neuroblasts expressing I-CreI, H2Av-RFP, Lamin-GFP, and HP1a RNAi treated with DMSO + Binucleine-2 (see Movie S6). (E) Comparison of the percentage of neuroblast divisions in which nuclear envelope channels were observed when I-CreI-expressing neuroblasts were treated with DMSO or DMSO + Binucleine-2 (asterisk indicates statistically significant to I-CreI; DMSO neuroblasts; P = 0.02, determined by a χ2 test) and when I-CreI- and HP1a RNAi-expressing neuroblasts were treated with DMSO or DMSO + Binucleine-2 (asterisk indicates statistically significant to I-CreI; DMSO + Binucleine-2 neuroblasts; P = 0.02, determined by a χ2 test). (F) Comparison of the percentage of neuroblast divisions in which micronuclei were observed when I-CreI-expressing neuroblasts were treated with DMSO or DMSO + Binucleine-2 (asterisk indicates statically significant to I-CreI; DMSO neuroblasts; P = 0.0005, determined by a χ2 test), and I-CreI- and HP1a RNAi-expressing neuroblasts were treated with DMSO or DMSO + Binucleine-2 (asterisk indicates statistical significance to I-CreI; DMSO + Binucleine-2 neuroblasts; P = 0.0007, determined by a χ2 test). Time is written as min:sec after anaphase onset. Bar, 2 μm. See also Figure S2 and Figure S3. NS indicates no statistical significance.
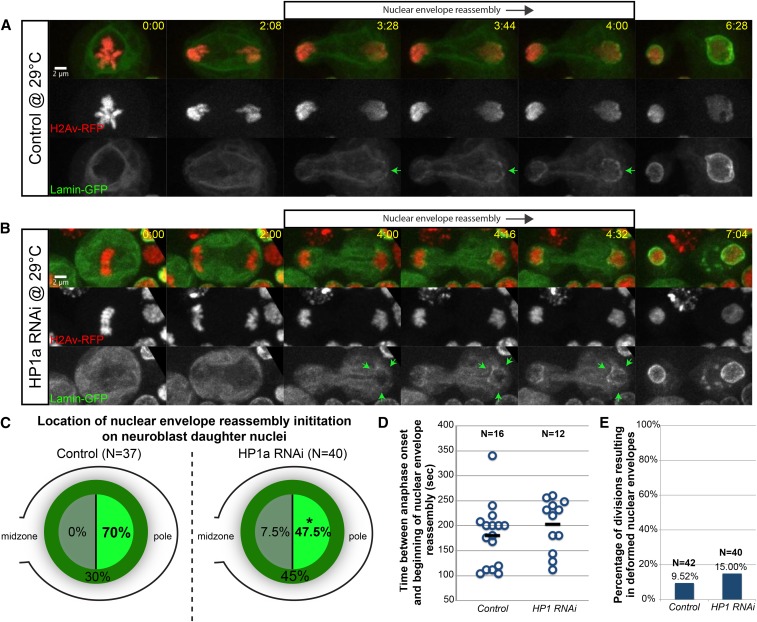
Figure 5.
Preferential initiation of nuclear envelope reassembly at the poleward face of segregating chromosomes requires HP1a. (A) Stills from a time-lapse movie of a mitotic neuroblast expressing H2Av-RFP (red fluorescent protein, red) and Lamin-GFP (green) (see Movie S7). Nuclear envelope reassembly initiation is indicated by the green arrows. (B) Stills from a time-lapse movie of a mitotic neuroblast expressing H2Av-RFP (red), Lamin-GFP (green), and RNA interference (RNAi) against HP1a (see Movie S8). (C) Graph of location frequency for nuclear envelope reassembly initiation in wild-type (left) and HP1a RNAi (right) daughter neuroblasts. Asterisk indicates statistical significance by a χ2 test (P = 0.04). (D) Graph of the time interval between anaphase onset and initiation of nuclear envelope reassembly in wild-type and HP1a-depleted neuroblast divisions. Each circle represents one division. (E) Graph of the percentage of divisions resulting in nuclear envelope deformities in wild-type and HP1a-depleted divisions. Time is written as min:sec after anaphase onset. Bar, 2 μm. See also Figure S3.
Figure preparation
Figures were assembled using ImageJ software and Adobe Illustrator (Adobe, San Jose, CA). Graphs were assembled in Microsoft Excel (Microsoft, Redmond, WA). Selected stills from experiments involving live imaging were adjusted for brightness and contrast using ImageJ to improve clarity.
Data availability
All fly stocks are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental data have been deposited on Figshare. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6752816.
Results
Aurora B kinase preferentially phosphorylates H3(S10) on acentrics and on chromatin near sites of channel formation
Aurora B kinase (FlyBase ID: FBgn0024227), a component of the chromosome passenger complex, initially localizes to chromosomes in early mitosis and then localizes to the spindle midzone during anaphase (Carmena et al. 2012). Aurora B modifies mitotic chromatin by phosphorylating histone H3 on serine 10 (Hsu et al. 2000), and previous studies demonstrated that the midzone-based pool of Aurora B phosphorylates segregating chromosomes in a spatial manner so that lagging whole chromosomes have relatively high phospho-H3(S10) levels (Fuller et al. 2008). In Drosophila neuroblasts bearing I-CreI-induced acentrics, Aurora B kinase persists on DNA tethers stretching from the lagging acentrics to the newly formed daughter nuclei after Aurora B removal from the main mass of segregating chromosomes (Royou et al. 2010; Karg et al. 2015). This highly localized Aurora B activity mediates nuclear envelope channel formation to allow acentric entry into telophase nuclei (Karg et al. 2015). Thus, we hypothesized that the Aurora B localized on the acentric and its associated tether might locally prohibit nuclear envelope formation.
We tested if Aurora B-dependent phosphorylation of H3(S10) persists on acentrics, tethers, and channel sites, despite this mark having been removed from the rest of the main nuclei, by fixing I-CreI-expressing mitotic neuroblasts from female third-instar larvae and staining with an antibody that specifically recognizes phospho-H3(S10) (Figure 1). In neuroblasts fixed in metaphase, we observed phospho-H3(S10) present along the length of all chromosomes, consistent with data from (McManus and Hendzel 2006) (Figure 1A, left panel). In anaphase neuroblasts, we observed a weak phospho-H3(S10) signal on segregating intact chromosomes. In contrast, late-segregating acentrics that remained at or had just segregated from the metaphase plate (Figure 1A, middle panels, red arrows) exhibited a strong phospho-H3(S10) signal (Figure 1A, middle panels, cyan arrows). In neuroblasts fixed in telophase, the stage at which acentrics begin to rejoin daughter nuclei, we observed a continued strong phospho-H3(S10) signal on acentrics. The phospho-H3(S10) signal abruptly ended at the point of contact between the acentric and daughter nucleus (Figure 1A, right panel).
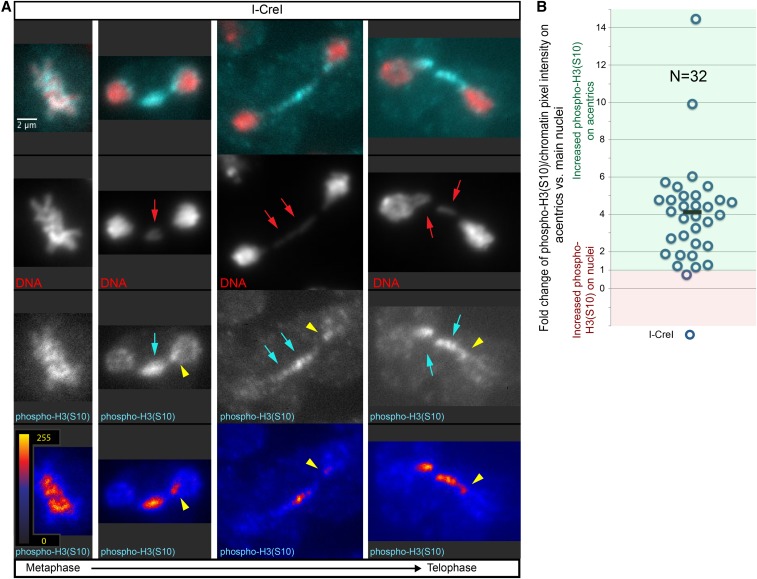
Figure 1.
H3(S10) is preferentially phosphorylated on acentrics and tethers. (A) Fixed mitotic neuroblasts expressing I-CreI in metaphase (left panel), anaphase (middle panels), and telophase (right panel) [red = DNA; blue = phospho-H3(S10)]. Acentrics are indicated by red arrows. Increased phosphorylation of H3(S10) is indicated by cyan arrows. phospho-H3(S10) hotspots are indicated by yellow arrowheads. (B) Graphical comparison of fold increases in the average phopsho-H3(S10)/DNA pixel intensity ratio for the areas of the acentrics vs. the areas of the main nuclei for I-CreI-expressing neuroblasts. Each circle represents one anaphase/telophase cell. Values > 1 (green box) indicate an increased phospho-H3(S10)/DNA pixel intensity ratio on acentrics. Values < 1 (red box) indicate increased phospho-H3(S10)/DNA pixel intensity ratio on main nuclei. Black bar indicates mean fold change. Bar, 2 μm. See also Figure S1.
To quantify these observations, we measured the fold change of the phospho-H3(S10)/DNA signal intensity from the main nuclei to the acentrics for each fixed neuroblast division imaged (Figure 1B). For 31 out of the 32 neuroblast divisions scored, we observed an increase in phospho-H3(S10)/DNA intensity on acentrics compared to main nuclei (mean fold change = 4.13; SD = 2.65; N = 32), consistent with previous reports (Fuller et al. 2008; de Castro et al. 2016).
Intriguingly, in a large proportion (47%; N = 19) of anaphase- or telophase-fixed neuroblast divisions in which at least one acentric remained distinct from the main nuclei, we clearly detected localized “hotspots” of strong phospho-H3(S10) intensity on one of the newly formed nuclei at presumptive sites of acentric entry. These hotspots correspond to the location where tethers contact the nuclei and nuclear envelope channel formation is generally observed (Figure 1A, yellow arrowheads). Taken together, these data demonstrate that acentrics, tethers, and the chromatin at sites of channel formation remain preferentially phosphorylated on H3(S10), even though phosphorylation of H3(S10) is broadly reduced on the chromatin in newly formed telophase nuclei.
To determine whether Aurora B kinase activity is responsible for the observed preferential phosphorylation of H3(S10) on acentrics and tethers, we compared the fold changes of phospho-H3(S10)/DNA intensity from the main nuclei to the acentrics between neuroblasts with normal and RNAi-reduced levels of Aurora B. In I-CreI-expressing neuroblasts fixed in anaphase and telophase (Figure S1A), we observed an average fold change of phospho-H3(S10)/DNA signal from main nuclei to acentrics of 6.39 (SD = 6.56; N = 23), with 78% of divisions showing a greater than twofold increase of phospho-H3(S10)/DNA intensity on acentrics compared to the main nuclei (and 57% of divisions showing a greater than fourfold increase of phospho-H3(10)/DNA intensity on acentrics compared to the main nuclei) (Figure S1C).
In contrast, upon reduction of Aurora B (Figure S1B), we observed a decreased average fold change of phospho-H3(S10)/DNA signal on acentrics compared to main nuclei of 2.34 (SD = 2.15; N = 20) (statistically significant by two-sided independent Student’s t-test, P = 0.01), with only 40% of divisions showing a greater than twofold increase of phospho-H3(S10)/DNA intensity on acentrics to main nuclei (compare to 78% of acentrics in wild-type conditions, significant by χ2 test, P = 0.0105) (and only 20% of divisions showing a greater than fourfold increase of phospho-H3(S10)/DNA intensity on acentrics compared to the main nuclei) (Figure S1C). These findings indicate that Aurora B kinase is responsible for the observed persistent phosphorylation of H3(S10) on acentric fragments during anaphase and telophase.
Aurora B kinase activity blocks HP1a association on late-segregating acentrics
Since phosphorylation of H3(S10) by Aurora B kinase is known to prevent H3 interaction with the heterochromatin component HP1α (Su(var)205, the mammalian ortholog of HP1a) (Fischle et al. 2005; Hirota et al. 2005), we hypothesized that the observed increase in phosphorylation of H3(S10) on acentrics with respect to the main nuclei would lead to an Aurora B-dependent preferential exclusion of HP1a on late-segregating acentrics. To test this hypothesis, we performed live imaging of dividing neuroblasts from female larvae expressing I-CreI, H2Av-RFP, and GFP-HP1a in control conditions (DMSO-treated neuroblasts) and conditions in which Aurora B kinase activity was partially inhibited through introduction of the Aurora B-specific small molecule inhibitor Binucleine-2 (Smurnyy et al. 2010) (Figure 2).
In control DMSO-treated mitotic neuroblasts (Figure 2A, see Movie S1), we observed the following patterns of HP1a association: in metaphase, little or no HP1a was detected on the chromosomes, and in anaphase, a strong HP1a signal was detected on the main segregating chromosomes, primarily on the pericentric heterochromatin (Figure 2A, green arrowheads). These results are consistent with previous data showing that a large proportion of HP1α/HP1a dissociates from chromosomes in early mitosis and reassociates with segregating chromosomes in anaphase (Fischle et al. 2005; Hirota et al. 2005; Dormann et al. 2006; Poleshko et al. 2013). Interestingly, in contrast to the anaphase recruitment of HP1a on the main chromosomes, little or no HP1a was detected on late-segregating acentrics, despite the expected high heterochromatin content of I-CreI-induced acentrics (Figure 2A, red arrows). In total, we only clearly detected HP1a on acentrics in ∼22% of neuroblast divisions that we imaged (N = 37) (Figure 2C).
In mitotic neuroblasts that were treated with the Aurora B inhibitor Binucleine-2 dissolved in DMSO (Figure 2B, see Movie S2), we observed a similar pattern of HP1a association on the main intact nuclei: little or no HP1a on metaphase chromosomes followed by anaphase recruitment of HP1a on the main segregating chromosomes (Figure 2B, green arrowheads). However, in contrast to DMSO-treated neuroblasts, in which HP1a was not detected on late-segregating acentrics, we observed strong HP1a association on anaphase acentrics in neuroblasts treated with DMSO + Binucleine-2 (Figure 2B, green arrows). In all the DMSO + Binucleine-2-treated neuroblasts imaged, clear HP1a association was observed 55% of the time (N = 31) (Figure 2C) (compare to 22% of the time in control divisions, a statistically significant difference determined by a χ2 test, P = 0.005).
To more stringently quantify the levels of HP1a associated with segregating acentrics in wild-type and Aurora B-inhibited conditions, we measured the pixel intensity of GFP-HP1a on acentrics as they began to segregate poleward in both DMSO- and DMSO + Binucleine-2-treated neuroblasts imaged using spinning disk confocal microscopy (Figure 2D). This analysis revealed two key differences: (1) in Aurora B-inhibited neuroblasts (red line), segregating acentrics were associated with higher levels of HP1a than acentrics from control neuroblasts (blue line); and (2) HP1a association with acentrics from Aurora B-inhibited neuroblasts increased over time while HP1a association with acentrics from control neuroblasts slightly decreased. Taken together, these results indicate that Aurora B activity preferentially excludes HP1a from late-segregating acentrics.
Additionally, we observed that acentrics from DMSO-treated control neuroblasts segregated normally and rejoined daughter nuclei in telophase (Figure 1A). We observed micronucleation in 40% of divisions (N = 30) (Figure 2E). Micronuclei were defined as acentrics that failed to enter daughter nuclei, remaining either physically distinct from nuclei or directly adjacent to nuclei but moving independently. However, In DMSO + Binucleine-2-treated neuroblasts, acentrics mostly failed to rejoin daughter nuclei, instead forming micronuclei (Figure 2B, red arrowheads). When we inhibited Aurora B activity, we observed micronucleation 71% of the time (N = 28) (Figure 2E) (a statistically significant increase compared to the 40% micronucleation observed in control divisions, determined by a χ2 test, P = 0.02).
We next determined whether HP1a association with acentrics was correlated with micronucleation. We scored individual acentrics for the presence of HP1a and whether the acentric formed a micronucleus (Figure 2F). We grouped the scored acentrics into four categories: (1) acentrics that were HP1a-free and formed micronuclei (Figure 2F, blue box), (2) acentrics that were HP1a-free and rejoined daughter nuclei (Figure 2F, green box), (3) acentrics that were HP1a-coated and formed micronuclei (Figure 2F, red box), and (4) acentrics that were HP1a-coated and rejoined daughter nuclei (Figure 2F, yellow box). Overall, we found that 79% of acentrics that were HP1a-free (N = 58) rejoined daughter nuclei, while only 16% of acentrics that were HP1a-coated were able to rejoin daughter nuclei (N = 32) (statistical significance determined by χ2 test, P = 5.34 × 10−9). The remaining 84% of acentrics that were HP1a-coated formed micronuclei. Thus, the absence or presence of HP1a is a strong predictor of the fate of the I-CreI-induced acentric, either entering the daughter nucleus or forming a micronucleus.
Aurora B kinase activity promotes nuclear envelope channel formation through HP1a exclusion from acentrics and tethers
Due to the correlation of HP1a–acentric association and the formation of micronuclei, as well as the ability of HP1α/HP1a to interact with and recruit the nuclear envelope (Ye and Worman 1996; Kourmouli et al. 2000), we tested whether Aurora B-mediated HP1a exclusion from acentrics might be a key factor in channel formation through localized inhibition of nuclear envelope reassembly. We performed live imaging of mitotic neuroblasts from female larvae expressing I-CreI, H2Av-RFP, and the nuclear envelope marker Lamin-GFP, and asked whether depletion of HP1a rescued the ability to form nuclear envelope channels when Aurora B was inhibited. The rationale is that while chromatin containing H3-HP1a is a strong promoter of nuclear envelope assembly, chromatin containing H3 by itself is only a neutral substrate for assembly, and thus prevention of H3-HP1a formation on the acentric and tether, either through Aurora B-mediated phosphorylation of H3(S10) or through depletion of HP1a, would be conducive to channel formation (Figure S2).
HP1α/HP1a is essential: homozygous null mutants result in embryonic lethality in Drosophila (Kellum and Alberts 1995). Therefore, to address the role of HP1a in nuclear envelope channel formation, we made use of the transgenic UAS/Gal4 system [for review, see Duffy (2002)] to deplete HP1a through RNAi. In our setup, we expressed UAS-HP1a-dsRNA in the larval central nervous system. One of the additional benefits of using the transgenic UAS/Gal4 system is the ability to fine-tune the degree of Gal4 activity by altering the temperature at which flies are grown (Duffy 2002). Using this property, we found that HP1a depletion was stronger when larvae were grown at 29° as opposed to the milder depletion observed when larvae were grown at room temperature (measured as 22°) (Figure S3). When larvae were grown at 29°, we observed an increase in chromosome segregation errors (Figure S3, A and A’), a previously observed phenotype in HP1a mutants (Kellum and Alberts 1995), and a decrease in survivability (Figure S3B) compared to larvae grown at 22°.
To determine the role of Aurora B-mediated exclusion of HP1a from acentrics/tethers in nuclear envelope channel formation, we performed live imaging on neuroblasts from female larvae in which levels of Aurora B and HP1a were modulated, and compared the rates of nuclear envelope channel formation and micronucleation. All larvae were grown in conditions of mild HP1a depletion (22°). In DMSO-treated neuroblasts (Figure 3, A and A’, see Movie S3), we observed that late-segregating acentrics (red arrows) entered daughter nuclei through channels in the nuclear envelope (green arrows) and successfully rejoined the main nuclear mass, consistent with previously reported data (Karg et al. 2015). In DMSO + Binucleine-2-treated neuroblasts (Figure 3, B and B’, see Movie S4), we observed that late-segregating acentrics (red arrows) failed to form channels in the daughter nuclear envelope and were subsequently locked out of the nuclei to form micronuclei (red arrowheads), as previously reported (Karg et al. 2015). In DMSO-treated HP1a RNAi-expressing neuroblasts (Figure 3, C and C’, see Movie S5), we observed that late-segregating acentrics (red arrows) entered daughter nuclei through channels in the nuclear envelope (green arrows), successfully rejoining the intact DNA. However, in contrast to the decreased rates of nuclear envelope channel formation and increased micronucleation observed upon inhibition of Aurora B in neuroblasts with wild-type HP1a levels, in DMSO + Binucleine-2-treated HP1a RNAi-expressing neuroblasts (Figure 3, D and D’, see Movie S6), we observed that late-segregating acentrics (red arrows) were once again capable of forming nuclear envelope channels (green arrows), through which acentrics rejoined daughter telophase nuclei.
In total, 67% of DMSO-treated control neuroblasts divisions (N = 21) resulted in visible nuclear envelope channels, and inhibition of Aurora B resulted in a decrease in divisions with visible nuclear envelope channels (36%; N = 45) (significance determined by a χ2 test, P = 0.02) (Figure 3E). In contrast, RNAi depletion of HP1a alone (73%; N = 22) or in combination with Aurora B inhibition (63%; N = 35) showed no difference in the percentage of divisions with visible nuclear envelope channels (no statistical significance determined by a χ2 test, P = 0.44) (Figure 3E). In accord with these observations, we also measured an increase in micronucleation from DMSO-treated control neuroblasts (26%; N = 19) to Aurora B-inhibited neuroblasts (74%; N = 42) (statistical significance determined by a χ2 test, P = 0.0005), while there was no difference in the rate of micronucleation when HP1a was depleted alone (33%; N = 18) or in combination with Aurora B inhibition (34%; N = 32) (no statistical significance determined by a χ2 test, P = 0.94) (Figure 3F). Taken together, these results indicate that forming an H3-HP1a complex along the acentric and tether promotes local nuclear envelope assembly, and preventing the formation of this complex, either through Aurora B-dependent H3 phosphorylation or depletion of HP1a, reduces nuclear envelope assembly and facilitates channel formation.
HP1a exclusion and Aurora B-mediated channel formation are important to prevent micronuclei formation in response to irradiation-induced acentrics
All of the above experiments relied on I-CreI-induced double-stranded breaks specifically in the centric heterochromatin of the X chromosome (Rong et al. 2002; Maggert and Golic 2005; Paredes and Maggert 2009; Golic and Golic 2011). We next sought to determine whether similar results would be obtained regarding HP1a–acentric association and channel formation when the acentrics were generated through X-irradiation (Figure 4). Unlike I-CreI-induced double-stranded breaks, ionizing radiation produces single- and double-stranded DNA breaks in both euchromatin and heterochromatin to yield late-segregating acentrics that vary both in size and chromatin composition (Bajer 1957; Roots et al. 1985; Puerto et al. 2001). Despite these differences between I-CreI and irradiation-induced acentrics, acentrics generated by X-irradiation also form BubR1-coated tethers, undergo delayed poleward segregation, and enter daughter telophase nuclei through nuclear envelope channels (Royou et al. 2010; Karg et al. 2015).
Figure 4.
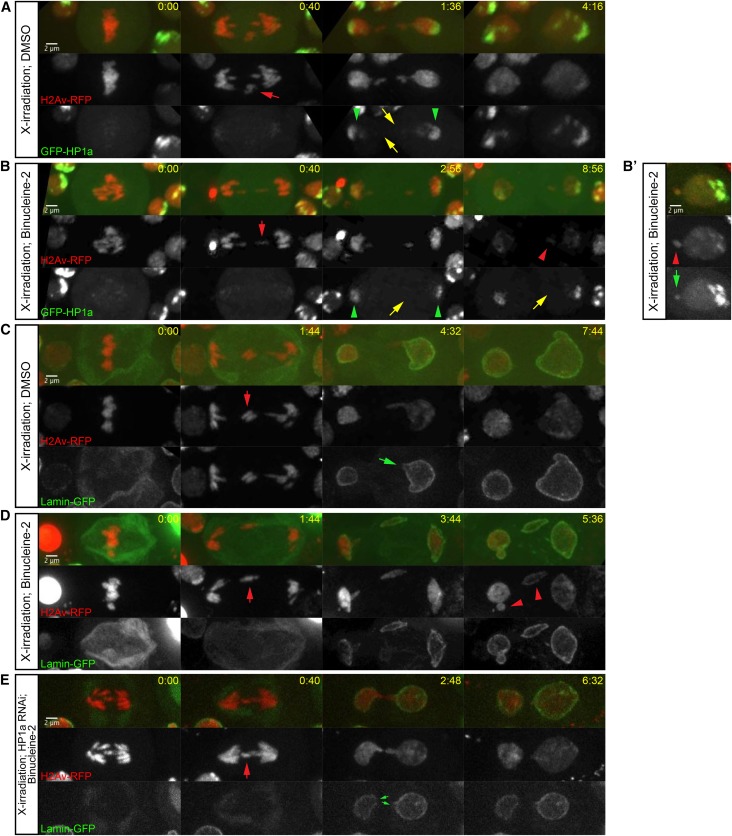
X-irradiation-induced acentrics fail to recruit HP1a and reenter daughter nuclei through Aurora B-mediated nuclear envelope channels. (A) Stills from a time-lapse movie of an X-irradiated neuroblast expressing H2Av-RFP (red fluorescent protein, red) and GFP-HP1a (green), and treated with DMSO. Acentrics are indicated by red arrows. HP1a association with the main daughter nuclei is indicated by green arrowheads. Lack of HP1a on segregating acentrics is depicted by yellow arrows. (B and B’) Stills from a time-lapse movie of an X-irradiated neuroblast expressing H2Av-RFP (red) and GFP-HP1a (green), and treated with DMSO + Binucleine-2. (B) A micronucleus is indicated by a red arrowhead. (B’) A micronucleus is indicated by a red arrowhead and coated with GFP-HP1a (indicated by a green arrow). (C) Stills from a time-lapse movie of an X-irradiated neuroblast expressing H2Av-RFP (red) and Lamin-GFP (green), and treated with DMSO. (D) Stills from a time-lapse movie of an X-irradiated neuroblast expressing H2Av-RFP (red) and Lamin-GFP (green), and treated with DMSO + Binucleine-2. (E) Stills from a time-lapse movie of an X-irradiated neuroblast expressing H2Av-RFP (red), Lamin-GFP (Green), and RNA interference (RNAi) against HP1a and treated with DMSO + Binucleine-2. Time is written as min:sec after anaphase onset. Bar, 2 μm.
To test if Aurora B activity preferentially inhibited HP1a recruitment to X-irradiation-induced acentrics and their tethers, we X-irradiated H2Av-RFP- and GFP-HP1a-expressing female larvae. We observed a similar pattern of HP1a–acentric dynamics during neuroblast division as we did upon I-CreI expression. In control conditions (DMSO-treated neuroblasts), we observed GFP-HP1a exclusion (yellow arrows) from X-irradiation-induced late-segregating chromatin (red arrows) despite its recruitment to main nuclei (green arrowheads) in eight out of eight divisions (Figure 4A), with two out of eight divisions resulting in micronuclei. Upon Aurora B inhibition (DMSO + Binucleine-2-treated neuroblasts), we observed a small increase in GFP-HP1a association [3/11 divisions with detectable GFP-HP1a on acentrics (green arrows)] and a decreased ability for acentrics to rejoin daughter nuclei (six out of nine divisions resulting in micronuclei) (red arrowheads) (Figure 4, B and B’). It should be noted that the sample of X-irradiated acentric data is much smaller than that of I-CreI-induced acentrics, because the ionizing radiation was much less efficient at generating double-strand breaks that would result in detectable acentrics in dividing neuroblasts than I-CreI expression.
Intriguingly, we observed one way in which the recruitment of HP1a to X-irradiation-induced acentrics differed from that of I-CreI-induced acentrics: whereas I-CreI-induced acentrics recruited HP1a upon Aurora B inhibition in a majority of divisions (55%, N = 31) (Figure 4C), X-irradiation-induced acentrics only recruited HP1a upon Aurora B inhibition in a minority of divisions (27%, N = 11) (Figure 4B’ green arrow). Nevertheless, these X-irradiation-induced HP1a-free acentrics still formed micronuclei (four out of six observed micronuclei were HP1a-free) (Figure 4B red arrowhead), suggesting that exclusion of HP1a from acentrics is one of several pathways through which Aurora B mediates channel formation and acentric entry into daughter nuclei.
We additionally monitored nuclear envelope channel formation in response to X-irradiation-induced acentrics by X-irradiating H2Av-RFP- and Lamin-GFP-expressing neuroblasts. In X-irradiated DMSO-treated neuroblasts, we observed X-irradiation-induced lagging chromatin (red arrow) reintegrate into telophase daughter nuclei by passing through channels in the nuclear envelope (green arrow) in six out of nine divisions (Figure 4C), with only three out of eight divisions resulting in micronuclei, similar to previous results (Karg et al. 2015). Treatment of neuroblasts with DMSO + Binucleine-2 to inhibit Aurora B resulted in decreased channel formation (four out of nine divisions with detectable channels) and increased micronucleation (five out of nine divisions resulting in micronuclei) (red arrowheads) (Figure 4D). Furthermore, upon reduction of HP1a by RNAi and Aurora B inhibition (DMSO + Binucleine-2-treated neuroblasts), we once again observed increased channel formation (9/12 divisions with detectable channels) (green arrows) and reduced micronucleation (2/12 divisions resulting in micronuclei), suggesting that Aurora B-mediated HP1a exclusion from lagging chromatin to promote channel formation is not simply limited to I-CreI-induced acentrics.
HP1a specifies preference for nuclear envelope reassembly initiation on the leading edge of segregating chromosomes of the self-renewing neuroblast daughter nucleus
Given that the association of HP1a on acentrics influences local nuclear envelope reassembly at the site of nuclear envelope channels, we hypothesized that HP1a might also play a direct role in global nuclear envelope reassembly in wild-type Drosophila neuroblast divisions. Support for this idea comes from studies demonstrating a requirement for HP1α/HP1a to tether heterochromatin to the nuclear envelope following mitosis (Poleshko et al. 2013) and in which the expression of a truncated form of HP1 disrupts artificial nuclear envelope assembly in mammalian cells (Kourmouli et al. 2000), leading to speculation that HP1a may promote nuclear envelope reassembly in vivo (Schooley et al. 2012).
To test this hypothesis, we performed live analyses of neuroblast divisions from female larvae and found that GFP-HP1a was always recruited to segregating chromosomes before the initiation of nuclear envelope reassembly in the daughter neuroblast (Figure S4, A and A’). This is consistent with previous reports of HP1α/HP1a behavior in mitosis (Sugimoto et al. 2001; Poleshko et al. 2013). On average, we observed that GFP-HP1a was recruited to segregating chromosomes 150 sec (SD = 50 sec; N = 16) after anaphase onset and 210 sec (SD = 100 sec; N = 15) before nuclear envelope reassembly (Figure S4B). Furthermore, we observed that GFP-HP1a was recruited to the leading edge of segregating chromosomes (Figure S4C). In Drosophila neuroblast divisions, which give rise to a self-renewing neuroblast daughter and a ganglion mother cell daughter (GMC), nuclear envelope reassembly initiates on the pole-proximal side of chromosomes segregating to the neuroblast daughter (Katsani et al. 2008; Karg et al. 2015). Therefore, HP1a is located at the proper place and time to mediate nuclear envelope reassembly in neuroblast daughters.
To test the role of HP1a in global nuclear envelope reassembly, we performed live imaging of mitotic neuroblasts from female larvae expressing H2Av-RFP and Lamin-GFP, and monitored nuclear envelope reassembly of neuroblast daughters in wild-type conditions or conditions in which HP1a was strongly depleted through RNAi (Figure 5). As previously reported, in wild-type divisions, we observed a dramatic asymmetry in nuclear envelope reassembly initiation on the self-renewing neuroblast daughter cell (Katsani et al. 2008). Nuclear envelope reassembly first initiated on the pole-proximal edge of chromosomes segregating to the neuroblast daughter (Figure 5A, green arrows, see Movie S7) before completion on the midzone-proximal face of the segregated chromosomes. As this asymmetry was not as distinct in the differentiating GMC, we focused our studies on the self-renewing neuroblast daughter.
In wild-type neuroblast daughters, we observed nuclear envelope reassembly initiate on the pole-proximal edge of chromosomes 70% of the time, on the midzone-proximal edge of chromosomes 0% of the time, and around all sides of the chromosomes at once 30% of the time (N = 37) (Figure 5C). In contrast, in HP1a-depleted neuroblast daughters (Figure 5B, see Movie S8), we observed nuclear envelope reassembly initiate on the pole-proximal edge of chromosomes only 47.5% of the time (statistical significance determined by a χ2 test, P = 0.04), on the midzone-proximal edge of chromosomes 7.5% of the time, and around all sides of the chromosome at once (Figure 6B, green arrows) 45% of the time (N = 40) (Figure 5C).
Figure 6.
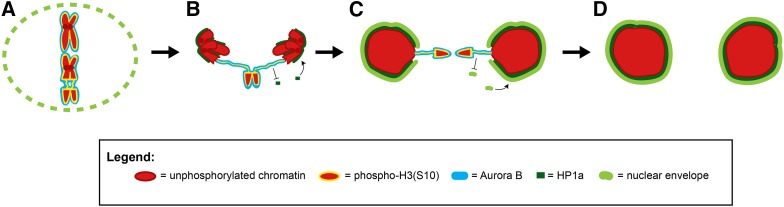
Model for Aurora B-mediated nuclear envelope channel formation. (A) During metaphase, Aurora B (blue) phosphorylates H3(S10) (yellow) on chromosomes (red), acentrics are pushed to the edge of the metaphase plate, and the nuclear envelope (lime green) is partially disassembled. (B) During anaphase, Aurora B, a component of the chromosome passenger complex, is removed from the main chromosomes and relocalizes to the spindle midzone. Phospho-H3(S10) marks on the main nuclei are removed and HP1 (dark green) is recruited to the main nuclei. However, persistent Aurora B on the acentric and tether continues to phosphorylate H3(S10) and inhibits HP1a recruitment to the acentric/tether. (C) During telophase, nuclear envelope components reform connections with chromatin through HP1a and nuclear envelope reassembly proceeds. However, the exclusion of HP1a from Aurora B-coated tether/acentrics prevents the accumulation of nuclear envelope components on the tether/acentrics and at the site where the tether contacts the main nucleus, leading to local delays in nuclear envelope reassembly and the formation of nuclear envelope channels. (D) Successful incorporation of late-segregating acentrics into telophase nuclei through nuclear envelope channels results in euploid daughter cells.
In spite of this difference in nuclear envelope reassembly initiation preference, we observed no change in the timing of nuclear envelope reassembly initiation (Figure 5D) (no statistical significance determined by an independent two-sided Student’s t-test, P = 0.312) or the amount of nuclear envelope deformities (Figure 5E) (no statistical significance determined by a χ2 test, P = 0.45) upon HP1a depletion. Taken together, these results suggest that HP1a specifies the pole-proximal location of nuclear envelope reassembly initiation in self-renewing neuroblast daughter cells.
Discussion
Despite the inability of acentric chromosomes to form kinetochore–microtubule attachments, studies show that while some acentrics fail to segregate properly (Fenech et al. 2011), others are capable of efficient poleward segregation (Bajer 1957; Malkova et al. 1996; Ahmad and Golic 1998; Galgoczy and Toczyski 2001; Titen and Golic 2008; Royou et al. 2010; Bretscher and Fox 2016; Karg et al. 2017). Failure of acentrics to reincorporate into daughter telophase nuclei leads to the formation of micronuclei, which result in aneuploidy or DNA damage, and are a hallmark of cancer (Santos et al. 2010; Bonassi et al. 2011; Crasta et al. 2012; Zhang et al. 2015; Vázquez-Diez et al. 2016; Ly et al. 2017). In Drosophila, I-CreI-induced acentrics avoid this fate by passing through Aurora B-mediated channels in the nuclear envelope (Karg et al. 2015).
Here, we observed that late-segregating acentrics in anaphase and telophase neuroblast divisions are marked with a strong phospho-H3(S10) signal despite the removal of the majority of this mark from the main nuclei (Figure 1). Consistent with previous reports (Fuller et al. 2008), the strength of this signal is dependent upon Aurora B (Figure S1). Intriguingly, in addition to the high phospho-H3(S10) signal on the acentrics, we observed high levels of phospho-H3(S10) on areas of the main nuclei at sites where tethers normally contact the main nuclei. During late anaphase, when the normal chromosomes begin to reassemble a nuclear envelope, acentrics and the sites of acentric entry into daughter nuclei remain nuclear envelope-free (Karg et al. 2015), suggesting an inverse correlation between nuclear envelope reassembly and phospho-H3(S10) modification. This finding is consistent with studies suggesting that, in addition to the phosphorylation state of nuclear envelope components (Steen et al. 2000; Onischenko et al. 2005), nuclear envelope reassembly is also regulated by various chromatin remodeling events, including the removal of phospho-H3(S10) (Vagnarelli et al. 2011; Schooley et al. 2015).
Our studies also demonstrate that HP1a is excluded from late-segregating I-CreI-induced acentrics despite HP1a recruitment to the main nuclei (Figure 2). We note that the chromatin makeup of I-CreI-induced acentrics should be sufficient to recruit HP1a, as I-CreI creates double-stranded breaks in the pericentric region of the X chromosome, resulting in an acentric fragment that contains a large portion of heterochromatin (Rong et al. 2002; Maggert and Golic 2005). Presumably, the difference in recruitment of HP1a to acentrics and the main nuclei is due to the difference in the phosphorylation state of H3(S10) on acentrics and the main nuclei, as Aurora B-mediated phospho-H3(S10) is prohibitive to HP1α/HP1a binding (Fischle et al. 2005; Hirota et al. 2005). In support of this view, inhibition of Aurora B kinase activity resulted in increased HP1a association with acentrics (Figure 2). However, no increase in HP1a at tether contact sites on the main nuclei was detected upon Aurora B inhibition, possibly due to a limited ability to consistently observe tethers at our imaging resolution.
Furthermore, we found that acentrics possessing high levels of HP1a are largely unable to reintegrate into daughter nuclei and instead form micronuclei (Figure 2). Under the same Aurora B inhibition conditions in which we detected HP1a association with acentrics and micronucleation, we observed increased nuclear envelope reassembly around acentrics and decreased nuclear envelope channel formation on the main nuclei (Figure 2 and Figure 3). We hypothesized that HP1a presence on I-CreI-induced acentrics was prohibitive to acentric entry into daughter nuclei, possibly due to the inability to form a nuclear envelope channel. This hypothesis predicts that: (1) in wild-type conditions, Aurora B activity would inhibit formation of an H3-HP1a complex on the acentric and tether and lead to slow nuclear envelope assembly at these sites and the formation of a channel; (2) when Aurora B is inhibited, H3 on the acentric and tether would bind to HP1a, which would stimulate nuclear envelope assembly at these sites and prevent channel formation; and (3) when Aurora B is inhibited and HP1a is depleted, no H3-HP1a complex would form on the acentric and tether, leading to slow nuclear envelope assembly at these sites and the formation of a channel, reminiscent of wild-type conditions (Figure S2). Our data show that upon codepletion of HP1a with Aurora B inhibition, nuclear envelope reassembly on acentrics is reduced and channel formation occurs at frequencies similar to those detected in wild-type Aurora B and HP1a conditions (Figure 3). Essentially, depletion of HP1a masks the phenotype of Aurora B inhibition, evocative of a classic epistatic relationship in which Aurora B mediates nuclear envelope channel formation by preferentially excluding HP1a from acentrics and their tethers.
Therefore, we propose a model for nuclear envelope channel formation in which highly localized concentrations of Aurora B kinase phosphorylate H3(S10) specifically on acentrics, their associated tethers, and at sites where the tethers contact the main nuclei. This prevents local heterochromatic recruitment of HP1a and subsequent recruitment of nuclear envelope lamina on the acentrics and at the sites where acentrics rejoin daughter nuclei, leading to the formation of channels through which acentrics pass to maintain genome integrity (Figure 6).
Interestingly, channel formation mediated by HP1a exclusion is similar to the mechanism of human polyomavirus egress from its host nucleus. Viral angoprotein binds to HP1α/HP1a and disrupts its binding with the inner nuclear membrane protein LBR, causing sections of weakened nuclear envelope through which virions leave the nucleus (Okada et al. 2005). Thus, regulation of chromatin–HP1a–nuclear envelope interactions may represent a conserved method for bypassing the barrier of the nuclear envelope.
By generating acentrics through X-irradiation, we found that the general pattern of HP1a recruitment driving nuclear envelope reassembly on acentrics was not dependent upon the system used to generate acentrics or due to the exact physical nature of an I-CreI-induced acentric (Figure 4). However, we note that while micronuclei derived from I-CreI-induced acentrics were generally HP1a-coated, a higher proportion of micronuclei derived from irradiation-induced acentrics were HP1a-free. It is tempting to speculate that these HP1a-free micronuclei were derived from largely euchromatic acentrics that simply lack a sufficient amount of heterochromatin to recruit HP1a. This observation suggests that Aurora B may mediate acentric entry into daughter nuclei through multiple pathways of which preferential exclusion of HP1a is one. It is possible these HP1a-free acentrics remain capable of recruiting a nuclear envelope through an HP1a-independent pathway, perhaps involving an interaction between LAP2/emerin/MAN1 (LEM) domain-containing inner nuclear membrane proteins and the DNA-cross-bridging factor barrier-to-autointegration factor (Haraguchi et al. 2001; Samwer et al. 2017).
To support our finding that HP1a promotes nuclear envelope assembly in vivo, we examined its role in assembling the nuclear envelope on normal intact chromatin. We observed HP1a localize to the leading edge of segregating chromosomes before nuclear envelope reassembly (Figure S4). As previously reported, we observed nuclear envelope reassembly initiate on the leading edge of chromosomes segregating to daughter neuroblasts, proceeding to wrap around and complete reassembly on the midzone-proximal face of the nascent nucleus (Robbins and Gonatas 1964; Katsani et al. 2008; Karg et al. 2015). However, reducing HP1a levels disrupts the preferential nuclear envelope assembly on the pole-proximal face of the segregating chromosomes (control = 70% pole-proximal initiation; HP1a depletion = 47.5% pole-proximal initiation) (Figure 5). This result is complementary to a growing body of evidence demonstrating that HP1 proteins may play key roles in nuclear envelope reassembly. For example, in mammalian cells, HP1α/HP1a recruits PRR14 to segregating chromosomes where it tethers heterochromatin to the nuclear envelope (Poleshko et al. 2013), and in vitro experimentation shows HP1β/HP1b to be important for recruiting nuclear envelope components to interphase-like chromatin (Kourmouli et al. 2000). In addition, depletion of the PP1γ subunit Repo-Man leads to retained phospho-H3(S10) marks on chromatin, loss of HP1α/HP1a recruitment to mitotic chromosomes, and defects in nuclear envelope reassembly (Vagnarelli et al. 2011). One mechanism by which HP1a might bias nuclear envelope reassembly to initiate on the leading edge of segregating chromosomes is by enhancing the natural ability of nuclear envelope components to bind to chromatin, possibly through its interaction with the inner nuclear membrane protein LBR (Ye and Worman 1996; Ulbert et al. 2006).
Despite the clear preference for the initiation of nuclear envelope reassembly on the leading edge of chromosomes segregating to daughter neuroblasts, we observed no such preference on the chromosomes segregating to the daughter GMC (Figure 5). It is possible this difference is due to the relatively small size of the GMC daughter, or that in Drosophila neuroblast divisions, the endoplasmic reticulum, from which the nuclear envelope extends during mitotic exit, is asymmetrically localized to the spindle pole of the neuroblast daughter (Smyth et al. 2015).
Our data also address a key question regarding nuclear envelope channel formation: how do late-segregating acentrics near the spindle midzone act at a distance to influence nuclear envelope reassembly dynamics on main nuclei near the poles? In our system, we believe there are two pools of Aurora B: a constitutive midzone-based pool (Fuller et al. 2008; Afonso et al. 2014) and a tether-based pool, which stretches from the acentric to the main nucleus (Royou et al. 2010). Given that nuclear envelope channels are only observed on the main nuclei when acentrics and the tether-based pool of Aurora B are present, we proposed that the pool of Aurora B responsible for channel formation is the tether-based Aurora B (Karg et al. 2015). Our observation of phospho-H3(S10) hotspots on the main nuclei at sites closest to acentrics is consistent with the hypothesis that tether-based Aurora B activity controls channel formation (Figure 2A, yellow arrowheads). Since the midzone pool of Aurora B is confined away from the main telophase nuclei, it is probable that these hotspots are due to the activity of Aurora B along tethers, which stretch from acentrics and contact main nuclei at sites closest to the acentrics. These phospho-H3(S10) hotspots could then locally prevent HP1a association and nuclear envelope reassembly. Thus, while nuclear envelope reassembly can still proceed around the rest of the nucleus, it is inhibited at the site of these hotspots, resulting in channels.
In summary, our results reveal a novel mechanism by which genome integrity is maintained. Late-segregating acentric fragments pose a significant hazard, as they are at high risk of forming micronuclei that induce dramatic rearrangements in the genome (Fenech 2000; Zhang et al. 2015; Ly et al. 2017). Consequently it is likely that cells have evolved mechanisms to prevent the formation of micronuclei. Here, we provide evidence for one such mechanism, in which Aurora B-mediated inhibition of HP1a-chromatin association during anaphase/telophase prevents the formation of micronuclei from late-segregating acentric fragments.
Acknowledgments
We thank W. Saxton and S. Strome for use of their equipment, B. Abrams for additional help with microscopy, the members of the C. Forsberg laboratory for their assistance with irradiation experiments, and G. Karpen and P. O’Farrell for their helpful advice. This work was funded by National Institutes of Health (NIH) grant R01-GM-120321 awarded to W.S.; B.W. was also supported by NIH grant 5T32-GM-008646-18.
Author contributions: B.W. and W.S. conceived experiments and wrote the manuscript. B.W. performed experiments.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6752816.
Communicating editor: J. Bateman
Literature Cited
- Abbas T., Keaton M. A., Dutta A., 2013. Genomic instability in cancer. Cold Spring Harb. Perspect. Biol. 5: a012914 10.1101/cshperspect.a012914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso O., Matos I., Pereira A. J., Aguiar P., Lampson M. A., et al. , 2014. Feedback control of chromosome separation by a midzone Aurora B Gradient. Science 345: 332–336. 10.1126/science.1251121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K., Golic K. G., 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148: 775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Hetzer M. W., 2008. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182: 911–924. 10.1083/jcb.200805140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajer A., 1957. Cine-micrographic studies on chromosome movements in β-irradiated cells. Chromosoma 9: 319–331. 10.1007/BF02568084 [DOI] [PubMed] [Google Scholar]
- Baur T., Ramadan K., Schlundt A., Kartenbeck J., Meyer H. H., 2007. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J. Cell Sci. 120: 2895–2903. 10.1242/jcs.010181 [DOI] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M. G., Gatti M., 2000. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2: 54–56. 10.1038/71378 [DOI] [PubMed] [Google Scholar]
- Bonassi S., El-Zein R., Bolognesi C., Fenech M., 2011. Micronuclei frequency in peripheral blood lymphocytes and cancer risk: evidence from human studies. Mutagenesis 26: 93–100. 10.1093/mutage/geq075 [DOI] [PubMed] [Google Scholar]
- Bretscher H. S., Fox D. T., 2016. Proliferation of double-strand break-resistant polyploid cells requires Drosophila FANCD2. Dev. Cell 37: 444–457. 10.1016/j.devcel.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M. E., Karess R. E., 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15: 856–861. 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Doe C. Q., 2009. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17: 134–141. 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Carmena M., Wheelock M., Funabiki H., Earnshaw W. C., 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13: 789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Siriaco G., Raffa G. D., Kellum R., Gatti M., 2003. The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5: 82–84. 10.1038/ncb902 [DOI] [PubMed] [Google Scholar]
- Chaudhary N., Courvalin J., 1993. Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol. 122: 295–306. 10.1083/jcb.122.2.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Fioravanti D., Salmon E. D., Degrassi F., 2002. Merotelic kinetochore orientation vs. chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J. Cell Sci. 115: 507–515. [DOI] [PubMed] [Google Scholar]
- Crasta K., Ganem N. J., Dagher R., Jantermann A. B., Ivanova E. V., et al. , 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., et al. , 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154: 71–84. 10.1083/jcb.200101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro I. J., Gokhan E., Vagnarelli P., 2016. Resetting a functional G1 nucleus after mitosis. Chromosoma 125: 607–619. 10.1007/s00412-015-0561-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derive N., Landmann C., Montembault E., Claverie M. C., Pierre-Elies P., et al. , 2015. Bub3-BubR1-dependent sequestration of Cdc20Fizzy at DNA breaks facilitates the correct segregation of broken chromosomes. J. Cell Biol. 211: 517–532. 10.1083/jcb.201504059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann H. L., Tseng B. S., Allis C. D., Funabiki H., Fischle W., 2006. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle 5: 2842–2851. 10.4161/cc.5.24.3540 [DOI] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., et al. , 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180: 857–865. 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S., 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274: 1664–1672. 10.1126/science.274.5293.1664 [DOI] [PubMed] [Google Scholar]
- Fenech M., 2000. The in vitro micronucleus technique. Mutat. Res. 455: 81–95. 10.1016/S0027-5107(00)00065-8 [DOI] [PubMed] [Google Scholar]
- Fenech M., Kirsch-Volders M., Natarajan A. T., Surralles J., Crott J. W., et al. , 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26: 125–132. 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., et al. , 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Fuller B. G., Lampson M. A., Foley E. A., Rosasco-Nitcher S., Le K. V., et al. , 2008. Midzone activation of aurora B in anaphase produces intracellular phosphorylation gradient. Nature 453: 1132–1136. 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy D. J., Toczyski D. P., 2001. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 21: 1710–1718. 10.1128/MCB.21.5.1710-1718.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic M. M., Golic K. G., 2011. A simple and rapid method for constructing ring-X chromosomes in Drosophila melanogaster. Chromosoma 120: 159–164. 10.1007/s00412-010-0297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Koujin T., Segura-Totten M., Lee K. K., Matsuoka Y., et al. , 2001. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114: 4575–4585. [DOI] [PubMed] [Google Scholar]
- Hirota T., Lipp J. J., Toh B. H., Peters J. M., 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180. 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., et al. , 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291. 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Huang Y., Jiang L., Yi Q., Lv L., Wang Z., et al. , 2012. Lagging chromosomes entrapped in micronuclei are not ‘lost’ by cells. Cell Res. 22: 932–935. 10.1038/cr.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D., 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurons and glial cells. Development 124: 761–771. [DOI] [PubMed] [Google Scholar]
- Kanda T., Wahl G. M., 2000. The dynamics of acentric chromosomes in cancer cells revealed by GFP-based chromosome labeling strategies. J. Cell. Biochem. Suppl. 35: 107–114. [DOI] [PubMed] [Google Scholar]
- Karg T., Warecki B., Sullivan W., 2015. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol. Biol. Cell 26: 2227–2241. 10.1091/mbc.e15-01-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg T., Elting M. W., Vicars H., Dumont S., Sullivan W., 2017. The chromokinesin Klp3a and microtubules facilitate acentric chromosome segregation. J. Cell Biol. 6: 1597–1608. 10.1083/jcb.201604079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani K. R., Karess R. E., Dostatni N., Doye V., 2008. In vivo dynamics of Drosophila nuclear envelope components. Mol. Biol. Cell 19: 3652–3666. 10.1091/mbc.e07-11-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. A., Melo J. A., Cheung S. K., Vaze M. B., Haber J. E., et al. , 2004. DNA breaks promote genomic instability by impeding proper chromosome-segregation. Curr. Biol. 14: 2096–2106. 10.1016/j.cub.2004.10.051 [DOI] [PubMed] [Google Scholar]
- Kellum R., Alberts B. M., 1995. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 108: 1419–1431. [DOI] [PubMed] [Google Scholar]
- Kotadia S., Montembault E., Sullivan W., Royou A., 2012. Cell elongation is an adaptive response for clearing long chromatid arms from the cleavage plane. J. Cell Biol. 199: 745–753. 10.1083/jcb.201208041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N., Theodoropoulos P. A., Dialynas G., Bakou A., Politou A. S., et al. , 2000. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J. 19: 6558–6568. 10.1093/emboj/19.23.6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain J. R., Jr., Oldenbourg R., Cole R. W., Rieder C. L., 2001. Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell 12: 4054–4065. 10.1091/mbc.12.12.4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. M., Goodman C. S., 1994. Ectopic and increased expression of Fasciclin II alters motorneuron growth cone guidance. Neuron 13: 507–523. 10.1016/0896-6273(94)90022-1 [DOI] [PubMed] [Google Scholar]
- Lu L., Ladinsky M. S., Kirchhausen T., 2011. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol. 194: 425–440. 10.1083/jcb.201012063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P., Teitz L. S., Kim D. H., Shoshani O., Skaletsky H., et al. , 2017. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 19: 68–75. 10.1038/ncb3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert K. A., Golic K. G., 2005. Highly efficient sex chromosome interchanges produced by I–CreI expression in Drosophila. Genetics 171: 1103–1114. 10.1534/genetics.104.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Ivanov E. L., Haber J. E., 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93: 7131–7136. 10.1073/pnas.93.14.7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus K. J., Hendzel M. J., 2006. The relationship between histone H3 phosphorylation and acetylation throughout the mammalian cell cycle. Biochem. Cell Biol. 84: 640–657. 10.1139/o06-086 [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Cole R. W., Rieder C. L., 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12: 1797–1806. 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Newport J. W., Wilson K. L., Dunphy W. G., 1990. A lamin-independent pathway for nuclear envelope assembly. J. Cell Biol. 111: 2247–2259. 10.1083/jcb.111.6.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Suzuki T., Sunden Y., Orba Y., Kose S., et al. , 2005. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: role in nuclear egress of viral particles. EMBO Rep. 6: 452–457. 10.1038/sj.embor.7400406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y., Hodgson L., Mantell J., Verkade P., Carlton J. G., 2015. ESCRT-III controls nuclear envelope reformation. Nature 522: 236–239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E. A., Gubanova N. V., Kiseleva E. V., Halberg E., 2005. Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol. Biol. Cell 16: 5152–5162. 10.1091/mbc.e05-07-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S., Maggert K. A., 2009. Expression of I–CreI endonuclease generates deletions within the rDNA of Drosophila. Genetics 181: 1661–1671. 10.1534/genetics.108.099093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A., Uzawa S., Perry P. E., Cande W. Z., Allshire R. C., 2000. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 113: 4177–4191. [DOI] [PubMed] [Google Scholar]
- Poleshko A., Mansfield K. M., Burlingame C. C., Andrake M. D., Shah N. R., et al. , 2013. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 5: 292–301. 10.1016/j.celrep.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudaki H., Kourmouli N., Drosou V., Bakou A., Theodoropoulos P. A., et al. , 2001. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2: 920–925. 10.1093/embo-reports/kve199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerto S., Ramírez M. J., Marcos R., Creus A., Surrallés J., 2001. Radiation-induced chromosome aberrations in human euchromatic (17cen-p53) and heterochromatic (1cen-1q12) regions. Mutagenesis 16: 291–296. 10.1093/mutage/16.4.291 [DOI] [PubMed] [Google Scholar]
- R Core Team, 2014 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
- Ramadan K., Bruderer R., Spiga F. M., Popp O., Baur T., et al. , 2007. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450: 1258–1262. 10.1038/nature06388 [DOI] [PubMed] [Google Scholar]
- Robbins E., Gonatas N. K., 1964. The Ultrastructure of a mammalian cell during the mitotic cycle. J. Cell Biol. 21: 429–463. 10.1083/jcb.21.3.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., et al. , 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581. 10.1101/gad.986602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roots R., Kraft G., Gosschalk E., 1985. The formation of radiation-induced DNA breaks: the ratio of double-strand breaks to single-strand breaks. Int. J. Radiat. Oncol. Biol. Phys. 11: 259–265. 10.1016/0360-3016(85)90147-6 [DOI] [PubMed] [Google Scholar]
- Royou A., Macias H., Sullivan W., 2005. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr. Biol. 15: 334–339. 10.1016/j.cub.2005.02.026 [DOI] [PubMed] [Google Scholar]
- Royou A., Gagou M. E., Karess R., Sullivan W., 2010. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell 140: 235–245. 10.1016/j.cell.2009.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos S. A., Ranatunga N. S., Yuan J. P., Green M. D., Forsburg S. L., 2015. Replication stress in early S phase generates apparent micronuclei and chromosome rearrangement in fission yeast. Mol. Biol. Cell 26: 3439–3450. 10.1091/mbc.e15-05-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer M., Schneider M. W. G., Schmalhorst P. S., Jude J. G., Zuber J., et al. , 2017. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170: 956–972.e23. 10.1016/j.cell.2017.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R. A., Teixeira A. C., Mayorano M. B., Carrara H. H. A., Andrade J. M., et al. , 2010. Basal levels of DNA damage detected by micronuclei and comet assays in untreated breast cancer patients and healthy women. Clin. Exp. Med. 10: 87–92. 10.1007/s10238-009-0079-4 [DOI] [PubMed] [Google Scholar]
- Schellhaus A. K., De Magistris P., Antonin W., 2015. Nuclear reformation at the end of mitosis. J. Mol. Biol. 428: 1962–1985. 10.1016/j.jmb.2015.09.016 [DOI] [PubMed] [Google Scholar]
- Schooley A., Vollmer B., Antonin W., 2012. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma 121: 539–554. 10.1007/s00412-012-0388-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley A., Moreno-Andrés D., De Magistris P., Vollmer B., Antonin W., 2015. The lysine demethylase LSD1 is required for nuclear envelope formation at the end of mitosis. J. Cell Sci. 128: 3466–3477. 10.1242/jcs.173013 [DOI] [PubMed] [Google Scholar]
- Smurnyy Y., Toms A. V., Hickson G. R., Eck M. J., Eggert U. S., 2010. Binucleine 2, an isoform-specific inhibitor of Drosophila Aurora B kinase, provides insights into the mechanism of cytokinesis. ACS Chem. Biol. 5: 1015–1020. 10.1021/cb1001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J. T., Schoborg T. A., Bergman Z. J., Riggs B., Russan N. M., 2015. Proper symmetric and asymmetric endoplasmic reticulum partitioning requires astral microtubules. Open Biol. 5: 150067 10.1098/rsob.150067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen R. L., Martins S. B., Taskén K., Collas P., 2000. Recruitment of protein phosphatase 1 to the nuclear envelope by A-kinase anchoring protein AKAP149 is a prerequisite for nuclear lamina assembly. J. Cell Biol. 150: 1251–1262. 10.1083/jcb.150.6.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Tasaka H., Dotsu M., 2001. Molecular behavior in living mitotic cells of human centromere heterochromatin protein HP1α ectopically expressed as a fusion to red fluorescent protein. Cell Struct. Funct. 26: 705–718. 10.1247/csf.26.705 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner A., Hawley R. S., 2000. Drosophila Protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Titen S. W., Golic K. G., 2008. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics 180: 1821–1832. 10.1534/genetics.108.093625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S., Platani M., Boue S., Mattaj I. W., 2006. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173: 469–476. 10.1083/jcb.200512078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Ribeiro S., Sennels L., Sanchez-Pulido L., de Lima Alves F., et al. , 2011. Repo-man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 21: 328–342. 10.1016/j.devcel.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Diez C., Yamagata K., Trivedi S., Haverfield J., FitzHarris G., 2016. Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. Proc. Natl. Acad. Sci. USA 113: 626–631. 10.1073/pnas.1517628112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M., Schink K. O., Campsteijn C., Wegner C. S., Schultz S. W., et al. , 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522: 231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Ye Q., Worman H. J., 1996. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem. 271: 14653–14656. 10.1074/jbc.271.25.14653 [DOI] [PubMed] [Google Scholar]
- Ye Q., Callebaut I., Pezhman A., Courvalin J. C., Worman H. J., 1997. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 272: 14983–14989. 10.1074/jbc.272.23.14983 [DOI] [PubMed] [Google Scholar]
- Zhang C. Z., Spektor A., Cornilis H., Francis J. M., Jackson E. K., et al. , 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All fly stocks are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental data have been deposited on Figshare. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6752816.