Abstract
We investigated the expression of anthocyanin structural genes and transcription factors (TFs) associated with varying anthocyanin content during different developmental stages (S1–S4) of the gerbera cultivars ‘Nathasha’ and ‘Rosalin’. Accumulation of anthocyanin started at S1 and reached a maximum at S3 in both cultivars. Enhancement of anthocyanin content in ‘Nathasha’ was associated with upregulation of ANS and MYB10, whereas in ‘Rosalin’, upregulation was associated with CHS1, MYB10, and MYC1. Low-temperature exposure (6 °C) enhanced anthocyanin content to a greater extent than that at 22 °C via stronger upregulation of CHS1 and MYB10 in ‘Nathasha’ and CHS1 in ‘Rosalin’, irrespective of flower developmental stage. However, differences in anthocyanin content between the two cultivars were found to be influenced by the expression levels of all structural genes and TFs, irrespective of flower developmental stage and temperature conditions. We suggest that differences in the regulation mechanisms of anthocyanin biosynthesis and coloration pattern between ‘Nathasha’ and ‘Rosalin’ are related to differences in the expression patterns of structural genes and TFs; however, further functional studies of the key genes in anthocyanin biosynthesis are needed.
Keywords: Anthocyanin, Flower stage, Gerbera, Gene expression, Temperature
Introduction
Gerbera hybrids are among the most important cut flowers globally and are commonly produced by crossing two wild species, Gerbera jamesonii and Gerbera viridifolia (Hansen 1999). These are extremely popular flowers, and given the availability of multiple flower colors and continuous year-round production in greenhouses, they ranked fourth in popularity in Dutch auctions, after roses, chrysanthemums, and tulips (Plasmeijer 2012). The richness of their flower colors makes gerbera an attractive candidate for anthocyanin biosynthesis research. In most plant species, flower colors ranging from red to blue are attributed to anthocyanins (Quattrocchio et al. 2006) resulting from different branches of the anthocyanin biosynthesis pathway (Chandler and Tanaka 2007; Wessinger and Rausher 2014).
The structural genes involved in anthocyanin synthesis, including phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), flavanone-3-hydroxylase (F3H), dihydroflavonol-4-reductase (DFR), and anthocyanidin synthase (ANS), have been well characterized in many species (Schwinn and Davies 2004; Grotewold 2006; Guo et al. 2014). Transcriptional regulation of these structural genes is considered the major mechanism responsible for the diversity of flower color in plants (Quattrocchio et al. 1998; Yamagishi et al. 2010) and is achieved mainly by a complex of R2R3-MYB and basic helix–loop–helix (bHLH) transcription factors (TFs) associated with WD40 repeat proteins (Broun et al. 2005; Koes et al. 2005; Petroni and Tonelli 2011). R2R3-MYB TFs are believed to play a key role by binding directly to the promoters of structural genes via highly-conserved DNA-binding domains (Takos et al. 2006; Zhang et al. 2014; Tian et al. 2015).
Although previous studies have provided valuable insights into the mechanisms responsible for flower coloration in many plant species, the mechanisms underlying the regulation of anthocyanin biosynthesis, and which factors contribute to the diversity in flower coloration among plant species, remain unclear. Similarly, although some studies have investigated the relationships between the regulation of anthocyanin biosynthesis and flower colors in gerbera, the mechanisms determining color within the genus remain largely unknown. The R2R3-MYB TF GMYB10 is believed to regulate anthocyanin structural genes in gerbera by interacting with the bHLH TF GMYC1 (Elomaa et al. 1998, 2003). In addition, ectopic expression of GMYB10 has been shown to markedly enhance anthocyanin content in the transgenic gerbera cultivar Terraregina (Laitinen et al. 2008). Moreover, CHS-encoding genes such as GCHS1 and GCHS4 have been shown to influence anthocyanin accumulation in gerbera vegetative and floral organs (Helariutta et al. 1995; Laitinen et al. 2005). Loss of one of two enzymatically functional DFR forms has also been shown to result in white-colored petals in the gerbera cultivar ‘Ivory’ (Bashandy et al. 2015). Furthermore, temperature has been shown to influence anthocyanin biosynthesis in some flowers, including chrysanthemum, lily, rose, and petunia (Biran and Halevey 1973; Biran et al. 1973; Shvarts et al. 1997; Dela et al. 2003; Nozaki et al. 2006; Lai et al. 2011), with low temperatures stimulating anthocyanin accumulation in petunia (Shvarts et al. 1997) and rose (Biran and Halevey 1973; Biran et al. 1973). To the best of our knowledge, the role of low temperature in anthocyanin biosynthesis in gerbera has not been investigated to date. Moreover, the regulatory mechanism underlying the effects of temperature on anthocyanin biosynthesis in flowers remains unknown. Hence, it is of interest to explore the mechanism whereby temperature affects anthocyanin biosynthesis in gerbera.
In this study, we isolated anthocyanin structural genes and transcription factors known to be associated with the mechanisms of anthocyanin regulation from two gerbera cultivars with distinctly different flower colors, ‘Nathasha’ and ‘Rosalin’, and investigated the expression patterns of these genes during different developmental stages. In addition, we also investigated the role of temperature in controlling anthocyanin biosynthesis in gerbera by determining and comparing expression patterns of the structural genes and TFs in gerbera plants exposed to low or normal temperatures.
Methods
Plant materials
Flowers of the gerbera cultivars ‘Nathasha’ and ‘Rosalin’ are very graceful and attractive to flower users and lovers; they are, therefore, popular and commercially important in the flower industry. In this study, we used two commercial gerbera cultivars with distinct flower colors, ‘Nathasha’ and ‘Rosalin’, grown at a commercial flower production farm in Youngju, Korea. Flowers were collected at different developmental stages (S1–S4, early flower to the fully-open flower stages) (Fig. 1). To determine anthocyanin content and expression of related structural genes and transcription factors (TFs) for each stage, approximately 2 g of petals (from three different flowers) was collected, immediately frozen in liquid nitrogen, and then stored at − 70 °C until they were used for isolation of total RNA and extraction of pigments. One biological sample contained three different flowers (plants), and analysis was performed for three different biological samples.
Fig. 1.
Flower samples collected from different developmental stages (S1–S4) of the gerbera cultivars ‘Rosalin and Nathasha’
Effect of temperature on the regulation of anthocyanin
To investigate the effect of temperature on anthocyanin regulation, 20 plants of the two cultivars at different flowering stages (S2 and S3) were exposed to two different temperatures (6 and 22 °C) under a 16-h photoperiod (photon flux density of 30 mmol m−2 s−1, cool white fluorescent lamps). For the determination of anthocyanin content and expression of related structural genes and TFs, approximately 2 g of petals (from three different flowers) was collected from plants (stages S2 and S3) subjected to temperatures of 6 and 22 °C for 3 days. Samples were immediately frozen in liquid nitrogen and then stored at − 70 °C until they were used for the isolation of total RNA and extraction of pigments, and the experiment was repeated for three different biological samples.
Analysis of total anthocyanin content
Total anthocyanin content in gerbera flowers collected at different developmental stages was analyzed, following previously described methods (Ai et al. 2016). Briefly, for each sample, ~ 500 mg of petals was ground to a fine powder and transferred to an extraction solution for anthocyanin extraction. The extract was incubated at 4 °C for 24 h, then centrifuged at 13,000 rpm at 4 °C for 20 min. The supernatant containing anthocyanin was analyzed spectrophotometrically (Shimadzu, Kyoto, Japan). Data for each sample represent the average of three biological replicates.
RNA extraction
Total RNA was isolated from 100 mg of petals (from one biological sample) from each of the two cultivars collected at different developmental stages using TRI Reagent™ Solution (Ambion, USA). RNA integrity was assessed via electrophoresis using 1% agarose gels, and concentrations of the extracted RNA were determined using a NanoDrop spectrophotometer (Thermo, USA). RNA extraction was performed for three different biological samples.
Expression analysis of anthocyanin structural genes and TFs using qRT-PCR
To determine the transcript levels of anthocyanin structural genes and TFs, cDNA was synthesized from total RNA isolated from petals of the two cultivars collected at different developmental stages. One microgram of total RNA and oligo dT20 primers were used for cDNA synthesis (ReverTra Ace-á, Toyobo, Japan). Transcript levels of the candidate genes (Table 1) were measured using a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Waltham, USA) following published methods (Ai et al. 2016). Gene expression levels were normalized to the actin gene to minimize variations in the cDNA template. The method used to calculate relative gene expression was quantitative–comparative CT (ΔΔCT). The primers and PCR conditions used for amplification of the detected genes are listed in Table 1. Data for each sample represent the average of three different biological replicates.
Table 1.
Primer sequences used for detection of genes related to anthocyanin biosynthesis by qRT-PCR
| Gene | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) |
|---|---|---|
| CHS1 | CGG CCG TTG TTT GAA ATG GT | ATC AAC CCA GGC ACG TCT TT |
| CHS4 | AAA CTA CGT GCC ACA CGA CA | AAA CCC AAA CAA AAC GCC CC |
| F3H | TCA CAT TGG GTC TGA AGC GG | AAT CCA ACT CTT GCC ACC GT |
| F3′H | GTC GGA AAC CTA CCG CAT CT | TCG CGT CGT GAG TCT TCA AA |
| DFR | ACT TTG GCT GAA AAA GCC GC | GCC CGT GAT CAA AGA AAG CG |
| ANS | TTA TCA ACG AGC GGC ATC CA | TTT TTC CGG GTC GTC GGA TT |
| MYB10 | TGG AAC ACC CAT CTT CGC TC | GCT TTG CCT GTT ACC GAA CC |
| MYC1 | CCG CAT TAT CCC CCG AAG AT | GAC GCG CTT TTT GCT AGG AG |
| Actin | AGG TTA CGC ACT TCC TCA CG | TGT CAC GAA CGA TTT CCC GT |
| 2-Pyrone synthase(2PS) | CAA AGA AGC CGC AGT CAA GG | AGC GTT TGA CTG AAG GGG AG |
PCR condition: 95 °C (10 min) → [95 °C (15 s)] × 40 cycles → 57 °C (1 min)] → 95 °C (15 s) → 59.3 °C (1 min)
Statistical analysis
The data were analyzed using SPSS ver. 11.09 (IBM Corporation, Armonk, NY, USA) and are presented as the means of three replicates. A least significant difference test (LSD test) was used to analyze the differences between means. The significance level was set at P < 0.05.
Results
Anthocyanin accumulation in two different gerbera cultivars
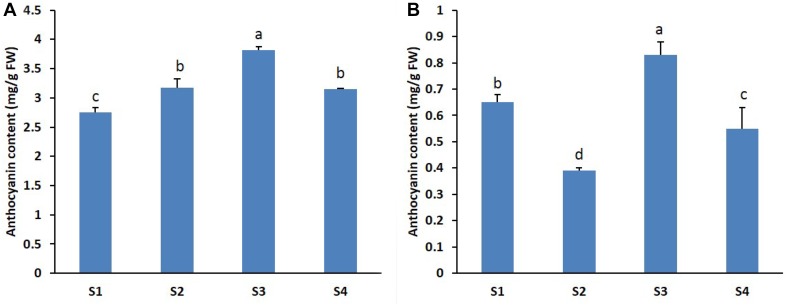
Anthocyanin content in petals of the two gerbera cultivars ‘Nathasha’ and ‘Rosalin’ was measured at different stages of flower development. Overall, the anthocyanin content varied between the cultivars. The anthocyanin content in ‘Nathasha’ was found to be higher than that in ‘Rosalin’ (Fig. 2a, b), which is consistent with their respective flower colors. However, the cultivars differed with respect to the stages at which the highest anthocyanin contents were produced, being at stages S2 and S3 for ‘Nathasha’ but only at stage S3 for ‘Rosalin’. Coloration patterns during the different developmental stages differed marginally between the two cultivars, reflected in anthocyanin content which was lowest at S1 for ‘Nathasha’ and at S2 for ‘Rosalin’, whereas both cultivars had peaks at S3 and both exhibited a subsequent decline at S4.
Fig. 2.
Anthocyanin contents of the two gerbera cultivars ‘Nathasha (a) and Rosalin (b)’ at different flower developmental stages. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average anthocyanin content
Expression patterns of anthocyanin structural genes in the two cultivars
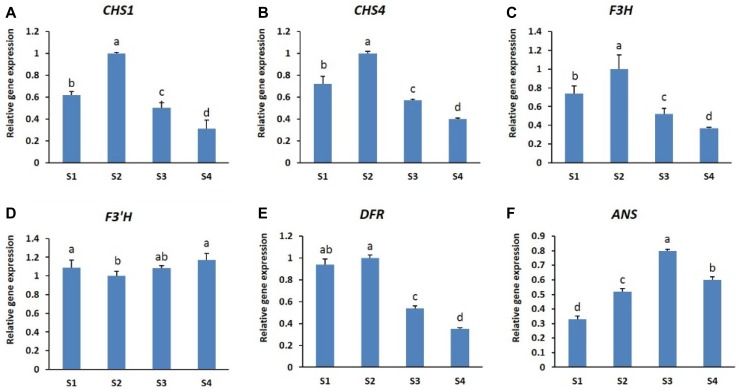
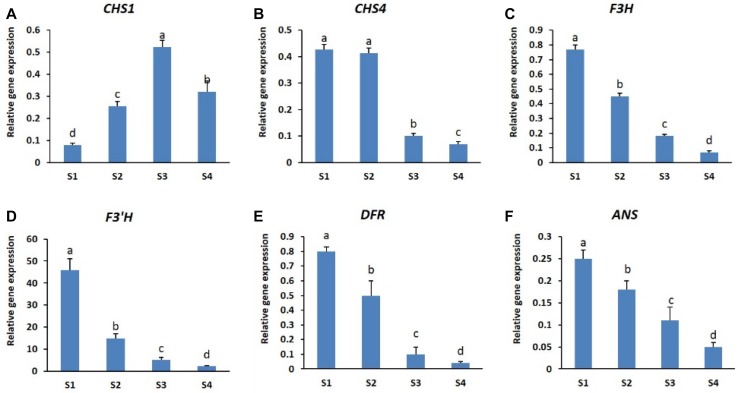
The results shown in Fig. 2a, b indicate that the anthocyanin content in petals of both cultivars differed at different stages of development. Therefore, we examined the transcript levels of genes involved in anthocyanin biosynthesis at these developmental stages (CHS1, CHS4, F3H, F3′H, DFR, and ANS) by real-time PCR. The results indicated that all the structural genes were expressed in both cultivars and at all the developmental stages studied; however, the expression patterns in the two cultivars differed at different developmental stages (Figs. 3, 4). In ‘Nathasha’, the expression levels of all structural genes, except F3′H and ANS, were found to be highest at stage S2 and declined thereafter (in stages S3 and S4), indicating similar expression patterns for most of the genes (Fig. 3a–f). However, the highest anthocyanin content was observed at stage 3, corresponding to the highest expression levels of ANS. In addition, the higher anthocyanin content observed at S4 than at S1 was also consistent with the expression levels of ANS. In ‘Rosalin’, the expression levels of all genes, except CHS1, were highest at S1 and decreased thereafter (from S2 to S4 stages), although CHS4 levels remained at their highest until stage S2 (Fig. 4a–f). In contrast to the other genes, the expression of CHS1 was found to be lowest at S1, and thereafter gradually increased from S1 to S3 before decreasing at S4 (Fig. 4b). The high expression of anthocyanin structural genes (CHS4, F3H, F3′H, DFR, and ANS) at S1 was consistent with the high anthocyanin content detected at this stage; however, these genes were expressed at relatively low levels at stage S3, when the highest amounts of anthocyanin were produced. Interestingly, only CHS1 was expressed at high levels at stage S3. When comparing gene expression between the cultivars, the expression levels of most structural genes at all stages were generally higher in ‘Nathasha’ than in ‘Rosalin’, which may be associated with the differences in anthocyanin levels observed at the different developmental stages.
Fig. 3.
Expression patterns of different anthocyanin structural genes (a–f) during different flower developmental stages of gerbera cultivar ‘Nathasha’. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average means
Fig. 4.
Expression patterns of different anthocyanin structural genes (a–f) during different flower developmental stages of gerbera cultivar ‘Rosalin’. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average means
Expression levels of transcription factors in the two cultivars
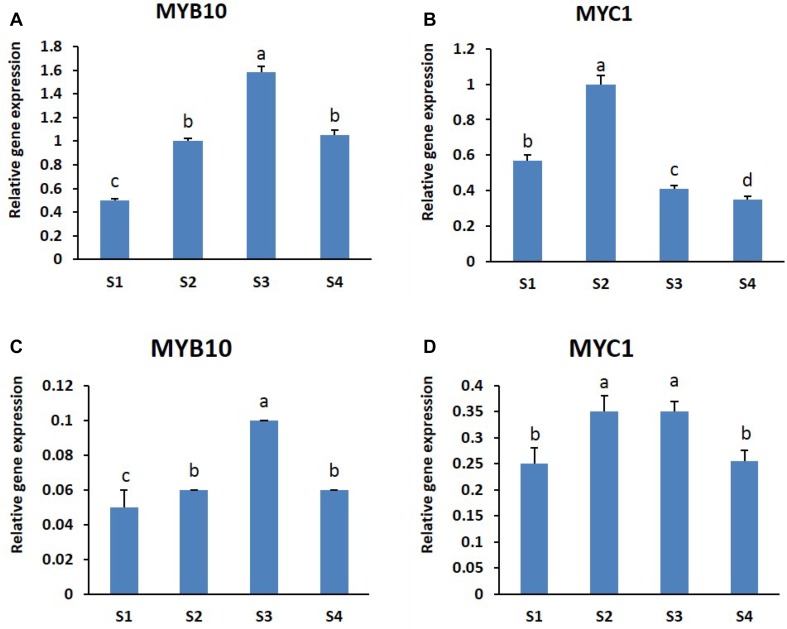
The expression levels of two transcription factors (TFs), MYB10 and MYC1, were measured using a real-time quantitative PCR (Fig. 5). The qRT-PCR results showed that both TFs were expressed at all developmental stages studied, in both cultivars. In ‘Nathasha’, the expression levels of MYB10 were low at the S1 stage and increased until S3, but showed a subsequent decline at stage S4 although expression levels at S4 were still higher than at S1. However, similar expression patterns were not observed for MYC1 (Fig. 5a, b). The expression pattern of MYB10 is consistent with the anthocyanin content observed at the different stages. Similarly, expression of MYB10 in ‘Rosalin’ was found to be more consistent with the corresponding changes in anthocyanin content at all stages than was the expression of MYC1 (Fig. 5c, d). However, we found higher expression levels of both TFs in the red-flowered ‘Nathasha’ than in the pink-flowered ‘Rosalin’.
Fig. 5.
Expression patterns of MYB10 and MYC1 in the two gerbera cultivars (a, b) ‘Nathasha’ and (c, d) ‘Rosalin’ at different flower developmental stages. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average means
Accumulation of anthocyanin in gerbera under low temperature
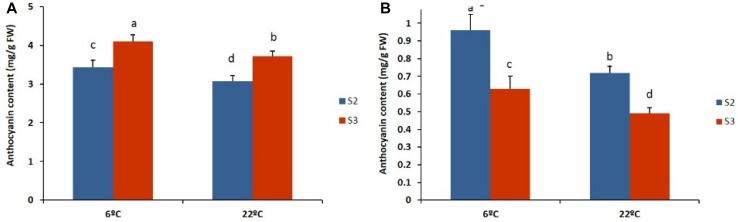
When flowers at developmental stages S2 and S3 were subjected to two different temperatures (6 and 22 °C), we observed differences in the anthocyanin content in both cultivars, with a significantly higher anthocyanin content being observed at 6 °C than at 22 °C (Figs. 6, 7).
Fig. 6.
Comparison of flower colors of the two different stages (S2 and S3) of the gerbera cultivars ‘Nathasha (a) and Rosalin (b)’ under the two different temperatures
Fig. 7.
Anthocyanin contents of the two different stages (S2 and S3) of the gerbera cultivars ‘Nathasha (a) and Rosalin (b)’ under the two different temperatures. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average anthocyanin content
Expression of anthocyanin biosynthesis genes exposed to low temperature
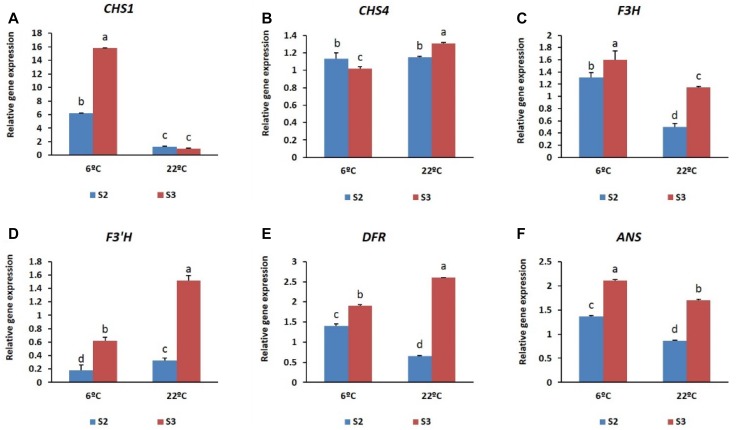
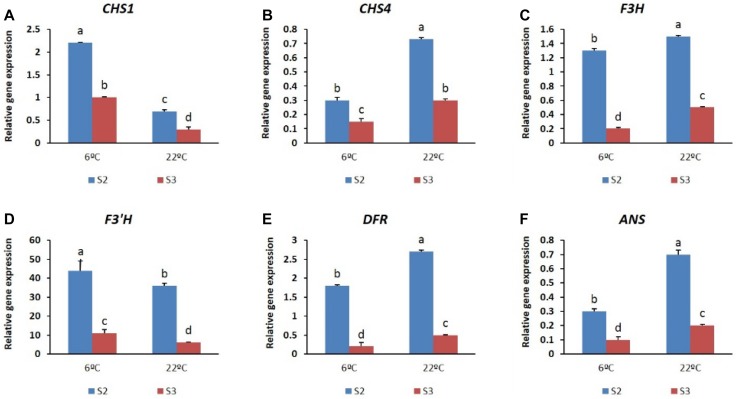
To measure the transcript levels of structural genes expressed at stages S2 and S3, the two cultivars were subjected to two temperatures (a low-temperature treatment at 6 °C and a high-temperature treatment at 22 °C). Differential expression of transcripts was evident from the qRT-PCR analysis. Briefly, in both the cultivars studied, all the structural genes analyzed were expressed at each of the different stages, and their expression was found to be temperature dependent. In ‘Nathasha’, the expression levels of CHS1, F3H, and ANS were found to be higher at 6 °C than at 22 °C at all stages, whereas the transcript levels of CHS4, F3′H, and DFR showed a stronger induction at 22 °C, particularly at the S3 stage (Fig. 8). As previously noted, the accumulation of anthocyanin was higher overall at 6 °C than at 22 °C and, based on these results, we suggest that the higher content of anthocyanin induced at 6 °C is associated with the higher transcriptional activation of CHS1, F3H, and ANS genes. Similarly, detection of higher anthocyanin content at S3 compared with S2 also reflects the observed gene expression patterns, as the transcript levels of the genes expressed at S3 were higher than at S2. In the case of ‘Rosalin’, plants exposed to a temperature of 6 °C during the flowering stages exhibited a higher anthocyanin content than did those exposed to a temperature of 22 °C. However, in plants subjected to 6 °C, higher amounts of anthocyanin were observed at S2 than at S3, which was associated with higher transcript levels of CHS1 expressed at S2 (Fig. 9). Overall, in both cultivars, our results show that temperature has an important role in the regulation of anthocyanin biosynthesis, which nevertheless was observed to be stage dependent.
Fig. 8.
Expression patterns of different anthocyanin structural genes (a–f) at the two different stages (S2 and S3) of gerbera cultivar ‘Nathasha’ under the two different temperatures. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average means
Fig. 9.
Expression patterns of different anthocyanin structural genes (a–f) at the two different stages (S2 and S3) of gerbera cultivar ‘Rosalin’ under the two different temperatures. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average means
Expression levels of the transcription factors under low temperature
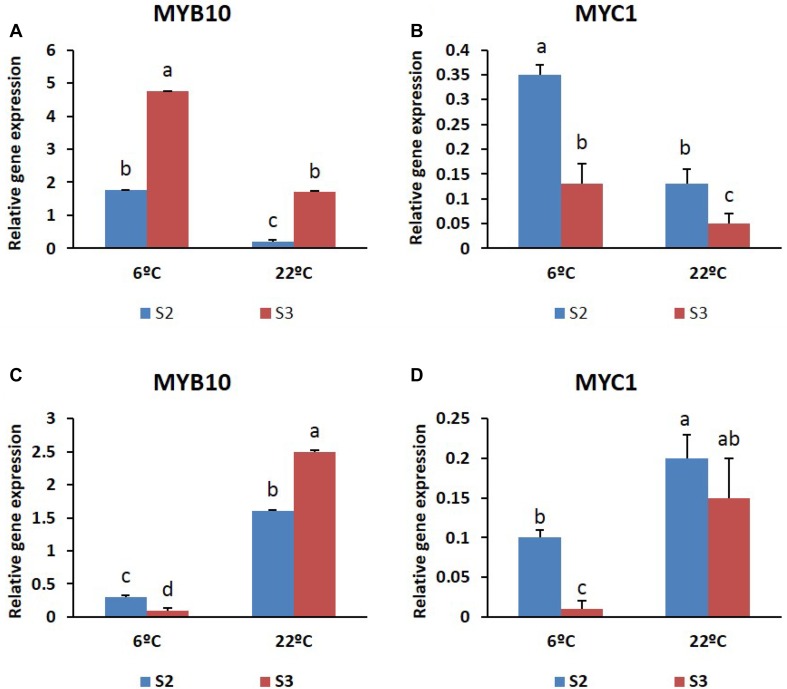
In this report, we observed temperature-dependent transcriptional activation of TFs; however, the responses tended to be flowering stage and cultivar dependent. For instance, in ‘Nathasha’, transcript levels of MYB10 and MYC1 were consistently higher at all stages when induced by low temperature (6 °C) (Fig. 10a, b). Accumulation of higher anthocyanin content in S3 at 6 °C was consistent with the higher expression levels of MYB10. Surprisingly, in ‘Rosalin’, transcript levels of the TFs were barely detectable at S2 or S3 at 6 °C, whereas high expression was observed at all stages at 22 °C (Fig. 10c, d). In addition, a relationship between transcriptional activation and anthocyanin content could not be established in this cultivar because, at 6 °C, transcript levels of the TFs were relatively low at S2, although the anthocyanin content was found to be high. This implies that the mechanisms underlying anthocyanin accumulation associated with transcriptional activation of the TFs are complex in the cultivar ‘Rosalin’.
Fig. 10.
Expression patterns of MYB10 and MYC1 at the two different stages (S2 and S3) of the two gerbera cultivars (a, b) ‘Nathasha’ and (c, d) ‘Rosalin’ under the two different temperatures. Data for each sample represent the average of three biological replicates, and bars indicate the standard deviation of the average means
Discussion
The diverse flower colors seen in many ornamental plants are generally influenced by differential expression of structural genes and TFs involved in the anthocyanin biosynthesis pathway (Nakatsuka et al. 2005; Zhao et al. 2012). Hence, analysis of the relevant differential expression patterns is an appropriate approach to understanding how flower color is associated with gene expression patterns in relation to developmental stages and environmental cues. In antirrhinum, petunia, and sacred lotus, the expression profiles and precise roles of structural genes and TFs determining flower pigmentation have been characterized (Martin et al. 1993; Quattrocchio et al. 1998; Collette et al. 2004; Wang et al. 2016). As gerbera has diverse flower colors, a number of reports have highlighted the use of gerbera as an Asteraceae model for studying the association between flower color and gene expression patterns during flower development (Helariutta et al. 1996; Elomaa et al. 1998; Laitinen et al. 2005; Deng et al. 2014; Bashandy et al. 2015). Although efforts have been made to elucidate the anthocyanin synthesis pathway in gerbera, the limited information currently available has hindered investigation of the complicated mechanisms underlying flower coloration. Therefore, in the present study, we examined the expression patterns of various structural genes (CHS1, CHS4, F3H, F3H, DFR, and ANS) and two TFs (MYB10 and MYC1) to determine their relationship with anthocyanin production in the two gerbera cultivars, ‘Nathasha’ and ‘Rosalin’. In addition, the effects of temperature on anthocyanin synthesis were also investigated in both cultivars by profiling the expression patterns of the same structural genes and TFs under different temperatures.
In this study, differences in anthocyanin content during flower development were observed in both cultivars. Overall, higher amounts of anthocyanin were recorded at the S3 flowering stage for ‘Nathasha’. The high anthocyanin content at this stage coincided with the detection of high transcript levels of ANS. Surprisingly, significantly lower amounts of anthocyanin were observed at stage S1 than at S3, despite higher expression levels of the structural genes (CHS1, CHS4, F3H, and DFR) at S1, which may be explained by the higher expression levels of ANS at S3 than at S1. Furthermore, the increase in anthocyanin content at S3 was marginally higher than at S4, which may also be due to the higher expression levels of ANS than of the other structural genes. The involvement of ANS in anthocyanin biosynthesis has been previously identified in gerbera (Wellmann et al. 2006) and other flowers (Rosati et al. 1999; Shimizu et al. 2011), although functional roles for CHS1, F3H, F3′H, and DFR in anthocyanin accumulation in gerbera have also been reported (Martens et al. 2002; Meng et al. 2004; Seitz et al. 2006; Deng et al. 2014; Bashandy et al. 2015). In ‘Rosalin’, all structural genes were expressed at all stages, although CHS1 showed a contrasting expression pattern, with an initial low expression at S1 that then increased linearly until S3, before decreasing at S4. A similar trend for CHS1 expression has been reported in the gerbera cultivars Terraregina and President (Deng et al. 2014), which have flower colors similar to those of ‘Rosalin’. Higher expression of all genes, except CHS1, at S1 is consistent with high anthocyanin content; however, their transcript levels at S3 were not consistent with the high anthocyanin content observed at this stage, and it is possible that the high anthocyanin content at S3 was due to the high expression of CHS1. The transcript levels of CHS4, F3H, F3′H, DFR, and ANS at S2 were significantly higher than those observed at S3, but the lower amounts of anthocyanin observed at S2 than at S3 may also be due to lower transcript levels of CHS1. Interestingly, although CHS1 and CHS4 are members of the CHS multigene family, CHS4 is unlikely to contribute to anthocyanin accumulation based on the comparison of their expression levels. Deng et al. (2014) also reported that although CHS4 was strongly expressed during petal development in gerbera, it did not contribute to pigmentation, and that instead it was CHS1 expression that determined pigmentation levels (Helariutta et al. 1993). In support of this, the temporal correlation between CHS1 expression during petal development and anthocyanin synthesis in gerbera has been reported (Helariutta et al. 1995). Hence, CHS1 may have a stronger influence on anthocyanin accumulation than other structural genes, at least in the ‘Rosalin’ cultivar. Taken together, our results suggest that ANS is more likely to be a key structural gene in ‘Nathasha’, whereas CHS1 may be more important in ‘Rosalin’. However, the accumulation of higher amounts of anthocyanin in ‘Nathasha’ may also be due to a cumulative effect of the high expression levels of all the other structural genes.
Analysis of the role of TFs in anthocyanin accumulation showed that, in ‘Nathasha’, the expression pattern of MYB10 was consistent with that of ANS at all stages; therefore, MYB10 may regulate ANS expression independently of other structural genes. Laitinen et al. (2008) also reported that overexpression of GMYB10 in gerbera upregulated late structural genes, including ANS. However, in ‘Rosalin’, MYB10 may directly regulate the expression of CHS1 upstream of anthocyanin synthesis, as suggested by the expression patterns of CHS1 and MYB10, particularly at S3 where the highest anthocyanin content was detected. As MYC1 was expressed at all stages in both cultivars, it may play a role in anthocyanin regulation by interacting with MYB10, as it has been shown that co-expression of MYB and MYC1 can regulate anthocyanin structural genes in gerbera (Elomaa et al. 1998, 2003).
The role of temperature in anthocyanin accumulation is well known in some flowers. High temperature is associated with a decrease in anthocyanin pigmentation in chrysanthemum (Nozaki et al. 2006), lily (Lai et al. 2011), and rose (Dela et al. 2003), whereas low temperatures have been reported to stimulate anthocyanin accumulation in petunia (Shvarts et al. 1997) and rose (Biran and Halevey 1973; Biran et al. 1973); however, the mechanism whereby temperature might influence anthocyanin accumulation in flowers is not known. To the best of our knowledge, the effects of low temperature on anthocyanin biosynthesis in gerbera have not been studied. In the present study, we investigated the effects of two different temperatures (6 and 22 °C) on anthocyanin pigmentation at two different stages (S2 and S3) of gerbera flower development to gain a better understanding of the mechanisms underlying anthocyanin accumulation in the cultivars ‘Nathasha’ and ‘Rosalin’. In both cultivars, exposure to low temperature (6 °C) led to higher anthocyanin accumulation than exposure to a higher temperature (22 °C). In ‘Nathasha’, the increase in anthocyanin levels in response to low-temperature exposure may be explained by the high expression levels of the CHS1, F3H, and ANS genes; however, the higher accumulation of anthocyanin at S3 compared with S2 may be attributable to the cumulative high expression of the structural genes (CHS1, F3H, F3′H, DFR, and ANS). Of the genes induced at 6 °C, CHS1 showed the highest induction, and its role in anthocyanin accumulation in gerbera has been previously documented. Strong induction of CHS by low-temperature treatment was reported in Arabidopsis and red orange (Leyva et al. 1995; Lo Piero et al. 2005), and upregulation of CHS under low temperature has also been reported for petunia flowers (Shvarts et al. 1997). Similarly, in ‘Rosalin’, an increase in anthocyanin content was observed at 6 °C, and this increase may be related to the high expression levels of CHS1. However, detection of higher anthocyanin levels at S2 than at S3 under the same temperature may also be explained by stronger activation of the other structural genes. Surprisingly, our results suggest that the higher expression of the genes induced at 22 °C did not affect anthocyanin content, which may be due to post-transcriptional regulation, as reported previously for gerbera (Deng et al. 2014). Overall, in both cultivars, low temperature-enhanced anthocyanin synthesis was associated mainly with CHS1, albeit likely complemented by the activity of other structural genes.
In ‘Nathasha’, enhancement of anthocyanin synthesis at low temperature may result from upregulation of TFs that bind to the promoters of anthocyanin structural genes. There is strong support for this argument because MYB10 and MYC1 showed stronger expression at 6 °C than at 22 °C, and the interactions between these TFs in anthocyanin accumulation has already been described for gerbera. In addition, upregulation of MYCs and MYBs has been shown to be modulated by temperature in various crops (Ban et al. 2007; Rowan et al. 2009; Lin-Wang et al. 2011). However, in ‘Rosalin’, the higher anthocyanin synthesis induced at 6 °C did not coincide with an increase in the expression of the two TFs, which was significantly lower at 6 °C. Accordingly, the molecular mechanisms by which TFs regulate anthocyanin biosynthesis and flower coloration in response to temperature remain largely unknown.
Conclusions
Accumulation of anthocyanin in the gerbera cultivars ‘Nathasha’ and ‘Rosalin’ starts at an early stage (S1) and reaches maximum levels during the middle stage (S3) of flower development. Differential expression of ANS and MYB10 during flower development was found to correlate with the varying anthocyanin content in ‘Nathasha’, whereas expression of CHS1, MYC1, and MYB10 was observed for ‘Rosalin’. The higher anthocyanin content in ‘Nathasha’ than in ‘Rosalin’ may be attributable to the cumulative expression levels of all structural genes and TFs. Our results showed that low-temperature exposure enhanced anthocyanin levels at all stages studied by upregulation of CHS1 and the TFs MYB10 and MYC1 in ‘Nathasha’, and CHS1 in ‘Rosalin’. In addition, the expression patterns of the structural genes and TFs induced by low temperature were cultivar and stage dependent. Although in this study we uncovered some of the roles of anthocyanin structural genes and TFs in anthocyanin biosynthesis during different developmental stages and under different temperatures, the potential mechanisms underlying the regulation of anthocyanin biosynthesis are likely to be complex. Further functional studies of key genes involved in anthocyanin biosynthesis are needed and should be explored using genetic transformation or gene silencing techniques.
Abbreviations
- PAL
Phenylalanine ammonia lyase
- CHS
Chalcone synthase
- F3H
Flavanone-3-hydroxylase
- F3′H
Flavonoid 3′ hydroxylase
- ANS
Anthocyanidin synthase
- DFR
Dihydroflavonol-4-reductase
- UFGT
UDP-glycose flavonoid 3-O-glycosyl transferase
- bHLH
Basic helix–loop–helix
- TF
Transcription factor
Author contributions
AHN designed the study, conducted the experiments, and wrote the manuscript. CKK supervised experiments at all stages and performed critical revisions of the manuscript. DYP and KIP were involved in qRT-PCR analysis and interpretation of the data. All authors read and approved the final manuscript.
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET), through the Agri-Bio Industry Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (Grant #: 315002-5). The funding body did not play a role in the design of the study and writing of the manuscript.
Compliance with ethical standards
Conflict of interest
There is no conflict of interests.
Contributor Information
Aung Htay Naing, Email: aunghtaynaing2005@gmail.com.
Chang Kil Kim, Email: ckkim@knu.ac.kr.
References
- Ai TN, Naing AH, Arun M, Kim CK. Sucrose-induced anthocyanin accumulation in vegetative tissue of Petunia plants requires anthocyanin regulatory transcription factor genes. Plant Sci. 2016;252:144–150. doi: 10.1016/j.plantsci.2016.06.021. [DOI] [PubMed] [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Hideo Bessho H, Moriguchi T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red colouration in apple skin. Plant Cell Physiol. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- Bashandy H, Pietiäinen M, Carvalho E, Lim K-J, Elomaa P, Martens S, Teeri TH. Anthocyanin biosynthesis in gerbera cultivar ‘Estelle’ and its acyanic sport ‘Ivory’. Planta. 2015;242:601–611. doi: 10.1007/s00425-015-2349-6. [DOI] [PubMed] [Google Scholar]
- Biran I, Halevy AH. Effect of short-term heat and shade treatments on petal colour of ‘baccara’ roses. Physiol Plant. 1973;31:180–185. doi: 10.1111/j.1399-3054.1974.tb03687.x. [DOI] [Google Scholar]
- Biran I, Enoch H, Zieslinand N, Helevy A. The influence of light intensity, temperature and carbon dioxide concentration on anthocyanin content and bluing of ‘baccara’ roses. Sci Hortic. 1973;1:157–164. doi: 10.1016/0304-4238(73)90026-5. [DOI] [Google Scholar]
- Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol. 2005;8:272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Chandler S, Tanaka Y. Genetic modification in floriculture. Crit Rev Plant Sci. 2007;26:169–197. doi: 10.1080/07352680701429381. [DOI] [Google Scholar]
- Collette VE, Jameson PE, Schwinn KE, Umaharan P, Davies KM. Temporal and spatial expression of flavonoid biosynthetic genes in flowers of Anthurium andraeanum. Physiol Plant. 2004;122:297–304. doi: 10.1111/j.1399-3054.2004.00402.x. [DOI] [Google Scholar]
- Dela G, Or E, Ovadia A, Nissim-Levi A, Weiss D, Oren-Shamir M. Changes in anthocyanin concentration and composition in ‘jaguar’ rose flowers due to transient high-temperature conditions. Plant Sci. 2003;164:333–340. doi: 10.1016/S0168-9452(02)00417-X. [DOI] [Google Scholar]
- Deng X, Bashandy H, Ainasoja M, Kontturi J, Pietiäinen M, Laitinen R, Albert VA, Valkonen JP, Elomaa P, Teeri TH. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytol. 2014;201:1469–1483. doi: 10.1111/nph.12610. [DOI] [PubMed] [Google Scholar]
- Elomaa P, Mehto M, Kotilainen M, Helariutta Y, Nevalainen L, Teeri TH. A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol- 4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae) Plant J. 1998;16:93–99. doi: 10.1046/j.1365-313x.1998.00273.x. [DOI] [PubMed] [Google Scholar]
- Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RAE, Teeri TH. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 2003;133:1831–1842. doi: 10.1104/pp.103.026039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annu Rev Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Guo N, Cheng F, Wu J, Liu B, Zheng SN, Liang JL, Wang XW. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014;15:426. doi: 10.1186/1471-2164-15-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HV. A story of the cultivated Gerbera. New Plantsman. 1999;6:85–95. [Google Scholar]
- Helariutta Y, Elomaa P, Kotilainen M, Seppeanen P, Teeri TH (1993) Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var Regina (Compositae). Plant Mol Biol. 22:183–93 [DOI] [PubMed]
- Helariutta Y, Elomaa P, Kotilainen M, Griesbach RJ, Schreoder J, Teeri TH. Chalcone synthase-like genes active during corolla development are differentially expressed and encode enzymes with different catalytic properties in Gerbera hybrida (Asteraceae) Plant Mol Biol. 1995;28:47–60. doi: 10.1007/BF00042037. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Kotilainen M, Elomaa P, Kalkkinen N, Bremer K, Teeri TH, Albert V. Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: evolution with substrate change and catalytic simplification. Proc Nat Acad Sci USA. 1996;93:9033–9038. doi: 10.1073/pnas.93.17.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lai YS, Yamagishi M, Suzuki T. Elevated temperature inhibits anthocyanin biosynthesis in the tepals of an Oriental hybrid lily via the suppression of LhMYB12 transcription. Sci Hortic. 2011;132:59–65. doi: 10.1016/j.scienta.2011.09.030. [DOI] [Google Scholar]
- Laitinen RAE, Immanen J, Auvinen P, Rudd S, Alatalo E, Paulin L, Ainasoja M, Kotilainen M, Koskela S, Teeri TH. Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae) Genom Res. 2005;15:475–486. doi: 10.1101/gr.3043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RA, Ainasoja M, Broholm SK, Teeri TH, Elomaa P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J Exp Bot. 2008;59:3691–3703. doi: 10.1093/jxb/ern216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A, Jarillo JA, Salinas J, Martinez-Zapater JM. Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995;108:39–46. doi: 10.1104/pp.108.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Micheletti D, Palmer J. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Env. 2011;34:1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x. [DOI] [PubMed] [Google Scholar]
- Lo Piero AR, Puglisi I, Rapisarda P, Petrone G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem. 2005;53:9083–9088. doi: 10.1021/jf051609s. [DOI] [PubMed] [Google Scholar]
- Martens S, Teeri T, Forkmann G. Heterologous expression of dihydroflavonol 4-reductases from various plants. FEBS Lett. 2002;531:453–458. doi: 10.1016/S0014-5793(02)03583-4. [DOI] [PubMed] [Google Scholar]
- Martin C, Gerats T. The control of pigment biosynthesis genes during petal development. Plant Cell. 1993;5:1253–1264. doi: 10.1105/tpc.5.10.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XC, Xing T, Wang X. The role of light in the regulation of anthocyanin accumulation in Gerbera hybrid. Plant Growth Reg. 2004;44:243–250. doi: 10.1007/s10725-004-4454-6. [DOI] [Google Scholar]
- Nakatsuka T, Nishihara M, Mishiba K, Yamamura S. Temporal expression of flavonoid biosynthesis related genes regulates flower pigmentation in gentian plants. Plant Sci. 2005;168:1309–1318. doi: 10.1016/j.plantsci.2005.01.009. [DOI] [Google Scholar]
- Nozaki K, Takamura T, Fukai S. Effects of high temperature on flower colour and anthocyanin content in pink flower genotypes of greenhouse chrysanthemum (Chrysanthemum morifolium Ramat.) J Hortic Sci Biotechnol. 2006;81:728–734. doi: 10.1080/14620316.2006.11512130. [DOI] [Google Scholar]
- Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Plasmeijer J, Yanai Y (2012) Floriculture Products Report 2012. Issue No. M12 (2011), 9 January 2012. Market News Service of International Trade Center
- Quattrocchio F, Wing JF, van der Woude K, Mol JNM, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998;13:475–488. doi: 10.1046/j.1365-313X.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati C, Cadic A, Duron M, Ingouff M, Simoneaub P. Molecular characterization of the anthocyanidin synthase gene in Forsythia × intermedia reveals organ-specific expression during flower development. Plant Sci. 1999;149:73–79. doi: 10.1016/S0168-9452(99)00146-6. [DOI] [Google Scholar]
- Rowan DD, Cao M, Lin-Wang K. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009;182:102–115. doi: 10.1111/j.1469-8137.2008.02737.x. [DOI] [PubMed] [Google Scholar]
- Schwinn KE, Davies KM. Flavonoids. In: Davies K, editor. Plant pigments and their manipulation. Oxford: Blackwell Publishing Ltd; 2004. pp. 92–149. [Google Scholar]
- Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol Biol. 2006;61:365–381. doi: 10.1007/s11103-006-0012-0. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Ohnishi N, Morikawa N, Ishigami A, Otake S, Rabah IO. A 94-bp deletion of anthocyanidin synthase gene in acyanic flower lines of lisianthus [Eustoma grandiflorum. (Raf.) Shinn] J Jpn Soc Hortic Sci. 2011;80:434–442. doi: 10.2503/jjshs1.80.434. [DOI] [Google Scholar]
- Shvarts M, Borochov A, Weiss D. Low temperature enhances petunia flower pigmentation and induces chalcone synthase gene expression. Physiol Plant. 1997;99:67–72. doi: 10.1111/j.1399-3054.1997.tb03432.x. [DOI] [Google Scholar]
- Takos AM, Jaffe´ FW, Jacob SR, Bogs J, Robinson SP, Walker AR. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Peng Z, Zhang J, Song T, Wan H, Zhang M, Yao Y. McMYB10 regulates coloration via activating McF3′H and later structural genes in ever-red leaf crabapple. Plant Biotech J. 2015;13:948–961. doi: 10.1111/pbi.12331. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shi S, Zhou Y, Zhou Y, Yang J, Tang X. Genomewide identification and characterization of GRAS transcription factors in sacred lotus (Nelumbo nucifera) Peer J. 2016;4:e2388. doi: 10.7717/peerj.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann F, Griesser M, Schwab W, Martens S, Eisenreich W, Matern U, Lukacin R. Anthocyanidin synthase from Gerbera hybrida catalyzes the conversion of (+)-catechin to cyanidin and a novel procyanidin. FEBS Letts. 2006;580:1642–1648. doi: 10.1016/j.febslet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Wessinger CA, Rausher MD. Predictability and irreversibility of genetic changes associated with flower color evolution in Penstemon barbatus. Evolution. 2014;68:1058–1070. doi: 10.1111/evo.12340. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K. Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 2010;51:463–474. doi: 10.1093/pcp/pcq011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Martin C. Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol. 2014;19:81–90. doi: 10.1016/j.pbi.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Zhao DQ, Tao J, Han CX, Ge JT. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.) Mol Biol Rep. 2012;39:11263–11275. doi: 10.1007/s11033-012-2036-7. [DOI] [PubMed] [Google Scholar]