Abstract
Using 5′ RACE, we have isolated four additional exons of the mu opioid receptor gene (Oprm), resulting in a gene spanning over 250 kb. The four new exons are contained within eight additional splice variants containing exon 11 at the 5′ terminus. Exon 11, which is under the control of a previously unknown upstream promoter, and exon 12 are located ≈10 kb and ≈8 kb upstream from exon 1, respectively. Exon 13 and 14 are located between exons 1 and 2. The regional distributions of the variants, as determined by reverse transcription-PCR, varied among themselves and were distinct from that of MOR-1, implying region-specific RNA processing. Three variants (MOR-1H, MOR-1I, and MOR-1J) contained two potential translational start points, with the translational start point in exon 1 producing proteins identical to the original MOR-1 protein. When expressed, the receptor binding of these three variants was indistinguishable from that of MOR-1. The remaining eight proteins using the translation start point in exon 11 were all truncated, with three (MOR-1G, MOR-1M, and MOR-1N) predicting proteins of only six transmembrane domains and the rest giving proteins under 10 kDa. Western blots with an exon 11-specific antiserum revealed bands consistent with the six transmembrane domain proteins within the brain, but the shorter proteins were not detected. Thus, the MOR-1 protein can be generated by four different splice variants of the Oprm gene under the control of two physically distinct promoters. Although the truncated proteins are expressed in brain with a unique regional distribution, their functional significance remains unknown.
Clinicians have long noted differences among the mu opioid analgesics (1, 2). Patients often respond differently to one mu opioid than another with regard to pain control and side-effects. Most impressive, however, is the presence of incomplete cross-tolerance, with highly tolerant patients often responding to a second drug at doses far lower than predicted from the relative potency of the drugs in naive patients. These clinical observations, coupled with preclinical studies, led to the suggestion of multiple mu receptors subtypes (3), a concept supported by subsequent molecular approaches (4–7).
Soon after the initial descriptions of the cloning of the mu opioid receptor MOR-1 (8–10), two groups reported alternative splicing of MOR-1 at the 3′ end of exon 3 (Fig. 1; refs. 11 and 12). MOR-1A lacks exon 4 and the predicted coding region extends 12 bases beyond the normal splice site in exon 3. MOR-1B contains an alternatively spliced exon 5 instead of the original exon 4. Although the binding selectivities of MOR-1B and MOR-1 are similar, their desensitization properties and regional distributions differ (12, 13). Subsequently, we reported five additional exons contained within the MOR-1 gene that combine to generate an additional four splice variants in which exon 4 is replaced by combinations of the new exons (refs. 14 and 15; Fig. 1). Immunohistochemical and confocal studies have documented cell-specific and region-specific expression of MOR-1, MOR-1C (16, 17), and MOR-1D (18), as well as the internalization of MOR-1C in vivo following administration of mu agonists, including morphine (19). The current studies describe an additional series of eight MOR-1 variants with splicing at the 5′ end of the mRNA.
Figure 1.
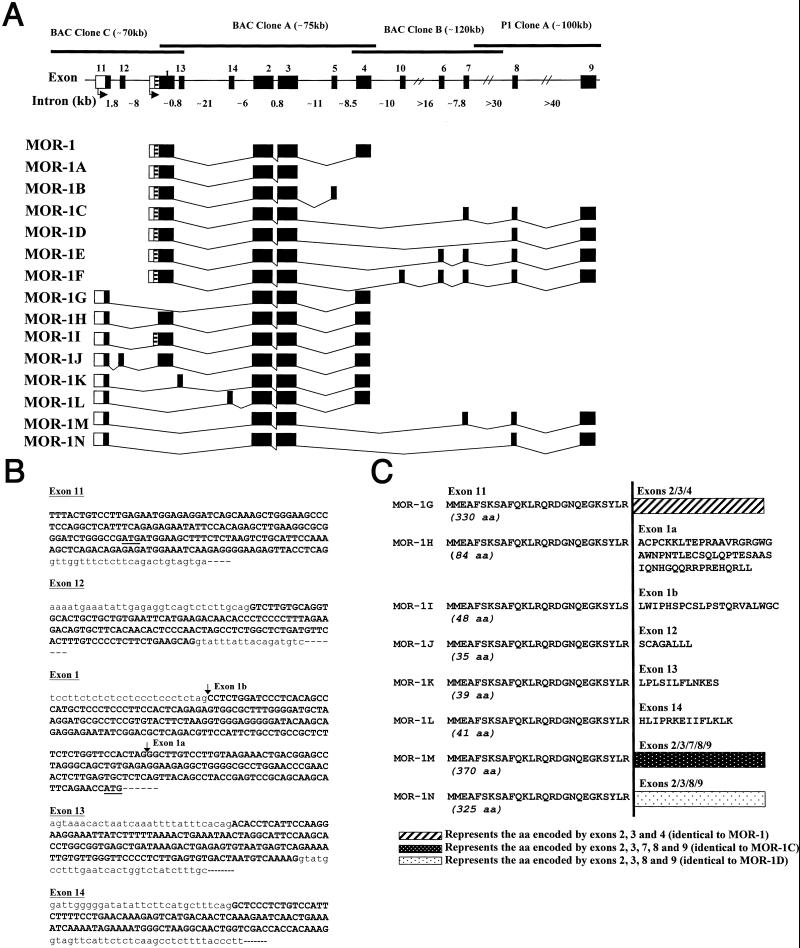
MOR-1 gene structure and alternative splicing. (A) Schematic of Oprm gene structure and alternative splicing. Coding exons are indicated by black boxes and 5′ flanking regions by blank boxes. Introns and bacterial artificial chromosome (BAC) clones are shown by light and heavy horizontal lines, respectively. The exons were numbered in the order in which they were discovered. Exon 1a (black) and exon 1b (striped) are portions of exon 1. (B) Partial genomic nucleotide sequences of exons 11, 12, 13, 14, and 1. The genomic sequences of exons 11, 12, 1, 13, and 14 have been deposited in the GenBank database (accession nos. AF260312–AF260314). Exon and partial intron sequences are shown in uppercase and lowercase, respectively. Putative translation start codon of exons 11 and 1 are underlined. The 5′ splice sites in exons 1a and 1b are indicated by vertical arrows. (C) Predicted amino acid sequences of the variants by using the exon 11 translation start point; and the amino acid sequences predicted from the variant cDNAs by using the exon 11 translation start codon. To save space, the partial amino acid sequences deduced from the exons 2 and further downstream are designated by the three boxes. Note: MOR-1H, MOR-1I, and MOR-1J also generate a protein identical to that of MOR-1. The glycine residue at position 18 is a potential N-myristoylation site. The cDNA and deduced amino acid sequences of the variants have been deposited in the GenBank database (accession nos. AF062755, AF260306–AF260311 and AF074972).
Materials and Methods
5′ RACE and PCR Cloning.
To identify previously unknown exons upstream from exon 1, total RNA extracted from CXBK and C57 Black/6 mouse brains were reverse-transcribed with random hexamers and oligo dT, hybridized with a 5′ biotinylated sense primer designed from exon 3 (5′-CATGGTACTGGGAGAACCTGCTCA-3′) and the cDNA captured and purified with magnetic beads covalently coupled to streptavidin (Dynabeads M280 Streptavidin). The purified cDNA was then ligated by using a T4 RNA ligase with an adapt primer (5′-CCCTTCTGTCGTCTTCTCGCAGCCGTA-3′) that had been modified by adding a phosphate group at the 5′ end and an amine group at the 3′ end to obtain an efficient unidirectional ligation. Because no bands could be observed with a single amplification, nested PCR was performed with the ligated cDNAs as template: first round PCR with a sense primer (5′-TACGGCTGCGAGAAGACGACAGAAGGG-3′) derived from the adapt primer and an antisense primer (5′-CCGGCATGATGAAGGCGAAGATGA-3′) from exon 3, followed by a second round with the same sense primer and a nested antisense primer (5′-CGGAAATCCAGGGCCTTGACCGG-3′) from exon 2. Several bands were obtained and cloned into pCRII-TOPO vector (Invitrogen). Sequence analysis revealed a new 184-bp sequence (exon 11) linked to the 5′ end of the exon 2 that predicted a previously uncharacterized amino acid sequence of 27 aa in-frame with the exon 2 sequence present in MOR-1.
To obtain full-length cDNAs that might be associated with the exon 11 sequence, two sense primers designed from the exon 11 (sense primer E11A, 5′-CCTCCAGGCTCATTTCAGAGAG-3′, and sense primer E11B, 5′-GGCGCGGGATCTGGGCCGATGATGGAAGCTTTCTCTAAGTCTGCATTC-3′), two antisense primers from the exon 4 (antisense primer E4A, 5′-CCAGGAAACCAGAGCCTCCCACAA-3′, and antisense primer E4B, 5′-GGCGTGGGACCCAGTTAGGGCA-3′), and two antisense primers from the exon 9 (antisense primer E9A, 5′-GAAAGGCATCTTCCCTCTCGCTGT-3′, and antisense primer E9B (5′-CCACACTGCTCACCAGCTCATCCC-3′) were used in nested-PCRs with the first-strand cDNA reverse-transcribed from mouse brain total RNA as templates. The PCR fragments were subcloned into pCRII-TOPO and sequenced in both directions with appropriate primers. Eight full-length cDNAs containing the exon 11 were identified and named MOR-1G, MOR-1H, MOR-1I, MOR-1J, MOR-1K, MOR-1L, MOR-1M, and MOR-1N (Fig. 1).
Northern Blot Analysis.
Northern blot analysis was performed as described (14). In brief, total RNA was isolated from mouse brain by a guanidine thiocyanate/phenol-chloroform extraction. Total RNA (100 μg/lane) was separated through a 0.8% formaldehyde agarose gel, and transferred to a GenePlus membrane. The membrane was probed with 32P-labeled fragments amplified by PCR in the presence of 32P-dCTP with the following primers: Exon 4 probe, sense primer, 5′-GCAGAAACTGCTCCATTGCCCTA-3′, and antisense primer, 5′-CAGGAAACCAGAGCCTCCACAA-3′; exon 11 probe, sense primer, 5′-GTCCTTGAGAATGGAGAGGATCAGC-3′, and antisense primer, 5′-CTCTTCCCCTCTTGATTTCCATCT-3′; exon 2/3 probe, sense primer, 5′-GCCTTAGCCACTAGCACGCTGCCC-3′, and antisense primer, 5′-GTGGATGGGGGTCCAGCAGACAA-3′. The Northern blots were repeated twice with similar results.
Regional Expression of the Variant mRNA.
Total RNA was isolated from different mouse brain regions as described and reverse-transcribed by a SuperScript II Reverse Transcriptase (GIBCO) with either antisense E4A or antisense E9A. The first-strand cDNAs were then used as templates in the first-round PCRs with the sense primer 11A and the antisense primer E4A or E9A. Because no visible bands on agarose gel were observed, second-round PCRs were performed by using the first-round PCRs as templates with the sense primer E11B and antisense primer E4B for MOR-1G, MOR-1H, MOR-1J, and MOR-1K, or E9B for MOR-1M and MOR-1N. The sense primer E11B was paired with an antisense E1A (5′-GTGGAACCAGAGAAGAGCGGCAGG-3′) or an antisense E14A (5′-CTTTGTGGTGGTCGACCAGTTGCC-3′) in the second-round PCRs to amplify MOR-1I and MOR-1L, respectively. A sense primer E1A designed from the exon 1b (5′-GGACGCTCAGACGTTCCATTCTGC-3′) and antisense E4A were used to amplify MOR-1 with the same templates. The agarose gel was stained with ethidium bromide and photographed with the Kodak DC120 Digital Camera/Imaging System. The predicted sizes of the PCR products for MOR-1G, MOR-1H, MOR-1I, MOR-1J, MOR-1K, MOR-1L, MOR-1M, and MOR-1N are 1,025 bp, 1,440 bp, 269 bp, 1,569 bp, 1,174 bp, 206 bp, 1,171 bp, and 1,082 bp, respectively. Each band was extracted from agarose gel, subcloned into pCRII-TOPOII vector, and confirmed by sequencing. RNA loading was estimated by a parallel PCR with a pair of β2-microglobulin (β2MG) primers (CLONTECH).
Western Blots.
Western blots were generated from mouse whole brain as reported (20). In brief, whole mouse brains were homogenized in Tris buffer with a mixture of proteinase inhibitors (Aprotinin, 2 μM; Leupeptin, 2 μM; Pepstatin, 2 μM; Bestatin, 2 μM; and PMSF, 0.5 mM). The homogenate was solubilized with loading buffer and 20 μg of the total brain protein was loaded and run on a 4–20% gradient SDS/polyacrylamide gel, and transferred to a poly(vinylidene difluoride) (PVDF) membrane. The membrane was blocked for 1 h in TTBS buffer (10 mM Tris⋅HCl, pH 7.4/150 mM NaCl/0.05% Tween 20, containing 5% nonfat dried milk), and incubated at room temperature for 1 h with the Exon 11 antibody (1:2,600 dilution; C. Abbadie, Y.-X.P., and G.W.P., unpublished data), which had been adsorbed with an acetone extract of Chinese hamster ovary (CHO) cell membranes. The same antibody also was preincubated at room temperature for 10 min with 10 μg of the peptide used to generate the antiserum as a control (preabsorbed). After washing with TTBS buffer, the membrane was incubated with peroxidase-conjugated goat anti-rabbit antibody (1:15,000 dilution, Vector Laboratories) in TTBS buffer at room temperature for 1 h. After washing with TTBS buffer, the signals were detected by using Renaissance chemiluminescence reagents (NEN).
Expression and Characterization of MOR-1H, MOR-1I, and MOR-1J in CHO Cells.
The variants MOR-1H, MOR-1I, and MOR-1J were subcloned into pcDNA3.1 (Invitrogen) and the resulting plasmids used to transfect CHO cells by LipofectAMINE reagent (GIBCO). Stable transformants were obtained 2 weeks after selection with G418. Either stable or transient transfections were screened in a [3H]DAMGO [D-Ala2,N-MePhe4,Gly5-ol]enkephalin binding assay, as described (15).
Results
Cloning a Novel Exon of MOR-1 Gene and Its Associated Splice Variants.
Using a modified 5′ RACE strategy, we isolated a previously uncharacterized exon (exon 11) sequence that was directly spliced to exon 2. To obtain full-length clones, we then did reverse transcription (RT)-PCR with sense primers designed from the exon 11 and antisense primers from exons 4 or 9, because all known MOR-1 variants contain either exon 4 or 9, but not both. The PCR with the exon 4 primer yielded six variants, and the exon 9 primer produced another two (Fig. 1A).
Several of these variants contained the 3′ end of exon 1, including the translation start point and the complete coding region. MOR-1H contained the 3′-terminal 415-bp fragment of exon 1 (exon 1a) inserted between exons 11 and 2, whereas MOR-1I contained the 3′-terminal 589-bp, comprised of exon 1a and an additional 174 bp immediately upstream (exon 1b). MOR-1J had a previously uncharacterized 129-bp insertion (exon 12) between exon 11 and exon 1a. The 5′ splice acceptor sites of exons 1a and 1b were located at 131 bp and 305 bp upstream, respectively, of the ATG translation start codon of MOR-1 (Fig. 1B). MOR-1K and MOR-1L had a 149-bp (exon 13) and a 108-bp (exon 14) insertion between exons 11 and 2, respectively.
Sequence analysis of exon 11 arbitrarily using the first of the two predicted methionine residues as a potential translation start codon predicted 27 aa (Fig. 1C; Table 1). In MOR-1G, the start codon in the exon 11 was in the same reading frame as exons 2–4 of MOR-1, resulting in a new variant in which the 96 aa encoded by exon 1 in MOR-1 were replaced by the 27 aa of exon 11. A similar relationship existed between MOR-1M (exons 11/2/3/7/8/9) and MOR-1C, and between MOR-1N (11/2/3/8/9) and MOR-1D. Unlike exon 1, which has a predicted transmembrane region, analysis of the 27 aa encoded by exon 11 did not predict a transmembrane region, implying that MOR-1G, MOR-1M, and MOR-1N contained only six transmembrane regions.
Table 1.
Summary of the structures of the protein products of the MOR-1 splice variants
| Clone | Predicted peptide products
|
Comments | ||
|---|---|---|---|---|
| aa/MW* | Exon start/stop sites† | TM‡ | ||
| MOR-1G | 330/38.0 | 11/4 | 6 | Amino terminus contains the 27 amino acids encoded by exon 11, followed by a sequence identical to the MOR-1 protein coded by exon 2 to end |
| MOR-1H | 84/9.6 | 11/1 | 0 | Amino terminus contains the 27 amino acids encoded by exon 11, but the remainder are unique |
| 398/44.4 | 1/4 | 7 | Identical to the MOR-1 protein | |
| MOR-1I | 48/5.5 | 11/1 | 0 | Amino terminus contains the 27 amino acids encoded by exon 11, but the remainder are unique |
| 398/44.4 | 1/4 | 7 | Identical to the MOR-1 protein | |
| MOR-1J | 35/3.9 | 11/12 | 0 | Amino terminus contains the 27 amino acids encoded by exon 11, but the remainder are unique |
| 398/44.4 | 1/4 | 7 | Identical to the MOR-1 protein | |
| MOR-1K | 39/4.6 | 11/13 | 0 | Amino terminus contains the 27 amino acids encoded by exon 11, but the remainder are unique |
| MOR-1L | 41/4.9 | 11/14 | 0 | Amino terminus contains the 27 amino acids encoded by exon 11, but the remainder are unique |
| MOR-1M | 370/42.3 | 11/9 | 6 | Amino terminus contains the 27 amino acids encoded by exon 11, followed by a sequence identical to the MOR-1C protein coded by exon 2 to end |
| MOR-1N | 325/37.5 | 11/8 | 6 | Amino terminus contains the 27 amino acids encoded by exon 11, followed by a sequence identical to the MOR-1D protein coded by exon 2 to end |
Predicted amino acid sequence (aa) and molecular weight (MW, in kDa).
Exon containing the translation initiation site and the exon containing the stop codon.
Predicted transmembrane domains.
MOR-1H, MOR-1I, and MOR-1J had two potential start codons, one in exon 11 and one in exon 1. The start codon from exon 1 predicted a protein identical to that encoded by MOR-1, although transcription is under the control of a different promoter. The start codon in exon 11 resulted in a frame shift that predicted short peptides of 84 aa for MOR-1H, 48 aa for MOR-1I, or 35 aa for MOR-1J because of early termination of translation within exons 1a, 1b, and 12, respectively (Fig. 1 B and C). Similarly, the exon 11 start codon predicted truncated peptides of 39 aa for MOR-1K and 41 aa for MOR-1L (Fig. 1 B and C). None of these short peptides had an identifiable transmembrane domain.
Isolation and Analysis of Genomic Clones.
Exon 11 was not present in the genomic clones previously isolated by our group (14, 15). Using PCR with exon 1 primers, we identified bacterial artificial chromosome (BAC) clone C (≈70 kb) from a mouse genomic BAC library (GenomeSystems, St. Louis). Analyses of the BAC clone C by Southern blotting, long PCR, and sequencing revealed overlap between BAC clones C and A. BAC clone C contained exons 11, 12, 1, and 13, but not exons further downstream (Fig. 1). Using these BAC clones, exons 11 and 12 mapped ≈10 kb and 8 kb upstream, respectively, of exon 1, whereas exons 13 and 14 were located ≈1 kb and 22 kb downstream of exon 1, respectively.
Partial sequences of BAC clones C and A indicated that gene sequences of the exons were identical to those identified in the original cDNAs and that all of the exons had flanking sequences consistent with consensus splice junctions (Fig. 1B). The splice sites of exon 1 were also in agreement with the GT/AG rule.
Expression of mRNA Transcripts of MOR-1 Variants in Mouse Brain.
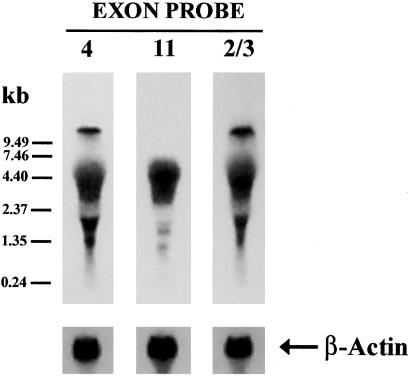
Northern blot analysis established the relative size and abundance of the new mRNA's in mouse brain (Fig. 2). Using a probe based on exon 4 or exons 2/3, we saw a well defined band corresponding to the previously reported 10.5-kb band seen with MOR-1 (8), as well as a diffuse band from 4–7 kb and several smaller bands from 1.5–2.5 kb, similar in size to those reported earlier (9). The exon 11 probe did not label the band corresponding to MOR-1 itself (10.5 kb), but did label the smaller bands.
Figure 2.
Northern blots of MOR-1 variants. Northern blot was performed with total RNA from mouse brain by using a 121-bp exon 4 probe, a 141-bp exon 11 probe, and a 546-bp exon 2/3 probe generated by PCR with appropriate primers, as described in Materials and Methods. Briefly, ≈100 μg per lane of total RNAs were loaded and separated on the same 0.8% formaldehyde agarose gel, and transferred to a GenePlus membrane. The membranes hybridized with the exon-specific probes were exposed to the Kodak BioMax MS film for 24 h. The membranes stripped off the probes were rehybridized with a 32P-labeled β-actin probe amplified by PCR with an amplimer set (CLONTECH) to estimate RNA loading.
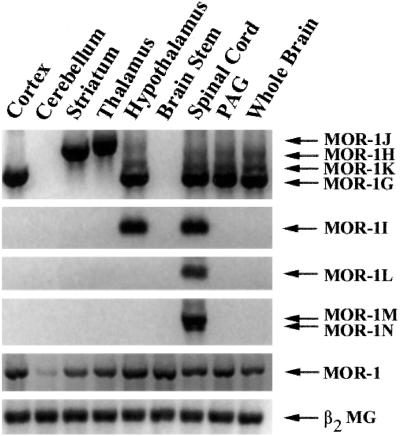
We next examined the regional distribution of mRNA expression by using RT-PCR (Fig. 3). The MOR-1 transcript driven by the original exon 1 promoter was observed after the first-round PCR and was expressed in all regions with relatively equal abundance, except for lower levels in the cerebellum. The expression of all of the other variants derived from the exon 11 promoter were detected only in the second round of PCR, implying lower levels within brain. MOR-1G was highly expressed in the cortex, hypothalamus, periaqueductal gray (PAG), and spinal cord, but not in the other regions. In contrast, MOR-1H was heavily expressed in the striatum and thalamus. MOR-1J expression was highest in the thalamus, whereas the expression of MOR-1I was highest in the hypothalamus and spinal cord. Low levels of MOR-1K were seen in hypothalamus, PAG, and spinal cord. In these studies, we detected MOR-1L, MOR-1M, and MOR-1N only in the spinal cord.
Figure 3.
Regional RT-PCR of the exon 11-containing variants. The regional distributions of the indicated MOR-1 variants were examined by RT-PCR on the indicated brain regions, as described in Materials and Methods.
When using the translational start site in exon 1, MOR-1H, MOR-1I, and MOR-1J all generate the same protein as MOR-1 itself. To compare the regional distribution of these variants compared with MOR-1 at the mRNA level, we used a relative quantitative RT-PCR approach (Table 2). The expression of the variants varied significantly (P < 0.005), both among regions (P < 0.002) and among variants (P < 0.02). Overall, expression of each of the variants within the whole brain was less than 10% that of MOR-1. However, the relative expression of some of the variants was far greater within specific regions, such as MOR-1H in the striatum (19%) and MOR-1J in the thalamus (16%). Although all of the variants used the same promoter for exon 11, their relative expression within regions varied markedly. For example, within the striatum the levels of MOR-1H mRNA were over 3-fold greater than those of MOR-1J, but only half in the thalamus. Thus, the processing of these variants is region dependent.
Table 2.
Expression of MOR-1H, MOR-1I, and MOR-1J in selected brain regions by relative quantitative RT-PCR
| Receptor | Relative abundance of splice variant, percent of MOR-1
|
||||
|---|---|---|---|---|---|
| Cortex | Striatum | Thalamus | Hypothalamus | Whole brain | |
| MOR-1H | 0 | 19.3 ± 1.8 | 7.6 ± 5.3 | 4.8 ± 2.6 | 9.0 ± 2.6 |
| MOR-1I | 0 | 1.2 ± 1.2 | 2.3 ± 1.5 | 6.4 ± 3.7 | 2.4 ± 2.6 |
| MOR-1J | 0 | 6.3 ± 2.1 | 16.4 ± 0.9 | 8.0 ± 1.4 | 7.7 ± 0.5 |
Relative quantitative RT-PCR was used to compare the variants in different brain regions. Total RNA was reverse-transcribed and the first-strand cDNA amplified by PCR either with a pair of primers from exons 11 and 1 (the sense primer from exon 11, 5′-CCAAAAGCTCAGACAGAGAGATGGAAATCAAGAGGGGAA GAG-3′, and the antisense primer from exon 1, 5′-CCAGGAAGTTTCCAAAGAGGCCCACTACACACACGATAGAATAGAGGG-3′) for amplifying MOR-1H, MOR-1I, and MOR-1J, or a pair of primers from exons 1 and 2 (the sense primer from exon 1, 5′-GGACGCTCAGACGTTCCATTCTGC-3′, and the antisense primer from exon 2, 5′-GGGCAGCGTGCTAGTGGCTAAGGC-3′) for amplifying MOR-1 in the presence of a β-actin internal standard (Ambion, Austin, TX) to control for RNA loading. The PCR products were quantified with a digital imaging system after separation and ethidium bromide staining. Relative percentages of expression of MOR-1H, MOR-1I, and MOR-1J versus MOR-1 were normalized by β-actin expression. Results are the means ± SEM of three separate determinations. The results were analyzed by two-way ANOVA (P < 0.005), which also showed differences among regions (P < 0.002) and variants (P < 0.02).
Protein Expression of the MOR-1 Variants.
In vitro translation studies (data not shown) supported the predicted protein products. In vitro translation of MOR-1G, MOR-1N, and MOR-1M all produced single bands in SDS/PAGE consistent with the predicted size for the six transmembrane proteins, whereas MOR-1K and MOR-1L yielded products between 4 and 5 kDa, as predicted. All these in vitro translated products were immunoprecipitated by an antiserum against an exon 11 epitope (C. Abbadie, Y.-X.P., and G.W.P., unpublished data). In vitro translation of MOR-1H, MOR-1I, and MOR-1J generated two proteins for each clone, a small protein predicted to contain the exon 11 epitope, and a larger one corresponding to MOR-1. The exon 11 antiserum immunoprecipitated the small proteins, but not the larger band corresponding in size to MOR-1. When expressed in CHO cells, these three variants displayed receptor binding characteristics indistinguishable from earlier studies of MOR-1 (Table 3).
Table 3.
Binding characteristics of MOR-1H, MOR-1I, and MOR-1J expressed in CHO cells
| Ligand | Dissociation constant, nM
|
|||
|---|---|---|---|---|
| MOR-1 | MOR-1H | MOR-1I | MOR-1J | |
| [3H]DAMGO (KD) | 6.4 ± 1.6 | 2.9 ± 1.3 | 4.3 ± 0.5 | 4.9 ± 0.7 |
| Competitor (Ki) | ||||
| DAMGO | 1.8 ± 0.5* | 6.2 ± 1.0 | 4.5 ± 0.1 | 3.9 ± 0.5 |
| Morphine | 5.3 ± 2.0* | 6.3 ± 2.4 | 5.9 ± 0.3 | 5.7 ± 0.4 |
| U50,488H | >500 | >500 | >500 | >500 |
| DPDPE | >500 | >500 | >500 | >500 |
KD values for [3H]DAMGO [d-Ala2,N-MePhe4,Gly5-ol]enkephalin were determined from saturation studies and were used to determine Ki values that were calculated from IC50 values, as described (31, 32). ANOVA revealed no significant differences among the KD values of all four variants for [3H]DAMGO (P = 0.39). Comparison of competition values for MOR-1H, MOR-1I, and MOR-1J with ANOVA revealed no significant differences for DAMGO (P = 0.22) or morphine (P = 0.96). The Ki values for morphine and DAMGO against MOR-1 are from the literature (15).
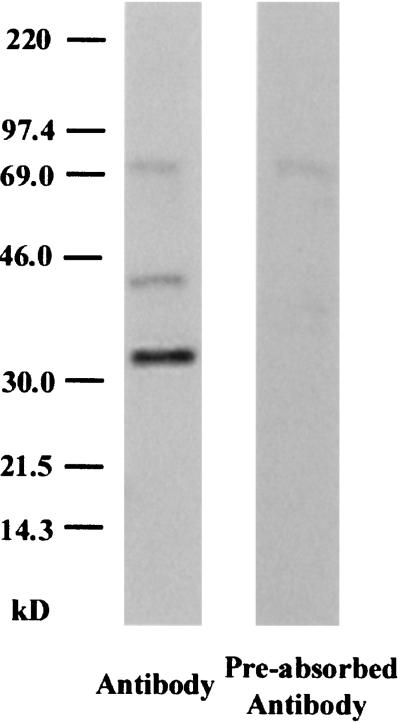
Western blots of brain by using an exon 11-selective antiserum revealed three bands (Fig. 4 Left). The smallest band was most intense and corresponded to the predicted size of MOR-1G (38 kDa) and/or MOR-1N (37.5 kDa). In some blots, the lower band appeared to be a doublet, raising the possibility that both variants were present. The middle band was less intense and corresponded to the predicted size of MOR-1M (42.3 kDa). Both bands were lost when the antibody was preadsorbed with the epitope-containing peptide and were not seen with preimmune serum, confirming their specificity. These bands in brain were similar to those seen on Western blots from HEK 293 cells transfected with the variants (data not shown). The largest band (≈70 kDa) was still detectable by the preadsorbed antibody, raising questions about its specificity. The lower molecular weight products (<10 kDa) seen with in vitro translation were not seen in these blots from brain tissue.
Figure 4.
Western blot of whole brain with an exon 11-specific antiserum. Western blots were performed as described in Materials and Methods. The lane on the Left used the antiserum, whereas the lane on the Right used antiserum that had been preadsorbed with the peptide used to generate the antibody and represents a control.
Discussion
Pharmacological studies have implied the possibility of multiple mu receptors (3, 4, 21, 22), a concept that was confirmed with the identification of seven splice variants of the mu opioid receptor gene MOR-1 (11, 12, 14, 15). The present study extends these earlier studies and reveals at least fourteen exons within the MOR-1 gene, Oprm, which spans 250 kb and generates fifteen splice variants. Unlike the earlier variants where the splicing involved the carboxy tip of the intracellular tail of the receptor with each conforming to the standard seven transmembrane domain structure of G-protein coupled receptors, the splicing of the eight new variants is located at the 5′ end of the mRNA and many of the variants are truncated. Equally important, the region upstream of exon 11 contains a new promoter responsible for the expression of these new variants (Y.-X.P., unpublished data). Thus, the mu receptor gene Oprm has two physically distinct promoters, one for exon 1 and one for exon 11. Unlike the protomor for exon 1, the exon 11 promoter has a TATA box as well as a different composition of potential cis regulatory sequences (Y.-X.P., unpublished data).
What makes this splicing intriguing is that three of the variants under the control of the exon 11 promoter generate the identical protein as MOR-1 itself, which is under the control of the exon 1 promoter. Despite yielding identical proteins to MOR-1, the regional distributions of MOR-1H, MOR-1I, and MOR-1J mRNA, as revealed by RT-PCR, differed from MOR-1 and from each other. Compared with the expression of MOR-1, the overall levels of the variants in the brain was under 10% those of MOR-1. However, within specific brain regions the abundance of some variants was far greater. More important, the relative abundance of the variant mRNAs differed markedly. The level of MOR-1H mRNA was 3-fold higher than MOR-1J in the striatum, but only half as much in the thalamus. Thus, there is differential regional processing even among these variants, as well as compared with MOR-1, which is under the control of the exon 1 promoter. Because Western blots did not detect the predicted small proteins (<10 kDa) seen with in viro translation of these three variants, we assume that they generated only the MOR-1 protein.
The remaining eight proteins raise others issues. All eight proteins use a translation start point within exon 11, were generated by in vitro translation and were immunoprecipitated by the exon 11-selective antiserum. Three proteins have six predicted transmembrane domains, whereas the other five are far smaller and have none. Western blots from brain tissue revealed only the bands corresponding to the six transmembrane domain products. The most intense band corresponded to MOR-1G and/or MOR-1N. In some Western blots this band appeared to be doublet, consistent with the expression of both variants. The intensity of the band corresponding to MOR-1M was far less intense. Structurally, none correspond to traditional G-protein coupled receptors, raising questions regarding their physiological relevance, if any. Truncated proteins have been reported for all of the other members of the opioid receptor gene family, including DOR-1, KOR-1 (23), and the orphanin FQ/nociceptin receptor (ORL1/KOR-3; ref. 24). However, their relevance also is uncertain (25, 26).
Exon 11-immunoreactivity has a regional distribution within the brain that differs from that seen with the exon 4 antisera used to define the distribution of MOR-1 (27, 28), the exon 7/8/9 antiserum used to define MOR-1C (16), and the exon 8/9 antiserum used to define MOR-1D (18), as well as earlier autoradiographic studies (ref. 29; C. Abbadie, Y.-X.P., and G.W.P., unpublished data). Whereas the exon 7/8/9 antiserum did not label the striatum and the exon 4 antiserum revealed clusters, the exon 11 antiserum showed a diffuse labeling in the striatum and the nucleus accumbens, with even more intense labeling in the olfactory tubercle. The loss of labeling following antisense treatment targeting exon 11 confirmed its specificity. Within the brainstem, exon 11 immunoreactivity was most pronounced within the substantia nigra. These regional differences suggest that these truncated proteins may be functionally relevant, but this remains to be proven. When expressed alone in CHO cells, the six transmembrane domain proteins did not display reproducible and robust opioid binding, although low levels could occasionally be seen. Lacking only the first transmembrane domain, the exon 11-containing variants may require an association with other G-protein coupled receptors or even other truncated receptor proteins, such as the MOR-1 variant in which exon 1 is directly spliced to exon 4 (30). This variant generates a single transmembrane domain corresponding to the first transmembrane domain of MOR-1, the one missing in all three of the proteins with six predicted transmembrane domains. Although highly speculative, it will be interesting to test this possibility in the future.
In conclusion, the murine mu opioid receptor gene Oprm undergoes extensive alternative splicing at both the 3′ and 5′ termini. There are now 15 variants composed of combinations of 14 exons extending over 250 kbp. There also is a second promoter located almost 10 kb upstream of the established exon 1 promoter that is responsible for the eight new splice variants, three of which are capable of generating proteins identical to that of MOR-1. The expression of these variants opens many new questions that need to be addressed in the future, including the need for four variants to make a single protein and the functional relevance of the truncated proteins.
Acknowledgments
This work was supported, in part, by the National Institute on Drug Abuse through Research Grant DA02615 (to G.W.P.), Mentored Scientist Award DA00296 (to Y.-X.P.), and Senior Scientist Award DA00220 (to G.W.P.), as well as a grant from the National Genetics Foundation (to G.W.P.) and a core grant to Memorial Sloan–Kettering Cancer Center (CA08748) from the National Cancer Institute.
Abbreviations
- MOR
mu opioid receptor
- RT
reverse transcription
- CHO
Chinese hamster ovary
- BAC
bacteria artificial chromosome
Footnotes
Portions of this work were presented at the International Narcotics Conference in Seattle, WA (July 2000), and the Society for Neuroscience Meeting in New Orleans, LA (November 2000).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Payne R, Pasternak G W. In: Principles of Drug Therapy in Neurology. Johnston M V, Macdonald R L, Young A B, editors. Philadelphia: F. A. Davis; 1992. pp. 268–301. [Google Scholar]
- 2.Foley K M. Sci Am. 1996;275:164–165. doi: 10.1038/scientificamerican0996-164. [DOI] [PubMed] [Google Scholar]
- 3.Wolozin B L, Pasternak G W. Proc Natl Acad Sci USA. 1981;78:6181–6185. doi: 10.1073/pnas.78.10.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasternak G W. Clin Neuropharmacol. 1993;16:1–18. doi: 10.1097/00002826-199302000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Rossi G C, Pan Y-X, Brown G P, Pasternak G W. FEBS Lett. 1995;369:192–196. doi: 10.1016/0014-5793(95)00757-z. [DOI] [PubMed] [Google Scholar]
- 6.Schuller A G, King M A, Zhang J, Bolan E, Pan Y-X, Morgan D J, Chang A, Czick M E, Unterwald E M, Pasternak G W, et al. Nat Neurosci. 1999;2:151–156. doi: 10.1038/5706. [DOI] [PubMed] [Google Scholar]
- 7.Pasternak G W. Trends Pharmacol Sci. 2001;22:67–70. doi: 10.1016/s0165-6147(00)01616-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang J B, Imai Y, Eppler C M, Gregor P, Spivak C E, Uhl G R. Proc Natl Acad Sci USA. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson R C, Mansour A, Akil H, Watson S J. Neuron. 1993;11:903–913. doi: 10.1016/0896-6273(93)90120-g. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Mestek A, Liu J, Hurley J A, Yu L. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- 11.Bare L A, Mansson E, Yang D. FEBS Lett. 1994;354:213–216. doi: 10.1016/0014-5793(94)01129-x. [DOI] [PubMed] [Google Scholar]
- 12.Zimprich A, Simon T, Hollt V. FEBS Lett. 1995;359:142–146. doi: 10.1016/0014-5793(95)00028-8. [DOI] [PubMed] [Google Scholar]
- 13.Schulz S, Schreff M, Koch T, Zimprich A, Gramsch C, Elde R, Höllt V. Neuroscience. 1998;82:613–622. doi: 10.1016/s0306-4522(97)00288-1. [DOI] [PubMed] [Google Scholar]
- 14.Pan Y-X, Xu J, Bolan E A, Chang A, Mahurter L, Rossi G C, Pasternak G W. FEBS Lett. 2000;466:337–340. doi: 10.1016/s0014-5793(00)01095-4. [DOI] [PubMed] [Google Scholar]
- 15.Pan Y-X, Xu J, Bolan E A, Abbadie C, Chang A, Zuckerman A, Rossi G C, Pasternak G W. Mol Pharmacol. 1999;56:396–403. doi: 10.1124/mol.56.2.396. [DOI] [PubMed] [Google Scholar]
- 16.Abbadie C, Pan Y-X, Pasternak G W. J Comp Neurol. 2000;419:244–256. doi: 10.1002/(sici)1096-9861(20000403)419:2<244::aid-cne8>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 17.Abbadie C, Gultekin S H, Pasternak G W. NeuroReport. 2000;11:1953–1957. doi: 10.1097/00001756-200006260-00029. [DOI] [PubMed] [Google Scholar]
- 18.Abbadie C, Pan Y-X, Drake C T, Pasternak G W. Neuroscience. 2000;100:141–153. doi: 10.1016/s0306-4522(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 19.Abbadie C, Pasternak G W. NeurorePort. 2001;12:3069–3072. doi: 10.1097/00001756-200110080-00017. [DOI] [PubMed] [Google Scholar]
- 20.Pan Y-X, Cheng J, Xu J, Rossi G C, Jacobson E, Ryan-Moro J, Brooks A I, Dean G E, Standifer K M, Pasternak G W. Mol Pharmacol. 1995;47:1180–1188. [PubMed] [Google Scholar]
- 21.Pasternak G W, Childers S R, Snyder S H. Science. 1980;208:514–516. doi: 10.1126/science.6245448. [DOI] [PubMed] [Google Scholar]
- 22.Pasternak G W, Childers S R, Snyder S H. J Pharmacol Exp Ther. 1980;214:455–462. [PubMed] [Google Scholar]
- 23.Gavériaux-Ruff C, Peluso J, Befort K, Simonin F, Zilliox C, Kieffer B L. Mol Brain Res. 1997;48:298–304. doi: 10.1016/s0169-328x(97)00109-5. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y-X, Xu J, Pasternak G W. Gene. 1996;171:255–260. doi: 10.1016/0378-1119(95)00890-x. [DOI] [PubMed] [Google Scholar]
- 25.Xie G X, Meuser T, Pietruck C, Sharma M, Palmer P P. Life Sci. 1999;64:2029–2037. doi: 10.1016/s0024-3205(99)00150-2. [DOI] [PubMed] [Google Scholar]
- 26.Wang J B, Johnson P S, Imai Y, Persico A M, Ozenberger B A, Eppler C M, Uhl G R. FEBS Lett. 1994;348:75–79. doi: 10.1016/0014-5793(94)00557-5. [DOI] [PubMed] [Google Scholar]
- 27.Arvidsson U, Riedl M, Chakrabarti S, Lee J-H, Nakano A H, Dado R J, Loh H H, Law P-Y, Wessendorf M W, Elde R. J Neurosci. 1995;15:3328–3341. doi: 10.1523/JNEUROSCI.15-05-03328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansour A, Fox C A, Burke S, Akil H, Watson S J. J Chem Neuroanat. 1995;8:283–305. doi: 10.1016/0891-0618(95)00055-c. [DOI] [PubMed] [Google Scholar]
- 29.Atweh S F, Kuhar M J. Brain Res. 1977;124:53–67. doi: 10.1016/0006-8993(77)90863-0. [DOI] [PubMed] [Google Scholar]
- 30.Du Y-L, Elliot K, Pan Y-X, Pasternak G W, Inturrisi C E. Soc. Neurosci. 1997. 23. [Google Scholar]
- 31.Cheng Y-C, Prusoff W H. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 32.Chou T-C. Mol Pharmacol. 1974;10:235–247. [PubMed] [Google Scholar]