Abstract
Aging associated DNA hypomethylation of LINE-1 and Alu retroelements may be a crucial determinant of loss of genomic integrity, deterioration and cancer. In peripheral blood LINE-1 hypomethylation has been reported to increase during aging, but other studies did not observe significant changes. We hypothesized that these apparently inconsistent reports might relate to differences between cellular and cell-free DNA. Using the technique of idiolocal normalization of real-time methylation-specific PCR (IDLN-MSP) for genetic imbalanced DNA specimens we obtained evidence that LINE-1 hypomethylation in cell-free DNA, but not cellular DNA from peripheral blood is an epigenetic biomarker for human aging. Furthermore, hypomethylation of cell-free DNA is more extensive in smokers, suggesting that it might be used as a surrogate marker for monitoring the improvement of smoking-induced adverse effects after cancelling smoking.
Keywords: Epigenetics, DNA methylation, Cell-free DNA, Retroelements, LINE-1
1. Introduction
Long Interspersed Nuclear Elements (LINEs) and Short Interspersed Nuclear Elements (SINEs) are crucial contributors to the dynamics, plasticity and integrity of the human genome. LINE-1s are the only currently active autonomous mobile DNA elements in humans with at least 100 potential mobile copies in any individual diploid genome (Goodier, 2016). They have massively expanded throughout evolution with roughly 500,000 copies occupying about 17 % of the human DNA; all but around hundred copies are inactivated by truncations or mutations (Goodier, 2016). Alu elements are small dimeric SINE retroelements with almost one million genomic copies. They lack a coding sequence and hence depend on an active LINE-1 retrotransposition machinery to become mobile (Dewannieux et al., 2003). Noteworthy active LINE-1s are able to mediate high rate Alu transposition (Dewannieux et al., 2003). LINE-1s retrotranspose by a “copy-and-paste” process which involves LINE-1 DNA endonuclease activity to nick target DNA at the site of insertion, thereby endangering genome integrity. Recombination events and insertions by activated LINE-1 and Alu elements are thought to contribute to a significant portion of human genetic diseases and cancer (Deininger and Batzer, 1999). To date, 124 LINE-1-mediated insertions have been reported that result in human genetic diseases. In addition a few LINE-1 insertions have been found which interrupt tumor suppressor genes in cancer. Consequently, it has been postulated that retroelements contribute to the development and progression of cancer (Hancks and Kazazian, 2016).
DNA methylation is an epigenetic mechanism capable of repressing gene expression, which might originally have been evolved as a host defense mechanism against transposable ”parasitic” genetic elements to protect functional integrity of the genome (Yoder et al., 1997). Indeed, more than 90% of all genomic differentially methylatable CpGs are located in CpG rich sequences of transposable repetitive elements, including LINE-1 and Alu sequences (Bollati et al., 2009). Methylation of CpGs in the LINE-1 promoter silences LINE-1 expression in differentiated somatic cells (Yoder et al., 1997, Steinhoff and Schulz, 2003, Hancks and Kazazian, 2012) and absence of DNA methylation is permissive for LINE-1 transcription (Bourchis and Bestor, 2004).
LINE-1 hypomethylation is common in cancer, an age related disease characterized by the accumulation of mutations and epimutations. For instance, it occurs in > 90% of urothelial carcinomas (UC), even at early stages (Neuhausen et al., 2006), going along with increased transcription of full-length LINE-1 elements (Kreimer et al., 2013). Furthermore, it is associated with chromosomal abnormalities developing in urothelial carcinoma and expanding during disease progression (Simon et al., 1998). Clear evidence from experimental tumor models demonstrates that LINE-1 retrotransposition indeed contributes to genetic instability in vivo (Symer et al., 2002) and that DNA hypomethylation is able to promote chromosomal instabilities and tumors (Gaudet et al., 2003).
A general decrease of methylcytosine also occurs with age (Wilson and Jones, 1983, Wilson et al., 1987) which cannot be explained by single-copy gene methylation, since these account for less than 5% of human DNA (Hoffmann and Schulz, 2005). Furthermore, it is inferred that such a general loss of 5-methylcytosine during aging may de-suppress silenced LINE-1s and increase genome instability (Gravina and Vijg, 2010). Accordingly, genome instability has been implicated as a cause of aging since the late 1940s (Vijg and Suh, 2013), and is a hallmark of cancer (Hanahan and Weinberg, 2011). Thus, it remains a question of crucial importance to which extent hypomethylation of LINE-1 and Alu elements develops in the course of aging since it may constitute a determinant for aging associated loss of genomic integrity, deterioration and cancer.
Previous studies on the association of age with Alu and LINE-1 methylation have yielded conflicting results (Bollati et al., 2009). In a comprehensive study using blood samples from 718 elderly male subjects between 55 and 92 years, Bollati et al. found that Alu methylation significantly decreases during aging whereas LINE-1 methylation does not. In a more recent study, LINE-1 sequences were found demethylated with age in white blood cells, whereas smoking and drinking status had no significant effect on LINE-1 hypomethylation (Cho et al., 2015).
Because of these apparent inconsistent results in previous studies and the high significance and important biological implications of aging-associated LINE-1 hypomethylation, we decided to measure LINE-1 and Alu methylation both in cellular and in cell-free DNA from peripheral blood. Cell-free DNA derives from apoptotic and necrotic cells (Gravina et al., 2016), and accordingly, this DNA fraction increases in cancer patients as a consequence of more frequent cell-death events (Stroun et al., 1989). Notably, we have previously observed significant differences between the LINE-1 methylation profiles of cell-free and cellular DNA even in healthy individuals (Ghanjati et al., 2014). We therefore hypothesized that the conflicting findings in previous studies might be explained, at least in part, by inter-cohort variations between the relative amounts of the two DNA fractions and their varied degree of LINE-1 hypomethylation.
This approach was facilitated by the technique of Idiolocal Normalized Methylation-Specific PCR (IDLN-MSP) through which we are able to compare DNA methylation differences in DNA samples with genomic variation (Ghanjati et al., 2014, Santourlidis et al., 2016), as mentioned above, another feature associated with human aging.
Here we present evidence for that LINE-1 and ALU DNA hypomethylation of cell-free DNA is an epigenetic biomarker of human aging.
2. Material and methods
2.1. Blood samples
All methods were carried out in accordance with relevant guidelines and regulations. We confirm that the experimental protocols were approved and informed consent was obtained from all participants. Whole blood samples for cellular DNA isolation and measurement of LINE-1 DNA methylation were collected from 166 healthy individuals, of which 104 were male and 62 female. They were aged between 23 and 61 years, with a mean age of 40 ± 9.2 (SD) years. For measurement of Alu DNA methylation of cellular DNA, 63 of the same whole blood samples were used from 38 male and 25 female probands, aged between 23 and 61 years, with a mean age of 40 ± 11 years.
Blood plasma samples for isolation of cell-free DNA and measurement of LINE-1 and Alu DNA methylation were collected from 62 probands of a different cohort. This comprised 42 male and 20 female probands, aged between 18 and 64 years, with a mean age of 39,39 ± 14,06 years.
From each proband, 7 ml blood was collected by venous phlebotomy in EDTA tubes. After a density gradient centrifugation, buffy coat and plasma were extracted and stored until DNA extraction. All samples were coded and frozen at -20 °C.
2.2. DNA isolation and bisulfite conversion
Cellular DNA isolation from the stored samples was performed using the QIAamp DNA blood kit (Qiagen, Hilden, Germany). Cell-free DNA Isolation from urine and blood samples was carried out by using the QIAamp Circulation Nucleic Acid Kit (Qiagen). Samples were centrifuged for 15 min at 4000 rpm before the isolation. Cell lysis was carried out at 60 °C for 40 min and the QIAGEN mini columns were incubated for 5 min at RT after addition of AVE buffer. For both methods the final elution was performed in 35 µl of elution buffer. For bisulfite conversion 500 ng cellular DNA of blood/urine samples and 100 ng of cell-free DNA from blood plasma/urine samples were used. Bisulfite conversion was carried out with the EpiTect Bisulfite Kit (Qiagen).
2.3. Relative Quantification of LINE-1 and Alu methylation in cellular and cell-free DNA
Converted DNA served as template for amplification of methylated LINE-1 and Alu sequences in a normalized real-time MSPCR approach for genetically imbalanced DNA specimens as described (Santourlidis et al. 2016). Quantification was done using Step One Plus Real Time PCR System (Applied Biosystems, Foster City, United States of America) with the following primers for LINE-1 (forward : 5′-GCGCGAGTCGAAGTAGGGC-3′; revers : 5′-CTCCGACCAAATATAAAATATAATCTCG-3′) and Alu (forward: 5′-ATTTTAGTATTTTGGGAGGTCGAGGC-3′; reverse: 5′-GCAATCTCGACTCACTACAAA CTCCG-3′). Amplification of an adjacent region without CpGs was used to normalize the amount of LINE-1 / Alu DNA methylation. The primers used for normalization were for LINE-1: forward: 5′-AGGTTTTATTTTTGGGGGTAGGGTATAG-3′; reverse: 5′-CCCCTACTAAAAAA TACCTCCCAATTAAAC-3′ and for Alu: forward: 5′-GGGTGGATTATGAGGTTAGGAGAT-3′; reverse: 5′-CATTCTCCTACCTCAACCTCCC-3′. The following annealing temperatures were chosen: LINE-1: 53 °C; LINE-1 control: 60 °C; Alu: 58 °C and Alu control: 61 °C. The amplification conditions were denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, TM°C for 40 s, and 72 °C for 15 s. DNA Methylation levels were calculated by the ΔΔCT method using StepOne Software 2.2 (Applied Biosystems).
2.4. Bisulfite genomic sequencing
For bisulfite sequencing of the LINE-1 promoter fragment the following primers were used: forward: 5′-GGT TTA TTT TAT TAG GGA GTG TTA G-3′ and reverse: 5′-ACA AAA ACA AAC AAA CCT CC-3′. The PCR cycling conditions were the same as described for real-time MSPCR with an annealing temperature of 51 °C. Amplification products were cloned into pCR2.1 vector using the TA Cloning Kit (Invitrogen, Carlsbad, United States) according to the manufacturer’s instructions. On average 30 clones were sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) on a DNA analyzer 3700 (Applied Biosystems) with M13 primer to obtain a representative methylation profile of each sample. The sequenced LINE-1 segment is part of the CpG rich LINE-1 promoter (Ref.: Human Transposon L1.2, NCBI Nucleotide Gen Bank accession number: M80343.1).
3. Results
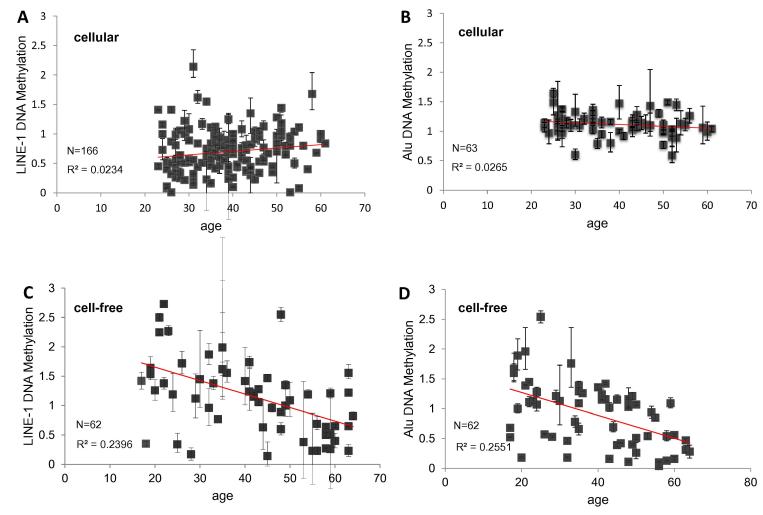
This study was carried out with blood samples of healthy donors from German population, aged between 23 and 61 years. The probands had no pre-existing conditions. LINE-1 and Alu DNA methylation of cellular and cell-free DNA was measured by Idiolocal Normalized real-time Methylation-Specific PCR as described (Santourlidis et al. 2016). Relative DNA methylation levels ranged from 0.01 to 2.14 for LINE-1 and from 0.59 to 1.64 for Alu elements in cellular DNA and between 0.14 to 2.73 for LINE-1 and 0.04 to 2.54 for Alu elements in cell-free DNA (Fig. 1).
Fig. 1.
Relative Quantification of LINE-1 and Alu DNA methylation in cellular and cell-free DNA. Correlation between age and DNA Methylation of LINE-1 and Alu retroelements in cellular DNA derived from PBMC samples (A, B) and cell-free DNA derived from blood plasma (C, D) of the indicated numbers of donors (N). The regression lines are colored in red and represent the Pearson’s correlation coefficients (R2).
3.1. No association between DNA methylation of LINE-1 and Alu retroelements in cellular DNA from peripheral blood and age
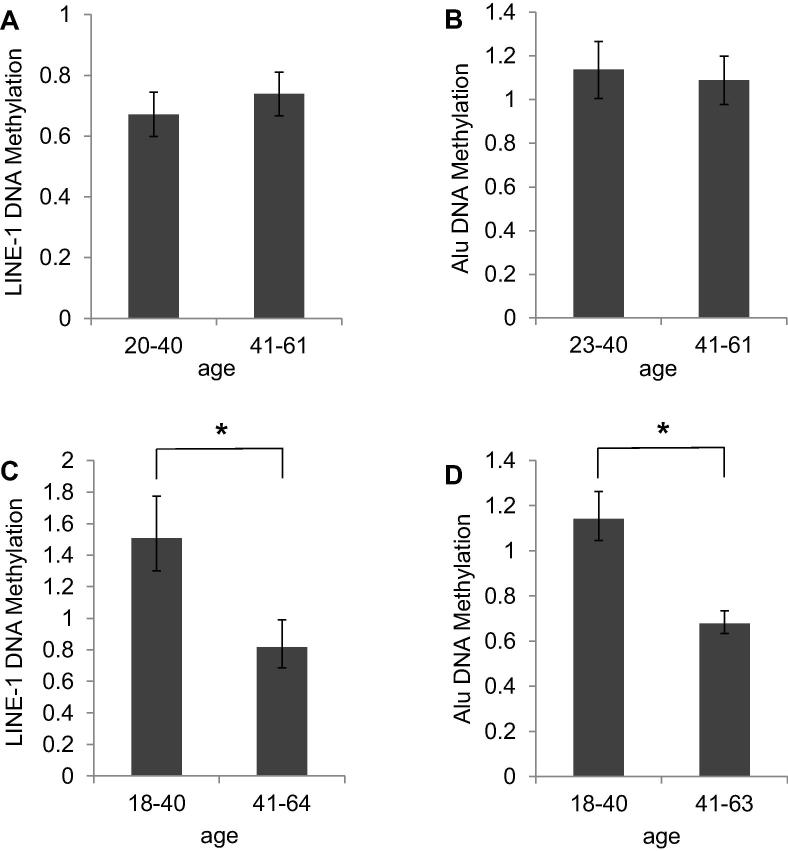
In cellular DNA, neither LINE-1 DNA methylation nor Alu DNA methylation were significantly correlated with age. The respective Pearson’s correlation coefficients were r2 = 0.0234 for LINE-1 and r2 = 0.0265 for Alu elements, respectively (Fig. 1, A/B). Also after separating the sample cohorts into two age subgroups (20–40, 41–61; 23–40, 41–61), no significant differences were observed (Fig. 2, A/B). For LINE1 methylation the mean values were 0.67 and 0.74 for the younger and older group, respectively (p = 0.21). For Alu methylation the respective mean values were 1.14 and 1.08 (p = 0.33). Thus DNA methylation of LINE-1 and Alu retrotransposons in cellular DNA of peripheral blood remains stable during aging.
Fig. 2.
Mean values of LINE-1 and Alu DNA Methylation within age subgroups. Age subgroups associated global DNA methylation at LINE-1 and Alu retrotransposons in cellular DNA of PBMCs (A, B) and peripheral blood cell-free DNA (C, D), respectively. The mean value of quantification has been applied in two-sample Student’s t-test with the significance threshold p > 0.05.
3.2. DNA methylation of LINE-1 and Alu retroelements decreases with age in cell-free DNA from peripheral blood
DNA methylation of LINE-1 and Alu retroelements in cell-free DNA correlated much better with age, with Pearson’s correlation coefficients of r2 = 0.24 and r2 = 0.26, respectively (Fig. 1C/D). The differences are accentuated by separation of the sample cohorts into two age subgroups (18–40, 41–64 years for LINE-1; 18–40, 41–63 years for Alu) (Fig. 2C/D). Mean values of LINE-1 DNA methylation levels were 1.44 for younger individuals and 0.85 for older probands aged between 41 and 64 years (p = 0.00002). Mean values of Alu DNA methylation were 1.14 for younger individuals and 0.68 for older probands (p = 0.02).
3.3. Age associated LINE-1 and Alu hypomethylation of cell-free DNA from peripheral blood plasma is more pronounced in smokers
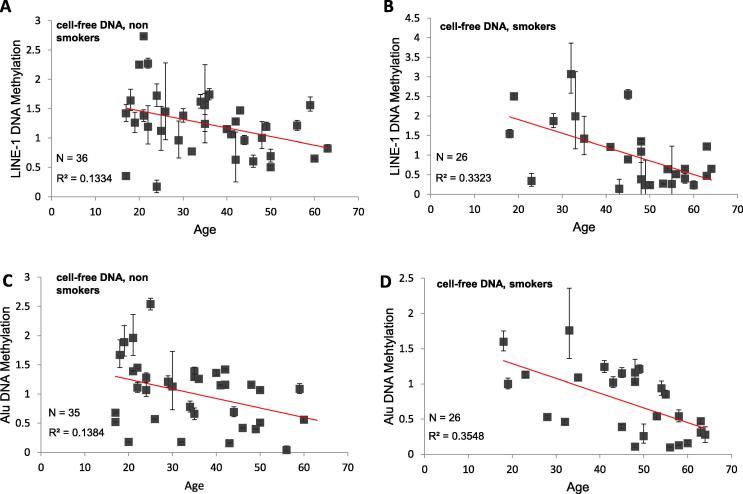
Our data show a more pronounced decrease of DNA methylation of LINE-1 and Alu retroelements with age in cell-free DNA of smokers, with Pearson’s correlation coefficients of r2 = 0.33 and r2 = 0.35, respectively (Fig. 3B/D).
Fig. 3.
Relative Quantification of LINE-1 DNA and Alu Methylation of cell-free blood plasma DNA in non-smokers and smokers. Correlation between age and DNA Methylation of LINE-1 and Alu in cell-free DNA of non smoking individuals (A/C) and smoking probands (B/D). The graphs show the correlation of DNA methylation in relation to subject’s age. The regression lines are colored in red and represent the Pearson’s correlation coefficients (R2).
3.4. LINE-1 DNA methylation differs between cellular and cell-free DNA in a healthy individual
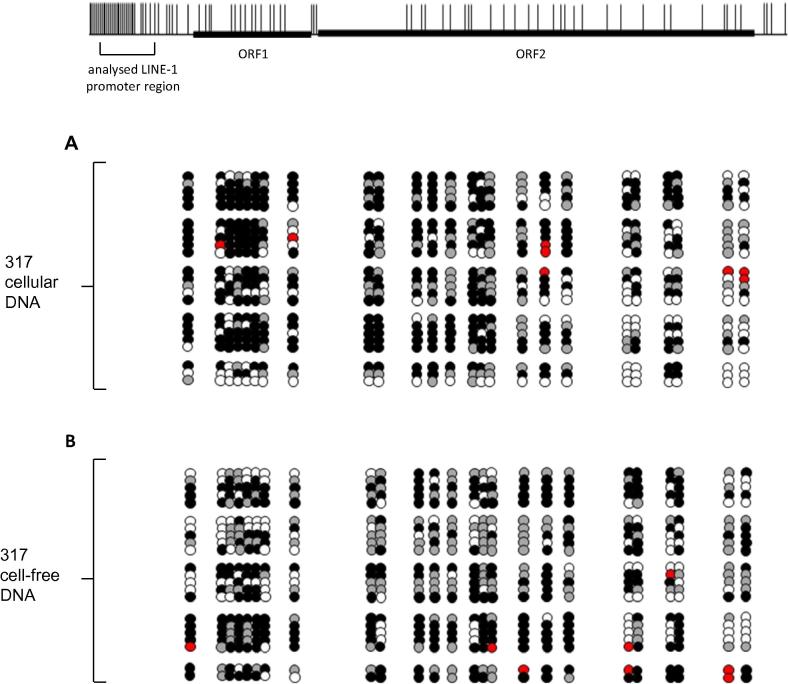
In order to validate the occurrence and define the pattern of DNA methylation differences in one retroelement between cellular and cell-free DNA we examined urinary cellular and cell-free DNA of a 60 year old healthy individual by bisulfite sequencing of LINE-1 promoter.
The analyzed CpG-rich LINE-1 promoter region contains 25 CpGs. To obtain a representative LINE-1 DNA methylation profile we bisulfite sequenced 23 clones from cellular DNA and 22 clones from cell-free DNA (Fig. 4). In both cases our analysis revealed dense CpG methylation within the analyzed 5′ regulatory region. However, in cellular DNA 326 of the 575 examined cytosines (56.7%) were found to be methylated, whereas in cell-free DNA in total 279 of the 550 examined cytosines (50.7%) were methylated. Noteworthy, the CpG positions −9, −16, −18, −19, −21, −22, −23, −25, −26, −28, −29 and −30 relative to the first ATG of LINE-1 open reading frame 1 (ORF1) appear to be more susceptible to demethylation in this cell-free DNA sample. In cellular DNA we detected one complete unmethylated LINE-1 sequence which is 98–99% identical to one known full-length LINE-1 sequence with intact open reading frames, as kindly provided by Dr. Goodier (Kazazian lab, Philadelphia). This detailed analysis exemplifying illustrates that differences of LINE-1 methylation between cellular and cell-free DNA exist in a healthy individual.
Fig. 4.
Detailed analysis of LINE-1 promoter CpG methylation derived from urinary cellular (A) and cell-free DNA (B) of a healthy individual. Above a schematic representation of a full-length LINE-1 retroelement including the distribution of CpG dinucleotides (short vertical lines), the open reading frames 1 and 2 (ORF1/2) and indicated by a square bracket the analyzed region is shown. Detailed CpG methylation profiles of the CpG rich 5′ regulatory region of LINE-1 gene are documented as they were revealed by bisulfite sequencing. Filled circles stand for methylated CpG dinucleotides. White circles stand for unmethylated CpGs. Grey circles stand for undefined CpG methylation status and red circles stand for an A or G nucleotide within the CpG position.
4. Discussion
Our data do not show a significant age associated hypomethylation of LINE-1 and Alu retroelements in cellular DNA from peripheral blood. This is in accordance with published data which show that age has no detectable effect on LINE-1 methylation in peripheral blood cells (El-Maarri et al., 2011). This cellular DNA fraction originates from leukocytes, i.e. neutrophils, lymphocytes, monocytes, eosinophils and basophiles (El-Maarri et al., 2011). We provide evidence that methylation of repetitive DNA elements is stable in these nucleated peripheral blood cells in individuals of 20–61 years of age. Thus this constitutes not an aging associated, alterable epigenetic component which might hypothetically contribute to immunosenescence. In our view this appears in accordance with the notion of highly differentiated and functionally specialized but relative short living cells which are continuously replaced by new ones. About 65% of them are thought to have life spans from few hours to weeks (El-Maarri et al., 2011). Nevertheless further studies should aim to show whether LINE-1 methylation also remains stable in very old, i.e. 70–90 years old individuals.
In contrast, we observed a clear age associated decline in the methylation of retroelements in cell-free DNA from peripheral blood. In healthy individuals this DNA fraction is suggested to derive from apoptotic cells, necrotic cells and from active cellular secretion of newly synthesized DNA (Bronkhorst et al., 2016). Based on this we conclude that cellular genome-wide hypomethylation occurs in an age-dependent fashion in endothelial, epithelial and other body cells from various organs. Subsequently, this event could lead to retrotransposon activation, induction of genomic instability (Ghanjati et al., 2014) and aberrant transcription patterns (Wolff et al., 2010). This may constitute one factor of aging associated increase of apoptosis which for example has been shown to occur in normal and pathological liver aging (Zhong et al., 2017). Genomic instability has long been recognized as a causal factor in aging. Noteworthy this hypothesis would imply that the amount of cell-free DNA in peripheral blood should increase with age and further studies should aim to show whether this could constitute a simple genetic marker of human aging.
In accordance with these observations we found increased hypomethylation of LINE-1 and Alu elements in the cell-free DNA of smokers. It is well established that smoking induces epigenetic alterations in global DNA methylation (Wangsri et al., 2012, Shigaki et al., 2012), e.g. LINE-1 hypomethylation in respiratory epithelia exposed to cigarette smoke (Liu et al., 2010). Furthermore, harmful components of smoke, e.g. nicotine, induce apoptosis in various cell types (Zeidler et al., 2007). Therefore it is a plausible assumption that smoking results in increased apoptosis and release of hypomethylated cell-free DNA into the blood stream, explaining our findings. We thus suggest that the amount and the degree of demethylation of cell-free DNA from peripheral blood are surrogate markers of smoking-induced adverse effects on human health that might be used for monitoring its improvement after cancelling smoking.
Consequently, we conclude here that hypomethylation of cell-free DNA is a biomarker of human age.
Acknowledgements
In Memoriam and in Gratitude to Professor Rolf Ackermann.
The authors are grateful to Wolfgang Schulz (Department of Urology, Medical Faculty, Heinrich Heine University, Düsseldorf, Germany) for revising the paper. They thank Johannes Fischer (Institute of Transplantation Diagnostics and Cell Therapeutics, Medical Faculty, Heinrich Heine University, Düsseldorf, Germany) for providing blood samples.
This study has been supported by:
Stiftung für Altersforschung, Heinrich-Heine-Universität Düsseldorf.
Prof. Dr. Rolf Ackermann, Wissenschaftliche Urologische Gesellschaft e.V.
M.J. A-B. has been supported by grants DFG10/15, DFG15/15 and DFG141/16 from Diputación Foral de Gipuzkoa, Spain, Ministry of Economy and Competitiveness, Spain, MINECO grants PI16/01430 and BFU 2016-7798-P and funds from Ikerbasque, Basque Foundation for Science, Spain.
The authors extend their appreciations to the Deanship of Scientific Research at King Saud University for funding the work through the research group project No. RGP-198.
We declare that the authors have no competing interests.
Footnotes
Peer review under responsibility of King Saud University.
References
- Bollati V., Schwartz J., Wright R., Litonjua A., Tarantini L., Suh H., Sparrow D., Vokonas P., Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech. Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourchis D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;2:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bronkhorst A.J., Wentzel J.F., Aucamp J., van Dyk E., du Plessis L., Pretorius P.J. Characterization of the cell-free DNA released by cultured cancer cells. BBA. 2016;1863:157–165. doi: 10.1016/j.bbamcr.2015.10.022. [DOI] [PubMed] [Google Scholar]
- Cho Y.H., Woo H.D., Jang Y., Porter V., Christensen S., Hamilton R.F., Jr., Chung H.W. The association of LINE-1 hypomethylation with age and centromere positive micronuclei in human lymphocytes. PLoS One. 2015;21:7. doi: 10.1371/journal.pone.0133909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- El-Maarri O., Walier M., Behne F., van Üüm J., Singer H., Diaz-Lacava A., Nüsgen N., Niemann B., Watzka M., Reinsberg J., van der Ven H., Wienker T., Stoffel-Wagner B., Schwaab R., Oldenburg J. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One. 2011;19:1. doi: 10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F., Hodgson J.G., Eden A., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;18:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Ghanjati F., Beermann A., Hermanns T., Poyet C., Araúzo-Bravo M.J., Seifert H.H., Schmidtpeter M., Goering W., Sorg R., Wernet P., Santourlidis S. Unreserved application of epigenetic methods to define differences of DNA methylation between urinary cellular and cell-free DNA. Cancer Biomark. 2014;14:295–302. doi: 10.3233/CBM-140407. [DOI] [PubMed] [Google Scholar]
- Goodier J.L. Restricting retrotransposons: a review. Mob. DNA. 2016;7:16. doi: 10.1186/s13100-016-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina S., Vijg J. Epigenetic factors in aging and longevity. Pflugers Arch. 2010;459:247–258. doi: 10.1007/s00424-009-0730-7. [DOI] [PubMed] [Google Scholar]
- Gravina S., Sedivy J.M., Vijg J. The dark side of circulating nucleic acids. Aging Cell. 2016;15:398–399. doi: 10.1111/acel.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;4:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hancks D.C., Kazazian H.H., Jr. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D.C., Kazazian H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.J., Schulz W.A. Causes and consequences of DNA hypomethylation in human cancer. Biochem. Cell Biol. 2005;83:296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- Kreimer U., Schulz W.A., Koch A., Niegisch G., Goering W. HERV-K and LINE-1 DNA methylation and reexpression in urothelial carcinoma. Front. Oncol. 2013;26:255. doi: 10.3389/fonc.2013.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen A., Florl A.R., Grimm M.O., Schulz W.A. DNA methylation alterations in urothelial carcinoma. Cancer Biol. Ther. 2006;5:993–1001. doi: 10.4161/cbt.5.8.2885. [DOI] [PubMed] [Google Scholar]
- Liu F., Killian J.K., Yang M., Walker R.L., Hong J.A., Zhang M., Davis S., Zhang Y., Hussain M., Xi S., Rao M., Meltzer P.A., Schrump D.S. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;24:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santourlidis, S., Ghanjati, F., Beermann, A., Hermanns, T., Poyet, C., 2016. IDLN-MSP: idiolocal normalization of real-time methylation-specific PCR for genetic imbalanced DNA specimens. Biotechniques 60, 84–87. [DOI] [PubMed]
- Shigaki H., Baba Y., Watanabe M., Iwagami S., Miyake K., Ishimoto T., Iwatsuki M., Baba H. LINE-1 hypomethylation in noncancerous esophageal mucosae is associated with smoking history. Ann. Surg. Oncol. 2012;19:4238–4243. doi: 10.1245/s10434-012-2488-y. [DOI] [PubMed] [Google Scholar]
- Simon R., Bürger H., Brinkschmidt C., Böcker W., Hertle L., Terpe H.J. Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J. Pathol. 1998;185:345–351. doi: 10.1002/(SICI)1096-9896(199808)185:4<345::AID-PATH109>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Steinhoff C., Schulz W.A. Transcriptional regulation of the human LINE-1 retrotransposon L1.2B. Mol. Genet. Genomics. 2003;270:394–402. doi: 10.1007/s00438-003-0931-2. [DOI] [PubMed] [Google Scholar]
- Stroun M., Anker P., Maurice P., Lyautey J., Lederrey C., Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;9:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- Vijg J., Suh Y. Genome instability and aging. Annu. Rev. Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- Wangsri S., Subbalekha K., Kitkumthorn N., Mutirangura A. Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia. PLoS One. 2012;7:9. doi: 10.1371/journal.pone.0045292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V.L., Jones P.A. DNA methylation decreases in aging but not inimmortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wilson V.L., Smith R.A., Ma S., Cutler R.G. Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- Wolff E.M. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;22:4. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zeidler R., Albermann K., Lang S. Nicotine and apoptosis. Apoptosis. 2007;12:1927–1943. doi: 10.1007/s10495-007-0102-8. [DOI] [PubMed] [Google Scholar]
- Zhong H.H., Hu S.J., Yu B., Jiang S.S., Zhang J., Luo D., Yang M.W., Su W.Y., Shao Y.L., Deng H.L., Hong F.F., Yang S.L. Apoptosis in the aging liver. Oncotarget. 2017;21:102640–102652. doi: 10.18632/oncotarget.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]