Abstract
Use of human pluripotent stem cells (hPSCs) and their differentiated derivatives have led to recent proof-of-principle drug discoveries, defining a pathway to the implementation of hPSC-based drug discovery (hPDD). Current hPDD strategies, however, have inevitable conceptual biases and technological limitations, including the dimensionality of cell-culture methods, cell maturity and functionality, experimental variability, and data reproducibility. In this review, we dissect representative hPDD systems via analysis of hPSC-based 2D-monolayers, 3D culture, and organoids. We discuss mechanisms of drug discovery and drug repurposing, and roles of membrane drug transporters in tissue maturation and hPDD using the example of drugs that target various mutations of CFTR, the cystic fibrosis transmembrane conductance regulator gene, in patients with cystic fibrosis.
Keywords: Pluripotent stem cells, differentiation, organoids, drug discovery, cystic fibrosis, ATP-binding Cassette, CFTR
Human Pluripotent Stem Cells and Drug Discovery Endeavors
Human pluripotent stem cells (hPSCs) are cells that possess the capacity to self-renew and to differentiate into all cell types of the adult body, which collectively include human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) [1–3]. The concept of hPSC-based drug discovery (hPDD) emerged soon after the first successful culture of hESC lines in vitro in 1998 [3]. Encouragingly, this hPDD strategy has been further emphasized and actively implemented since the establishment of iPSCs by genetically reprogramming adult mouse fibroblasts (in 2006) and human somatic cells (in 2007) to an embryonic stem cell-like state [1, 2, 4] (Figure 1).
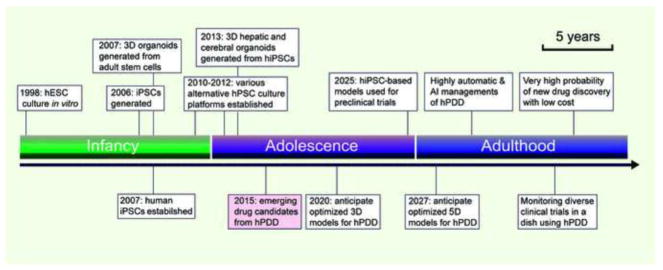
Figure 1.
Schema of drug developmental timeline, projected phases, and their corresponding or expected achievements through hPSC-based drug discovery (hPDD): (A) Stage of infancy (2000–2011): characterizing hPSC properties; improving hPSC growth, differentiation, cell maturation conditions; applying stem-cell niche concept for coculture, iPSC-disease modeling; and premature drug screening; (B) Stage of adolescence (2012–2025): Optimizing stem-cell niche signaling in vitro to guide multidimensional (e.g., 3D to 4D) organoid cultures; validating downstream functional assays; anticipating frequent reports of successful drug discovery and repurposing based on hPDD. (C) Stage of adulthood (2025 onwards): anticipation of optimized 5D-organoid models for hPDD, highly automatic and artificial intelligence (AI) managements; fast tracking preclinical trials in humanized models; monitoring diverse clinical trials (Phases I/II/III) in the laboratory with hPDD becoming a routine practice, and significantly enhancing probability of any new drug discovery or drug repurposing at a reasonable cost.
Although hPDD was widely used for drug discovery between the years 2000 and 2011, substantial experimental variability and reproducibility were encountered, hindering its application. Notable hPDD challenges have been: (i) choice of appropriate hPSC cultures and differentiation platforms due to a lack of understanding of the advantages and limitations of these platforms in hPDD (Figures 2 and 3) [5, 6], (2) heterogeneity and genomic instability arising from hPSC culture and differentiation [7–10], (3) that disease models established from hPSCs do not fully recapitulate diseases [11–13], and (4) problems with drug-screening designs (e.g., molecular target-based drug screens, a cutting-edge approach, have actually led to fewer approved drugs than original phenotypic screens in a recent evaluation [14]). As a result, no significant drug discoveries were made through hPDD in that period (Figure 1, Supplemental Table 1). Since 2012, understanding of hPSCs has increased, allowing efficient hPSC expansion, maintenance, and differentiation [5]. We predict that hPSC-derived systems will be routinely used to accurately model diseases and for effective evaluation of drug efficacy (Figure 1).
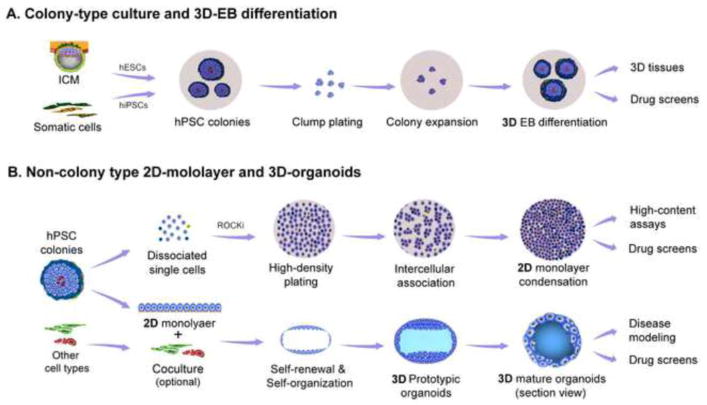
Figure 2.
Schema of 2D- and 3D-dimenisonal culture platforms. (A) Flow chart of hPSC colony-type culture and its downstream applications: hESCs [generated from the inner cell mass (ICM)] and hiPSCs (established by reprogramming somatic cells to pluripotent stem cells) are collectively named hPSCs. hPSCs conventionally grow as colonies on plastic dishes that are coated with extracellular metrices, form embryoid bodies (EBs) in suspension culture, and differentiate into 3D-tissues for downstream applications. (B) Flow chart of 2D-monolayer and 3D-organoid culture: (top panel) Heterogeneous hPSC colonies (top view of one representative colony in a culture dish), showing different cellular states in the periphery and center of the colony, can be homogenized as a 2D-monolayer for high-content imaging and high-throughput drug screening (HTS). (lower panel) 2D-monolayer differentiation, optionally cocultured with different cells (e, g., human umbilical vein endothelial cells) in the presence of proper extracellular matrices, can self-organize into 3D-organoids for disease modeling and drug screening.
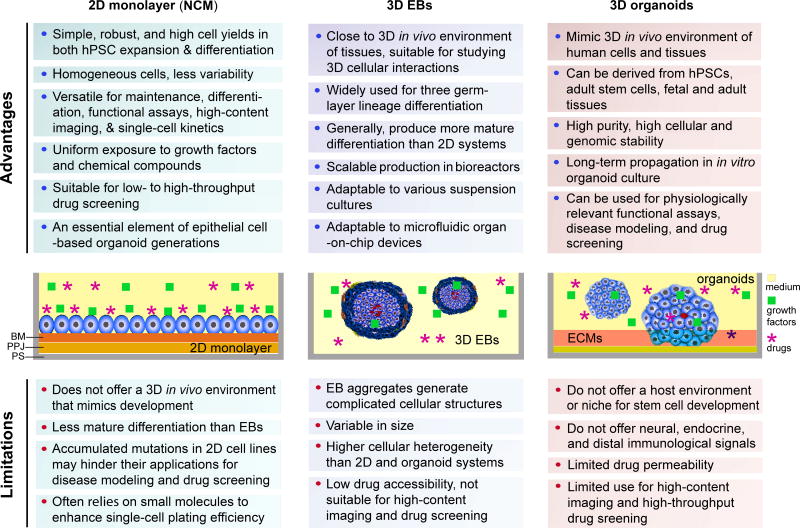
Figure 3.
Summary of advantages and limitations in 2D- and 3D-culture platforms in hPDD. The graphs depict a sectional view of cells and 2D- and 3D-culture environments supplemented with cell culture medium, growth factors, extracellular matrices, and drugs. Abbreviations: BM, basement membrane; D, dimension(al); EB, embryoid body; ECMs, extracellular metrices; NCM, non-colony type monolayer; PPJ, plastic-protein junction; PS, polystyrene surface of the cell culture dishes for hPSCs.
In this review, we do not intend to provide a comprehensive review of hPDD, as other similar reviews have already been published (Supplemental Table 1). Instead, we will discuss the heterogeneity and reproducibility of hPSCs and their differentiation platforms related to hPDD; roles of morphogenesis, organogenesis, and tissue integration; maturity and functionality of differentiated cells in multi-dimensional (e.g., 3D or 4D) cultures; and key factors affecting hPDD (e.g., the roles of membrane transporters in drug discovery). Lastly, we use the example of hPDD for cystic fibrosis (CF) to provide the molecular mechanism and resonate for the utility of hPDD in drug development.
Heterogeneity and Reproducibility of hPDD Studies
Role of 2D-monolayer Culture in Controlling hPSC Heterogeneity
Current technologies used for hPDD depend on various hPSC culture and differentiation platforms (e.g., colonies, 2D-monolayers, suspension, 3D-culture-based organoids) [5]. Experimental results obtained from in vitro hPSC culture and differentiation platforms are highly variable due to the existence of several pluripotent states (e.g., naive and primed) [15–17], and the use of heterogeneously differentiated and functionally immature cells. Thus, heterogeneity both in pluripotency and differentiation is one of the leading causes of experimental variability, reproducibility, and reliability. In fact, many uncontrollable variables in hPSC culture systems stem from an insufficient understanding of optimal growth conditions to support physiologically relevant hPSCs and differentiated cells in vitro. In general, hPSC-based models can be classified into four major categories: colonies, 2D-monolayers, suspension, and 3D-culture-based organoids. However, we will focus on 2D-monolayers, including single-cell based non-colony type monolayer (NCM) culture, and 3D-organoids used in hPDD (Figures 2–5).
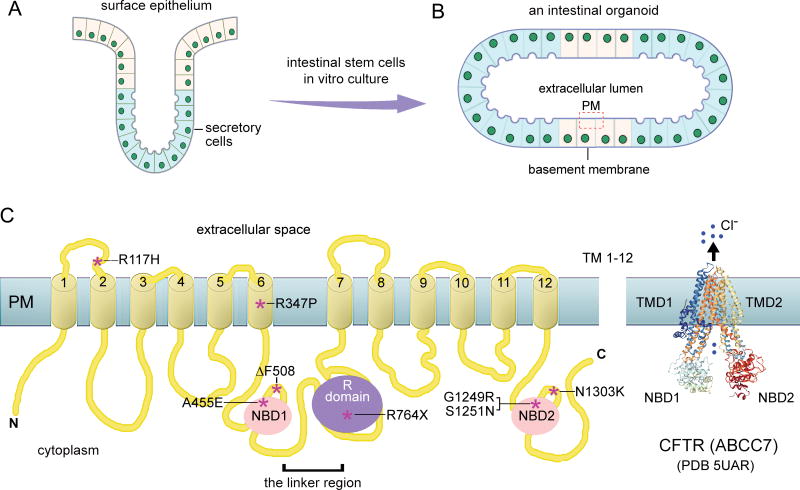
Figure 5.
ABC transporters as molecular targets of organoids derived from intestinal stem cells and hPSCs. (A) Cellular structure of adult intestinal cells, (B) an anatomical structure of cultured intestinal cell organoids and ABC transporter expression on the plasma membrane of the lumen; (C) 2D- and 3D- presentation of CFTR (ABCC7, a cystic fibrosis-associated chloride channel protein). The locations of CFTR mutations of interest, which are associated with cystic fibrosis, are indicated by asterisks.
Abbreviations: C, the carboxyl-terminus; N, the amino-terminus; NBD, nucleotide binding domain; PDB 5UAR: Protein Data Bank, dephosphorylated, ATP-free cystic fibrosis transmembrane conductance regulator (CFTR) from zebrafish; PM, plasma membrane; TM, transmembrane segment; TMD, transmembrane domain.
NCM culture was developed for homogeneous hPSC expansion, maintenance, and differentiation, aiming to minimize cellular heterogeneity and experimental variability [18]. It is based on dissociated single cells grown on suitable extracellular matrices (e.g., Matrigel) in the presence of Rho-associated protein kinase inhibitor (ROCKi) or Janus kinase 1 inhibitor (JAKi) (Figure 2B) [18–20]. The addition of these inhibitors enhances the initial 24-hour single-cell plating efficiency, thereby greatly reducing the heterogeneity of the culture, increasing hPSC expansion, and improving recovery rates after cryopreservation (Figure 3) [18, 21]. Hence, NCM culture significantly increases the reproducibility and efficiency of hPDD studies compared to colony-type culture and embryoid body (EB) suspension culture (Figures 2 and 3).
Role of 2D-monolayer Culture in Facilitating hPDD
Thus far, 2D-monolayer-based methods, including NCM, have been widely used for hPDD in various cellular and genetic models, including neuroectodermal disease [22–26], mesoderm-derived cardiomyocytes and cardiomyopathy [27–30], and several hepatic pathological conditions [31–33]. To better understand the role of 2D-monolayer culture in hPDD, here, we highlight a few representative cell types derived from three germ-layer lineages.
An early high-throughput drug screening (HTS) using hiPSC-derived neural crest cells was conducted in 2012 [22]. In their study, Studer, Lee, and colleagues screened 6812 compounds and achieved a target hit rate of 0.4%, which led to the identifications of 8 compounds that could rescue expression of IKBKAP, the gene that is responsible for familial dysautonomia [22]. Of particular interest, one small molecule (SKF-86466) was able to induce IKBKAP transcription through a molecular signaling pathway that involves modulation of intracellular cAMP levels and phosphorylation of the cAMP response element binding protein (CREB) [22]. Thus, this study highlights the utility of an hiPSC-based 2D-monolayer model in identifying potential drug candidates and revealing molecular mechanisms for the therapy of neuronal diseases.
The choice of hPSC culture platforms for mesoderm-derived cardiac tissues for hPDD is biased towards 3D systems. It is believed that conventional 3D-EB derived cardiomyocytes or cardiac tissues are better choices for hPDD than those of 2D-monolayers in terms of their cell maturity and electrophysiological and functional suitability [34–36]. Nevertheless, Lacone and coworkers implemented 2D-monolayer-based high-content phenotypic drug screening for diabetic cardiomyopathy using patient-specific hiPSC-derived cardiomyocytes. They screened 480 compounds in a chemical library, examined their effects on subcellular structure and sarcomeric integrity of cardiomyocytes, and attained a target hit rate of 5.8%, which produced at least 4 compounds that could reverse the diabetic cardiomyopathy phenotype [30]. This work empowers conventional phenotypic assays with a 2D-monolayer platform for therapeutic tactics to treat complicated metabolic diseases.
Moreover, 2D-monolayer culture, especially NCM, is particularly suitable for conducting low-throughput screening and up to HTS in endoderm-based epithelial cell models, since an epithelial monolayer is the essential element of many endodermal tissues. In addition, current hepatic differentiation protocols based on hPSC colony-type culture and 3D-EB differentiation do not easily produce homogeneous and fully mature hepatocytes in a small-well (e.g., 384-well) format, which hinders the use of HTS assays (which use small wells) for drug screening. In a recent study, Liang, Carpentier, and colleagues showed for the first time that 2D-NCM culture enables efficient and reproducible differentiation of functional hepatocyte-like cells in 384-well plates, with a coefficient of variation of albumin secretion in these wells below 15% [37]. Hence, this improved 2D-NCM protocol might facilitate compound screening to identify small molecules that regulate liver differentiation, metabolism, and response to hepatitis C virus infection [37]. Furthermore, 2D-NCM-based HTS was implemented to identify drugs that rescue alpha-1 antitrypsin deficiency, a liver disease without effective drugs or alternative therapies. Jang, Ye, and colleagues screened 3131 clinical compounds in an established chemical library and obtained a target hit rate of 8.3%, which led to 5 clinical drugs that could reduce mutant alpha-1 antitrypsin accumulation in hepatocyte-like cells derived from patient-specific hiPSCs [32].
Taken together, the above studies indicate the usefulness of 2D-monolayers in hPDD by offering proof-of-concept screens, confirming the activity of approved drugs in desired cells, demonstrating the predictive power of cytotoxicity of drugs, and identifying candidate drugs for therapeutics. Thus far, a limited number of clinical drugs obtained by 2D-monolayer systems are undergoing clinical trials [38]. One such drug, termed RG7800 [25] and intended to treat spinal muscular atrophy, has failed in a Phase I clinical trial (ClinicalTrials.gov Identifier: NCT02240355), due to “an unexpected toxicology finding observed in the 39-week monkey study.” Understanding the advantages and limitations of 2D-monolayer and conventional 3D-culture methods might be important to accelerate the success rate of clinical drug identification based on hPDD.
Limitations of 2D-monolayer Culture in hPDD and Prospective Resolutions
Compared with conventional 3D-culture systems (e.g., 3D-EBs), 2D-monolayer culture systems (including NCMs) have several major limitations that could potentially hamper their application in hPDD (Figure 3). These limitations include the absence of a 3D in vivo environment that mimics developmental and disease niches, less mature differentiated adult tissues (e.g., cardiac tissues) than 3D-culture-mediated differentiation [34, 35], accumulated chromosomal abnormalities and genomic mutations [reviewed in [6, 7, 39], and relying on small molecules such as ROCKi, which might interfere with the outcomes of drug screens, to enhance single-cell plating efficiency [18, 20, 21].
Nonetheless, the above mentioned major hurdles could be minimized or even circumvented. For example, it was shown that hESC culture at physiologic O2 (2%) reduces spontaneous chromosomal abnormalities without affecting hESC pluripotency, compared with hESC cultured under conventional normoxia (i.e., 21% O2) [40]. This study suggests that 2D-monolayer culture and hPDD should be carried out in physiological conditions (e.g., under hypoxic conditions). Regarding increasing cellular maturity, Herron et al. demonstrated a rapid maturation of hiPSC-derived cardiomyocytes, with a simple 2D-monolayer differentiation on polydimethylsiloxane (PDMS)-coated coverslips [41]. Recently, using a 2D-monolayer protocol based on the Wnt inhibitor, IWP2, Jeziorowska et al. obtained higher yields of hiPSC-derived cardiomyocytes as well as higher sarcomeric maturation than conventional 3D-EB-based methods [42]. Thus, these modifications are improving the development of functionally compatible adult tissues for hPDD. Finally, hPDD systems often use ROCKi and could be affected by its adverse effects, resulting from an unwanted pharmacological inhibition of ROCKs [reviewed in [43, 44]. Guided by this consideration, we have developed xeno-free and chemically defined conditions to avoid complicating interactions between ROCKi and drugs used for screening [45].
Other Roles of 2D-NCM: Genetic Engineering and Druggable RNAs and DNAs
The NCM-mediated platform facilitates genetic manipulations of hPSCs. hPSCs in conventional colony-type cultures (maintained in a primed pluripotent state) generate heterogeneous cellular states and have a very low DNA transfection or viral transduction efficiency. Hence, colony-type cultures are not suitable for genetic engineering by transfection with DNAs or RNAs via lipofectamine reagents or for transduction by viral particles [18, 45–47]. Conventionally, genetic analyses of hPSCs largely rely on isolating and characterizing individual stable clones, which takes several months. In contrast, NCM culture platforms considerably improve the efficiency of genetic engineering, permitting genetic manipulation of hPSCs and subsequent functional assays within 1 week [18, 48]. NCM-mediated platforms can be genetically manipulated using small hairpin RNAs (shRNAs), microRNAs, DNA oligonucleotides, plasmids, and lentiviral particles [18, 45, 48], potentially accelerating the discovery of druggable RNAs and DNAs.
In summary, 2D-monolayer systems, such as NCM culture, make it possible to reduce hPSC heterogeneity and experimental variability. These simple platforms have been widely used for high-content imaging and HTS despite their limitations. However, 2D-monolayer culture and differentiation also constitute the initial important steps necessary to generate 3D-organoids, based on epithelial layer formation (Figure 2B) [49, 50]. Precise integration of these 2D-monolayer units into a complex 3D-culture system (Figure 4), which can capitulate physiological morphogenesis and organogenesis (in a culture dish and a screen plate), would greatly expand avenues for hPDD.
Figure 4.
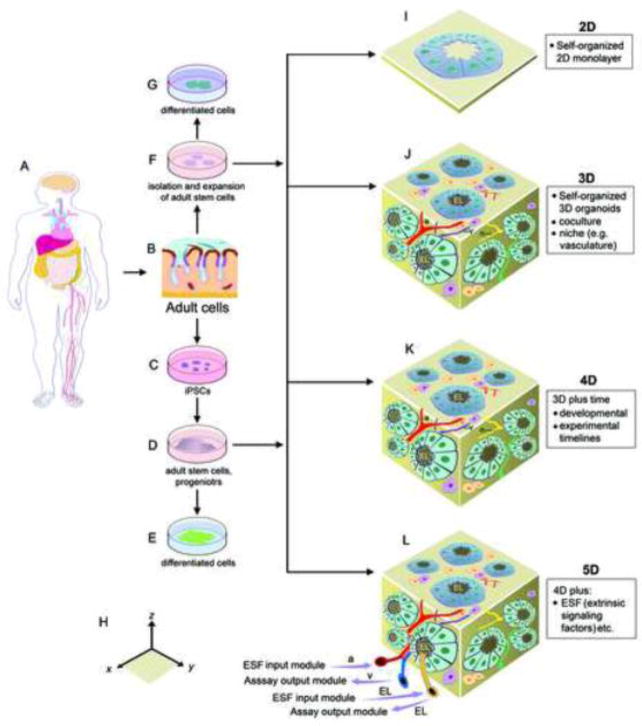
Schema of multi-dimensional culture, differentiation, and drug discoveries based on pluripotent and multipotent stem cell models: (A–E) Human somatic cells and adult stem cells (aSCs) as cell resources for reprogramming to generate iPSCs, followed by differentiation toward progenitors and functional mature cells; (A–G) Human adult cells or tissues as cell resources for isolation of progenitor and aSCs, followed by expansion and differentiation toward functional mature cells; (H) 3D coordinates based on x, y, and z axis. (I–L) 2D to the proposed 4D- and 5D-organoid cultures based on cell sources indicated (in C, D, and F). Of note, the dimension of time in 4D-organoids includes both the experimental (i.e., the time course of organoid culture under a specific condition) and the developmental (determined by intrinsic genetic and epigenetic programs) timelines of cells. 5D-organoids may be generated by introducing extrinsic signaling factors that occur in a human body, which are absent from 3D- and 4D-organoid culture, into 4D-organoid culture via small arterial and luminal input modules. Moreover, organoid-based assays could also be implemented via venous and luminal output modules as indicated. Abbreviations: a, small arteries; EL, epithelial lumen; ESF, extrinsic signaling factors that are absent in 3D- and 4D-organoid culture; v, small veins.
Role of Morphogenesis, Organogenesis, and Tissue Integration in hPDD
Brief Principles of Morphogenesis and Organogenesis
Applying the principles of developmental biology to hPSC culture may facilitate recapitulation of hPSC-based morphogenesis and organogenesis [7]. In an early study, differentiation of hESCs to cardiomyocytes was achieved by coculture with visceral endoderm-like cells [51]. To recapitulate physiologically relevant micro-environmental cues for hPDD, we need to optimize extracellular matrices, the concentration gradients of growth factors from cells or tissues, nutrients in growth medium, oxygen and carbon dioxide concentrations, and temperatures that cells would experience in vivo [7, 52]. Growth conditions that integrate autocrine, paracrine, and telecrine signals provide exogenous growth factors, ligands, and cell-permeable small molecules that can mimic in vivo microenvironments. These conditions may help facilitate growth of 3D-cocultures and the establishment of hPSC-derived organoids.
3D-coculture methods have been used to study the effects of chemical drugs on single hPSCs, single hPSC-derived colonies [53], and signaling trajectories among individual hPSCs and different types of differentiated cells [54, 55]. Supplementation of desired growth factors in the presence of pathway-specific small molecules significantly increases or decreases the maturation process of differentiated cells. Highly controllable 3D-organoids, using micro-bioreactor platforms including microfluidic bioreactors, are making hPDD possible at different scales.
Organoids and Disease Modeling
Organoids are now defined as stem-cell-derived 3D tissues with organ-specific cell types in vitro [reviewed in [6, 56–59]. In organoids, only stem cells are capable of self-renewal. In general, organoid cultures depend on the support form extracellular matrices and niche factors provide in the medium. Moreover, stem cells in organoids can differentiate toward adult tissues, which exhibit similar functionality as the tissue of origin, under proper differentiation conditions. There are diverse types of organoids based on their cell or tissue origins, which include hESC-, hiPSC-, adult stem cell (aSC)-, and primary tissue-derived organoids. Both hESC- and hiPSC-derived organoids are collectively named hPSC-derived organoids in the review.
Self-organized functional organoids have been generated using robust protocols for directed differentiation of hPSCs (Figure 2B). These organoids include: hepatic endoderm and functional liver buds with vasculature (i.e., miniature liver organoids) [49, 60], cerebral organoids (including cortical neurons, the 3D optic cup structure of the neural retina, and various brain regions containing progenitors) [61–63], hPSC-derived small intestine organoids [64], and hPSC-derived lung organoids [65].
Organoids retain the genetic and epigenetic signatures of original tissues or tumors, and as such, can be used to model human development and diverse types of diseases, including cystic fibrosis [reviewed in [6, 56–59]. Organoid systems have also been used to study infectious diseases to understand host–microbe interactions [57], such as replication of Norovirus in human intestinal stem cell-derived enteroid culture [66]. Brain organoids generated from hPSC-derived forebrain-specific regions and hPSC-derived neural stem cells can mimic the phenotypes of in vivo and in vitro Zika virus infection and damage [67, 68]. Moreover, forebrain organoids have been used to identify promising small-molecule inhibitors of Zika virus infection and induced neural cell death [69]. In addition, organoids derived from patients’ tumor specimens have been used to model different types of cancer, including colorectal carcinoma [70], pancreatic cancer [71], and liver cancer [72].
Thus, multi-dimensional organoids can be used to improve potential drug efficacy at the organ level. Furthermore, organoid conditions could be adapted to micro-bioreactor platforms, with various controllable parameters (e.g., cell densities, humidity, osmolality, acidity, and oxygen concentrations) and stiffness of extracellular matrices, to form 4D- or 5D-organoids (Figure 4). In brief, 3D-organoids are generated by self-organizing propagated and differentiated cells, including cocultured cells, in a three-dimensional geometric space (Figure 4J, 4H). 4D-organoids, as described here, are generated based on 3D-organoid culture, with an emphasis on the inclusion of the dimension of time (Figure 4K). Moreover, to enable the generation of physiologically-relevant organoids, we further propose the concept of 5D-organoids (Figure 4L). Theoretically, 5D-organoids could be established, based on 4D-organoid culture, by adding another dimensional complexity, which embraces many intercellular, immunological, and inflammatory signals that are absent from current 4D-organoid cultures [6, 7]. Thus, accurately providing additional extrinsic signaling factors (such as diverse types of cytokines) in 4D-organoid culture would facilitate the derivation of highly mature and physiologically-compatible 5D-organoids for regenerative medicine and drug screens. Clearly, the fidelity, maturity, and functionality of organoids are important for successful hPPD, and will be discussed next.
Evaluation of Maturity and Functionality of Differentiated Cells in Organoids
Although multi-dimensional hPSC platforms and 3D-cocultures are relatively easy to construct, such systems may not resemble live organisms in terms of physiological barriers, epigenetic factors, and immunomodulatory effects stimulated by in vivo environmental cues. Thus, they may not accurately represent human tissues in vivo. Immature cellular structures, similar to embryonic or neonatal tissues, are frequently encountered, which are largely associated with inadequate development of stem-cell niches. Therefore, these hPSC-derived models should be carefully evaluated and further optimized based on imitating stem-cell niche signals.
Stem-Cell Niche Concepts and Cellular Differentiation Commitments
Stem-cell niches are a specific microenvironment that allows stem cells to be maintained in an undifferentiated state [73–75]. Stem-cell niches function in a highly dynamic manner at different developmental stages of embryonic and postnatal tissues. We have recently proposed a paralogous stem-cell niche (PSN) concept that clarifies the coupling and uncoupling mechanisms between a specific stem-cell niche and its dynamic regeneration sites at different developmental stages [76]. Developmentally, PSNs are gradually transformed analogous niches within an individual species during development. After the first epithelial-mesenchymal transition (EMT) in the primitive streak, progressive variations of cell identity give rise to a cluster of adjacent stem-cell niches (i. e., PSNs) in order to foster specific types of progenitors and aSCs [76]. Notably, PSNs are distinct from each other in terms of their niche signaling pathways that are required for the commitment of a specific cell lineage. For example, long-term intestinal epithelial culture in vitro is made possible under a Wnt-dependent stem-cell niche [77]. In contrast, single Lgr5+ intestinal stem cells are able to form crypt-villus structures in vitro without the requirement of a “mesenchymal” niche, due to Wnt activation, mediated by the exogenous Wnt agonist R-spondin 1 in growth medium. Thus, Lgr5+ intestinal stem cells, which are functionally dependent on niche factors provided in the medium or produced by Paneth cells, can also self-organize epithelial structures [78]. Clearly, the above examples provide important insights into mechanisms regarding the dependency or independency of stem-cell niches in cellular differentiation and 3D-tissue organization. Therefore, it is important to provide basic niche components (e.g., extracellular matrices and proper niche-supporting cells) as well as critical signaling molecules (such as Wnt) to facilitate the generation of organoids with high fidelity (Figure 4K, 4L). Identification and implementation of the mechanisms behind faithful niche models may encourage maturity and functionality in hPSC-derived organoids for use in in vitro drug discovery.
Cell Maturity and Functionality at the Physiological Level
To assess the usefulness of a hPDD model, it is essential to examine the protein expression of multiple panels of tissue-specific regulators, enzymatic chemical metabolizers, and drug transporters. These regulatory proteins and drug transporters are key determinants of intracellular drug concentrations and activities. Critical proteins must also be expressed at the right time and in precise locations at physiologically relevant concentrations to faithfully implement their default functions. An unsatisfactory combination of any of the above parameters could undermine the success of any type of drug-discovery study. For example, defective or excessive expression of representative hepatic regulators, such as hepatocyte nuclear factor 4α (HNF4α), orphan nuclear receptor (PXR), oxysterol receptor LXR-α (LXR), bile acid receptor (FXR), and vitamin D3 receptor (VDR) [79, 80], might interfere with the function of generated hepatic organoids, thereby deterring their applications in hPDD. Moreover, compared with in vivo molecular standards, any altered expression of the predominant enzymes [e.g., cytochrome P450 family members (CYPs)] that metabolize drugs [81, 82] would significantly affect the metabolic profile of cells or hPSC-derived organoids. Similarly, altered ATP-binding cassette (ABC) transporters, which determine intracellular drug and ion concentrations [83–85], would dramatically affect the clearance of various drugs in cultured cells and hPSC-derived organoids.
Therefore, prior to any major drug-discovery projects, it is essential to determine the optimal maturity and functionality of differentiated cells and organoids in vitro and to characterize the hPSC-derived organoid tissues using native tissues as standards for comparison. Careful attention must be given to drug regulators and metabolizers (e.g., ABC transporters and P450 enzymes) that control the limiting steps of drug accumulation, excretion, and metabolism.
Prospective Roles of ABC Transporters in hPDD
Evaluation of Maturity and Functionality of Organoids with ABC Transporters
It is well established that ABC multidrug transporters play essential roles in both the distribution and efflux of drugs in a broad range of cell types and tissues under both physiological and pathological conditions [reviewed in [84–89]. Some ABC transporters are also critical components of the blood-brain, blood-testicular, and blood-placental barriers [85]. It is known that ABC transporters are major determinants of intrinsic and acquired cancer drug resistance [84, 88]. Altered or suboptimal expression of ABC transporters could hinder hPDD studies from achieving enhanced or decreased drug efflux in those hPSC-derived organoids of interest.
As an example of the role of ABC transporters with regards to the functional maturity of differentiated cells (e.g., hepatocytes), the presence of important hepatic transporters [e.g., ABCA1, ABCB1 (P-glycoprotein), ABCB4, ABCB11, ABCC2, ABCC7 (CFTR), ABCG2, ABCG5, and ABCG8] defines both the maturity and functionality of hepatocytes [83]. These ABC transporters have been shown to have a major effect on the drug-discovery pipeline using other cellular models at different stages. Thus, experimental data obtained from organoids with altered ABC transporter expression could potentially provide inaccurate information concerning preclinical drug testing, since these transporters affect accumulation of drugs in target tissues.
Evaluation of Prospective hPDD Data with ABC Transporters
Many pre-clinical drugs (such as a substantial percentage of effective kinase inhibitors) and drugs in the clinic are substrates of ABC transporters. These chemical substrates can be differentially exported from diverse types of cells and tissues by ABC transporters. This differential efflux of intracellular drugs may confuse the interpretation of drug-screening data and increase the risk of drug resistance in clinical trials. Clinical drug resistance to the potent tyrosine kinase inhibitor, STI571 (also known as imatinib mesylate) and subsequent treatment failure in patients is a clear example, as STI571 is a model substrate of both ABCB1 and ABCG2 [90, 91].
Furthermore, mutant cells sometimes bear critical ABC transporter mutations at drug-binding sites and pockets, or polymorphisms that affect substrate recognition [92–96]. Thus, ABC transporter-mediated drug assays should be compared between wild-type hPSC- and mutant hPSC-derived organoids. When using hPSC-derived 3D-organoids for drug screening and discovery, researchers should perform routine examinations to determine if preclinical drug candidates are bound by ABC transporters on cell surface membranes, are accumulated intracellularly, and are effluxed from various types of cells (including hepatocytes that are enriched with ABC transporter expression). In the following section, we will elucidate the role of ABC transporters in hPDD through a comprehensive analysis of CFTR mutations in CF organoids.
CF Organoids and Molecular Drug Discovery Based on CFTR
We have chosen to highlight CF-organoids in this review because drug-discovery studies using these organoids have confronted all the problems discussed in the previous sections. Significantly, CF-organoid studies have provided proof-of-concept examples from experimental investigation to clinical applications, and have thus accelerated the development of translational medicine. Therefore, CF-organoids serve as a good model for discussion.
CF is a life-threatening autosomal recessive disorder that affects approximately 70,000–100,000 individuals worldwide (www.cfww.org). The predominant causes of CF are various mutations found in the ATP-gated channel gene, CFTR. It is likely that patient groups with mixed drug responses may have considerable genetic heterogeneity, with rare mutations and complex alleles, all of which are difficult to recapitulate and assess pre-clinically in animal models. With regard to these complications, CF-organoid-based disease modeling offers an opportunity to identify responders for personalized medicine.
The Role of CF Organoids in Drug Discovery and Drug Repurposing
Simple CF intestinal organoids have a structure similar to that of a cellular cyst, in which the apical plasma membrane is located on the luminal side of the organoids (Figure 5A, 5B). Using these intestinal organoids, several powerful functional CFTR assays have been established [97]. One representative assay, termed forskolin-induced swelling (FIS), is based on adenylyl cyclase activator-mediated CFTR channel activation via increased cAMP concentrations. Through the FIS assay, CFTR activity can be directly determined by quantifying the amount of fluid secreted from the cells into the lumen and by measuring the size of the organoid lumen after adding the adenylyl cyclase activator, forskolin (Figure 5B). Hence, a rapid swelling phenotype is expected in healthy organoids after forskolin induction, whereas decreased or diminished swelling may be observed in CF organoids [97].
Through a CRISPR-meditated gene-editing approach, scientists were able to correct CFTR-ΔF508 mutations in CF-iPSCs derived from patients in in vitro cell culture and then differentiate these corrected cells to lung epithelial cells [98]. As a result, defective processing of CFTR to the cell membranes of these airway epithelial cells was reversed to the wild-type level. CFTR chloride current was observed in response to increased cAMP stimulation mediated by a cocktail including forskolin, similar to that observed in wild-type lung epithelial cells derived from hiPSCs. The rescue of chloride current was blocked by CFTRinh-172, a specific CFTR inhibitor [98]. With a similar CRISPR approach, Clevers and colleagues were also able to revert the defect of the rapid chloride-channel swelling response in intestinal organoids, derived from intestinal stem cells of CF patients, to normal conditions [99].
The FIS assay has revealed differential responses of primary intestinal organoids from CF patients to a number of CFTR correctors (such as C8, Corr-4a, and VRT-325) in addition to VX-809 and VX-770 [97]. The rapid swelling phenotype seen in healthy organoids was restored in CF patient organoids in the presence of both VX-809 and VX-770. Thus, the FIS assay is an accurate functional assay of CF drugs in organoids, which could be used for drug screens and drug repurposing efforts using patient-specific organoids in personalized medicine.
There are many advantages to using organoids derived from hPSCs and aSCs (e.g., intestinal stem cells) of patients for drug screening, particularly for CF patients with rare mutations and complex alleles [100]. Rectal organoids derived from CF patients with the common CF mutation ΔF508 (in one allele) combined with various variant classes, rare mutations and complex heterozygous alleles showed differential responses to the CF drugs tested (VX-809, VX-770, and Orkambi) [101]. In these assays, homozygous mutations with a specific splicing defect in the CFTR gene [102] showed no response to the CF drugs tested. This was apparently due to a defect in CFTR protein synthesis from immature transcripts. Thus, this genotype could be used as a negative control when performing this assay.
Compared with the homozygous ΔF508/ΔF508 genotype, the ΔF508/G1249R mutations are an extremely rare genotype, the functional impact of which on CF drug response was unknown. Two CF patients with the ΔF508/G1249R genotype were selected to test their response to CF drugs based on the results from their organoid assays [101]. Interestingly, the rectal organoids derived from these patients showed a strong response to VX-770 alone, but not to VX-809 or Orkambi in the FIS assay [101]. Notably, treatment of one patient with VX-770 rapidly relieved the patient’s symptoms, improved lung function, and elevated body weight [101]. This example highlights the clinical potential of an organoid-based assay for evaluation of drug efficacy and drug repurposing screen. We suggest that CFTR mutational organoids can be classified, patterned, assayed, and used for predicting the outcomes of CF drugs or drug combinations.
Empowering Organoid Studies with Molecular Insights
Despite the growing enthusiasm of applying organoids for hPDD, organoid-based technologies appear to have several inevitable limitations. The absence of immunological and inflammatory responses that occur in a human body may be a major limiting factor. There are also limitations in using organoids for HTS assays in their current platforms (due to spherical and often complicated cellular structures) (Figure 3 and see Outstanding Questions).
Outstanding questions.
Can human pluripotent stem cell (hPSC)-based drug discovery (hPDD) provide convincing platforms for in vitro drug discovery and drug repurposing with greatly reduced use of animal models? Could hPDD become a routine practice for any new drug discovery or drug repurposing at a reasonable cost?
Can experimental variability be minimized and avoided with better designed hPSC models? Can physiologically relevant organoids be generated in vitro for modeling of diverse diseases and for drug screens?
How can hPDD be optimized through existing coherent animal and human datasets, drug bank databases, and high-resolution protein structures to accelerate drug discovery for personalized medicine at a molecular level?
How can hPDD be used to complement or even replace preclinical drug development? Would it be possible to establish complicated organoid models to simulate human body physiology and to predict or monitor diverse clinical trials (e.g., Phases I/II/III) in the laboratory?
Many unique and unknown structural features of proteins (e.g., CFTRs) could be used to design new compounds and for drug screens in the context of 4D- or 5D-organoids due to their usefulness in human disease modeling and drug discovery (Figures 4 and 5). Indeed, this could become a robust area of research, which would likely facilitate the development of translational medicine and particularly the implementation of precision medicine at a molecular level (see Clinician’s Corner, Box 1. Moreover, this is multi-disciplinary work that requires cooperation between chemists, structural biologists, and stem-cell biologists, and physicians. In this collaboration team, chemists and structural biologists work on designing or modifying drugs based on high-resolution of desired protein structures. Stem-cell biologists focus on optimizing hPSC-derived organoid models for identifying lead compounds and testing their cytotoxicity and efficacy in animal models. Finally, clinicians are responsible for implementing clinical trials to determine drug safety and efficacy in patients and then to feedback this information to the collaboration team for further drug assessments.
Clinician’s corner (Box 1).
Human pluripotent stem-cell derived organoids faithfully retain genetic mutations of original tissues. Moreover, desired mutations in organoids can be generated by efficient gene-editing technologies such as CRISPR-Cas9. These organoids can be used to model human development and diverse types of diseases caused by mutations in one or multiple genes.
Multi-dimensional organoids can be used for in vitro drug screens and for evaluating drug efficacy at the organ level. There are many organs and tissues that express abundant ABC multidrug transporters. Physiologically or pathologically relevant organoids with optimal membrane expression of ABC transporters might provide accurate information about intracellular drug concentrations. Thus, organoid-based drug-transport assays may be particularly useful for determining whether pre-clinically developed drugs are substrates of one or more ABC transporters. This information may help clinicians overcome potential drug-transporter-mediated drug resistance in patients.
In clinics, patient groups often have considerable genetic heterogeneity, with various rare mutations and complex alleles that are difficult to assess pre-clinically in animal models. CFTR-organoid-based disease modeling and drug screens provide a convincing proof-of-concept evidence that mutational organoids from these patient groups might be classified, assayed, and used for predicting the outcomes of drugs or drug combinations and for identifying responders.
Different types of drug-response data from patients can be integrated with molecular and atomic structures of specific drug targets (e.g., CFTRs) to maximize a rational design of the next generation of clinical drugs. This is a multi-disciplinary effort that requires the cooperation between physicians, stem-cell biologists, and structural biologists, which would facilitate the implementation of precision medicine at a molecular level.
Concluding Remarks
hPDD studies have provided compelling platforms for in vitro drug discovery and drug repurposing with greatly reduced use of animal models and with the hope of complementing or even replacing some preclinical drug development. Experimental variability could be minimized and avoided with better designed hPSC models and physiologically relevant organoids. The availability of multi-dimensional coculture systems, hPSC-based disease modeling, hPSC- and aSC-derived organoids, and informative functional assays in vitro, offers the possibility to optimize hPDD through existing coherent animal and human datasets, the drug bank database, and high-resolution protein structures. We believe that the use of patient-relevant organoids as a disease model system will revolutionize the methods for drug discovery and development as evidenced in studies using mutation-specific CF-organoids for evaluation of drug efficacy.
Supplementary Material
Highlights.
Human pluripotent stem cells (hPSCs) and their differentiated derivatives represent valuable cell resources for organogenesis, disease modeling, and hPSC-based drug discovery (hPDD) in vitro.
Abundant contributing variables, including the dimensionality of cell-culture methods, cell maturity and functionality, and experimental variability, epitomize the major challenges of hPDD research.
Targeting various CFTR mutations in organoids derived from patients with cystic fibrosis provides a proof-of-principle drug discovery model, defining a potential pathway to the implementation of hPDD for personalized medicine.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) at the National Institute of Neurological Disorders and Stroke (K.G.C., B.S.M., and K.P.), the National Center for Advancing Translational Sciences (W.Z.), the National Cancer Institute (M.M.G.), and the National Institute of Dental and Craniofacial Research (P.G.R). Dr. Ronald D. McKay was supported by the Lieber Institute for Brain Development. We thank Dr. Richard Leapman and Dr. Pingyu Zhong for critical discussion and comments on the manuscript, and George Leiman for editorial assistance. We thank Dr. Alan Koretsky for his strong support of this work.
Glossary
- Morphogenesis
a process by which a cell, tissue, organ, or organism acquires its architectural characteristics. Morphogenesis is induced and controlled by many cellular and developmental programs
- Organogenesis
a process by which an organism develops and produces its body parts of specific tissue structures and functions (e.g., the liver and the limb) during development
- Tissue integration
distinct types of cell clusters that incorporate into a preexisting cell group or host tissues to integrate their cellular effects
- Naïve and primed
refer to naïve and primed pluripotent states established in the preimplantation embryos and the post implantation epiblast, respectively, during embryogenesis. This distinct pluripotency also extends to pluripotent stem cells that have identical pluripotent features under cell culture conditions
- 3D-cocultures
in vitro cell culture conditions that combine coculture elements with a three-dimensional environment
- Organoids
a stem-cell-derived 3D multicellular miniature of physiologically-relevant tissues in a culture dish, in which only stem cells are capable of self-renewal, self-organization, and differentiation toward adult tissues under corresponding conditions
- Micro-bioreactor platforms
a microscale version of conventional bioreactors (macrobioreactors), including microfluidic bioreactors, biochips, and cell-chip, which integrate numerous monitoring features used by macrobioreactors
- 5D-organoids
Compared to 4D-organoids, 5D-organoids have one-dimensional higher complexity, which is regulated by providing additional extrinsic signaling factors in 4D-organoids to generate highly physiologically-relevant organoids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Chen KG, et al. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14(1):13–26. doi: 10.1016/j.stem.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatehullah A, et al. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18(3):246–54. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 7.Chen KG, et al. Developmental insights from early mammalian embryos and core signaling pathways that influence human pluripotent cell growth and differentiation. Stem Cell Res. 2014;12(3):610–21. doi: 10.1016/j.scr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narva E, et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28(4):371–7. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 9.Draper JS, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22(1):53–4. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 10.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11(4):268–77. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104(4):e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee G, Studer L. Induced pluripotent stem cell technology for the study of human disease. Nat Methods. 2010;7(1):25–7. doi: 10.1038/nmeth.f.283. [DOI] [PubMed] [Google Scholar]
- 13.Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin Pharmacol Ther. 2011;89(5):655–61. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- 14.Zheng W, et al. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today. 2013;18(21–22):1067–73. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying QL, Smith A. The Art of Capturing Pluripotency: Creating the Right Culture. Stem Cell Reports. 2017;8(6):1457–1464. doi: 10.1016/j.stemcr.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 17.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–92. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Chen KG, et al. Non-colony type monolayer culture of human embryonic stem cells. Stem Cell Res. 2012;9(3):237–48. doi: 10.1016/j.scr.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Androutsellis-Theotokis A, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442(7104):823–6. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25(6):681–6. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 21.Kunova M, et al. Adaptation to robust monolayer expansion produces human pluripotent stem cells with improved viability. Stem Cells Transl Med. 2013;2(4):246–54. doi: 10.5966/sctm.2012-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G, et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol. 2012;30(12):1244–8. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoing S, et al. Discovery of inhibitors of microglial neurotoxicity acting through multiple mechanisms using a stem-cell-based phenotypic assay. Cell Stem Cell. 2012;11(5):620–32. doi: 10.1016/j.stem.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Charbord J, et al. High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells. 2013;31(9):1816–28. doi: 10.1002/stem.1430. [DOI] [PubMed] [Google Scholar]
- 25.Naryshkin NA, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345(6197):688–93. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann M, et al. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen. 2015;20(9):1101–11. doi: 10.1177/1087057115588287. [DOI] [PubMed] [Google Scholar]
- 27.Ma J, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301(5):H2006–17. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris K, et al. Comparison of electrophysiological data from human-induced pluripotent stem cell-derived cardiomyocytes to functional preclinical safety assays. Toxicol Sci. 2013;134(2):412–26. doi: 10.1093/toxsci/kft113. [DOI] [PubMed] [Google Scholar]
- 29.Doherty KR, et al. Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol Appl Pharmacol. 2015;285(1):51–60. doi: 10.1016/j.taap.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Drawnel FM, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014;9(3):810–21. doi: 10.1016/j.celrep.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 31.Sirenko O, et al. High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev Technol. 2014;12(1):43–54. doi: 10.1089/adt.2013.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi SM, et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57(6):2458–68. doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ware BR, et al. Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes. Toxicol Sci. 2015;145(2):252–62. doi: 10.1093/toxsci/kfv048. [DOI] [PubMed] [Google Scholar]
- 34.Mathur A, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D, et al. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34(23):5813–20. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinson JT, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349(6251):982–6. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carpentier A, et al. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Res. 2016;16(3):640–50. doi: 10.1016/j.scr.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullard A. Stem-cell discovery platforms yield first clinical candidates. Nat Rev Drug Discov. 2015;14(9):589–91. doi: 10.1038/nrd4708. [DOI] [PubMed] [Google Scholar]
- 39.Baker DE, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25(2):207–15. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 40.Forsyth NR, et al. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells. 2006;8(1):16–23. doi: 10.1089/clo.2006.8.16. [DOI] [PubMed] [Google Scholar]
- 41.Herron TJ, et al. Extracellular Matrix-Mediated Maturation of Human Pluripotent Stem Cell-Derived Cardiac Monolayer Structure and Electrophysiological Function. Circ Arrhythm Electrophysiol. 2016;9(4):e003638. doi: 10.1161/CIRCEP.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeziorowska D, et al. Differential Sarcomere and Electrophysiological Maturation of Human iPSC-Derived Cardiac Myocytes in Monolayer vs. Aggregation-Based Differentiation Protocols. Int J Mol Sci. 2017;18(6) doi: 10.3390/ijms18061173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loirand G. Rho Kinases in Health and Disease: From Basic Science to Translational Research. Pharmacol Rev. 2015;67(4):1074–95. doi: 10.1124/pr.115.010595. [DOI] [PubMed] [Google Scholar]
- 45.Chen KG, et al. Alternative cultures for human pluripotent stem cell production, maintenance, and genetic analysis. J Vis Exp. 2014;(89) doi: 10.3791/51519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braam SR, et al. Improved genetic manipulation of human embryonic stem cells. Nat Methods. 2008;5(5):389–92. doi: 10.1038/nmeth.1200. [DOI] [PubMed] [Google Scholar]
- 47.Liew CG, et al. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25(6):1521–8. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 48.Padmanabhan R, et al. Regulation and expression of the ATP-binding cassette transporter ABCG2 in human embryonic stem cells. Stem Cells. 2012;30(10):2175–87. doi: 10.1002/stem.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–4. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 50.Xia Y, et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol. 2013;15(12):1507–15. doi: 10.1038/ncb2872. [DOI] [PubMed] [Google Scholar]
- 51.Mummery C, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107(21):2733–40. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 52.Cimetta E, et al. Micro-bioreactor arrays for controlling cellular environments: design principles for human embryonic stem cell applications. Methods. 2009;47(2):81–9. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villa-Diaz LG, et al. Microfluidic culture of single human embryonic stem cell colonies. Lab Chip. 2009;9(12):1749–55. doi: 10.1039/b820380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moledina F, et al. Predictive microfluidic control of regulatory ligand trajectories in individual pluripotent cells. Proc Natl Acad Sci U S A. 2012;109(9):3264–9. doi: 10.1073/pnas.1111478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Meer AD, et al. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip. 2013;13(18):3562–8. doi: 10.1039/c3lc50435b. [DOI] [PubMed] [Google Scholar]
- 56.Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 57.Dutta D, et al. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol Med. 2017;23(5):393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 58.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 59.Koo BK, Huch M. Organoids: A new in vitro model system for biomedical science and disease modelling and promising source for cell-based transplantation. Dev Biol. 2016;420(2):197–198. doi: 10.1016/j.ydbio.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Takebe T, et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat Protoc. 2014;9(2):396–409. doi: 10.1038/nprot.2014.020. [DOI] [PubMed] [Google Scholar]
- 61.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771–85. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 64.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014;20(11):1310–4. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dye BR, et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015:4. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ettayebi K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353(6306):1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian X, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcez PP, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–8. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 69.Xu M, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22(10):1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Broutier L, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen KG, et al. Mouse Genetic Analysis of Bone Marrow Stem Cell Niches: Technological Pitfalls, Challenges, and Translational Considerations. Stem Cell Reports. 2017;9(5):1343–1358. doi: 10.1016/j.stemcr.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scadden DT. Nice neighborhood: emerging concepts of the stem cell niche. Cell. 2014;157(1):41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4(1–2):7–25. [PubMed] [Google Scholar]
- 76.Chen KG, et al. Concise Review: Conceptualizing Paralogous Stem-Cell Niches and Unfolding Bone Marrow Progenitor Cell Identities. Stem Cells. 2018;36(1):11–21. doi: 10.1002/stem.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15(6):701–6. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 79.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3(7):499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 80.Chawla A, et al. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–70. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 81.Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21(1):70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 82.Caswell JM, et al. Engineering and application of P450 monooxygenases in pharmaceutical and metabolite synthesis. Curr Opin Chem Biol. 2013;17(2):271–5. doi: 10.1016/j.cbpa.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 83.Wlcek K, Stieger B. ATP-binding cassette transporters in liver. Biofactors. 2014;40(2):188–98. doi: 10.1002/biof.1136. [DOI] [PubMed] [Google Scholar]
- 84.Gottesman MM, et al. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 85.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–92. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 86.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–42. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 87.Deeley RG, et al. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86(3):849–99. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 88.Chen KG, Sikic BI. Molecular pathways: regulation and therapeutic implications of multidrug resistance. Clin Cancer Res. 2012;18(7):1863–9. doi: 10.1158/1078-0432.CCR-11-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szakacs G, et al. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 90.Burger H, Nooter K. Pharmacokinetic resistance to imatinib mesylate: role of the ABC drug pumps ABCG2 (BCRP) and ABCB1 (MDR1) in the oral bioavailability of imatinib. Cell Cycle. 2004;3(12):1502–5. doi: 10.4161/cc.3.12.1331. [DOI] [PubMed] [Google Scholar]
- 91.Burger H, et al. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood. 2004;104(9):2940–2. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- 92.Chen G, et al. Multidrug-resistant human sarcoma cells with a mutant P-glycoprotein, altered phenotype, and resistance to cyclosporins. J Biol Chem. 1997;272(9):5974–82. doi: 10.1074/jbc.272.9.5974. [DOI] [PubMed] [Google Scholar]
- 93.Chen GK, et al. Loss of cyclosporin and azidopine binding are associated with altered ATPase activity by a mutant P-glycoprotein with deleted phe(335) Mol Pharmacol. 2000;57(4):769–77. doi: 10.1124/mol.57.4.769. [DOI] [PubMed] [Google Scholar]
- 94.Loo TW, Clarke DM. Recent progress in understanding the mechanism of P-glycoprotein-mediated drug efflux. J Membr Biol. 2005;206(3):173–85. doi: 10.1007/s00232-005-0792-1. [DOI] [PubMed] [Google Scholar]
- 95.Ambudkar SV, et al. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–98. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 96.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 97.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19(7):939–45. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 98.Firth AL, et al. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 2015;12(9):1385–90. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–8. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 100.Castellani C, et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros. 2008;7(3):179–96. doi: 10.1016/j.jcf.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dekkers JF, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8(344):344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 102.Dujardin G, et al. Splicing defects in the CFTR gene: minigene analysis of two mutations, 1811+1G>C and 1898+3A>G. J Cyst Fibros. 2011;10(3):212–6. doi: 10.1016/j.jcf.2010.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.