Abstract
Next-generation sequencing (NGS) is increasingly being used in a clinical setting for the molecular diagnosis of patients with heterogeneous disorders, such as premature ovarian insufficiency (POI). We performed NGS of ~1000 candidate genes in four unrelated patients with POI. We discovered the genetic cause of “isolated” POI in two cases, both of which had causative variants in surprising genes. In the first case, a homozygous nonsense variant in NBN was causative. Recessive function-altering NBN variants typically cause Nijmegen breakage syndrome characterized by microcephaly, cancer predisposition, and immunodeficiency, none of which are evident in the patient. At a cellular level, we found evidence of chromosomal instability. In the second case, compound heterozygous variants in EIF2B2 were causative. Recessive EIF2B2 function-altering variants usually cause leukoencephalopathy with episodic decline. Subsequent MRI revealed subclinical neurological abnormalities. These cases demonstrate that variants in NBN and EIF2B2, which usually cause severe syndromes, can cause apparently isolated POI, and that (1) NGS can precede clinical diagnosis and guide patient management, (2) NGS can redefine the phenotypic spectrum of syndromes, and (3) NGS may make unanticipated diagnoses that must be sensitively communicated to patients. Although there is rigorous debate about the handling of secondary/incidental findings using NGS, there is little discussion of the management of causative pleiotropic gene variants that have broader implications than that for which genetic studies were sought.
Introduction
Premature ovarian insufficiency (POI, OMIM #311360), affecting up to 1 in 100 women [1], is defined as cessation of menstruation with elevated follicle-stimulating hormone (FSH). POI etiology is diverse. It can be secondary to medical interventions, infections, or autoimmune disease. It can also be associated with metabolic and storage disorders, chromosomal instability disorders, and neurological disorders.
A number of studies have demonstrated a genetic component to POI pathogenesis; however, it is highly heterogeneous with no common genetic cause [2]. There are over 50 different genes implicated in the condition [2]. Many of these genes cause syndromic POI with clear genotype:phenotype associations. However, substantially less is known about the genetics of isolated POI. There are currently ~20 genes associated with isolated POI, but each explains only a small fraction of patients. Despite a clear genetic basis for POI and a large number of known POI-related genes, the majority of patients remain without a genetic diagnosis. The heterogeneity of POI and the incomplete understanding of POI genetics makes it suitable for studies using next-generation sequencing (NGS).
Our laboratory established a high-throughput NGS panel to sequence ~1000 genes known to be, or putatively, involved in disorders of sex development (DSD), including many POI-related genes [3]. We tested the utility of this panel for the genetic diagnosis of POI in a small pilot of four patients. We were able to make genetic diagnoses in two patients. One had function-altering (pathogenic/causative/damaging) variants in NBN encoding an important factor for the DNA damage response, while the other had causative variants in EIF2B2 encoding a subunit of the eukaryotic translation initiation factor 2B. Despite these being known POI-related genes, neither patient presented with the hallmark features related to variants in these genes. We used experimental and literature evidence to confirm variant impact, and to broaden and consolidate the accepted phenotypic spectrum of patients harboring variants affecting the function of these genes.
Our study confirms the utility of using NGS for the molecular diagnosis of women with POI, provides valuable insights into POI pathogenesis, highlights the blurred boundary between isolated and syndromic POI, and demonstrates benefits and sensitivities of using NGS for patient diagnosis.
Methods
Ethical adherence
Informed consent was obtained from all participants. All procedures were in accordance with the ethical standards of the Human Research Ethics Committee of the Royal Children’s Hospital, Melbourne.
Participants
The four patients included experienced POI in adolescence (diagnosis age range: 16–19 years). Apart from a congenital nerve defect reported for one patient, all others were diagnosed with isolated POI.
DNA preparation, Sanger sequencing, and RT-PCR
Genomic DNA was extracted from EDTA-blood samples by the Victorian Clinical Genetics Service (VCGS). Concentration and integrity were assessed by Qubit dsDNA BR Assay (Thermo Fisher Scientific) and TapeStation (Agilent), respectively. Selected SNVs were validated by Sanger sequencing using BigDye v3.1 Terminators (Applied Biosystems) and ABI 3130×, as per the manufacturers’ protocols. RNA was extracted from transformed lymphoblasts using TRIZOL Reagent (Invitrogen), and cDNA was generated using GoScript™ Reverse Transcription System (Promega) as per the manufacturers’ protocols. PCR primers were as previously described [4]. Data were submitted to ClinVar (SCV000692468–SCV000692470, https://www.ncbi.nlm.nih.gov/clinvar/).
HaloPlex NGS
A HaloPlex Custom Panel (Agilent Technologies) was designed to incorporate ~1000 genes known or predicted to be involved in DSD [3]. Procedures were as previously described [3].
Data analysis
The data from the sequencing run were analyzed using Cpipe [5]. Variants were filtered out if they had a frequency ≥ 0.01 in the 1000 genomes [6] or the NHLBI GO Exome Sequencing Project [7], or if they had been called ≥15 times in previous runs of the custom HaloPlex panel. Variant frequencies, including population-specific frequencies where relevant, were manually inspected using ExAC [8]. Noncoding variants (unless likely to affect splicing) and synonymous variants were filtered out (using snpeff annotation, http://snpeff.sourceforge.net/). The remaining variants were considered for their mode of inheritance and predicted impact on function based on four different prediction algorithms—Polyphen2 (genetics.bwh.harvard.edu/pph2), SIFT (sift.bii.a-star.edu.sg), LRT (http://www.genetics.wustl.edu/jflab/lrt_query.html), and MutationTaster (http://www.mutationtaster.org/).
Results
Patient 1 has a homozygous NBN nonsense variant
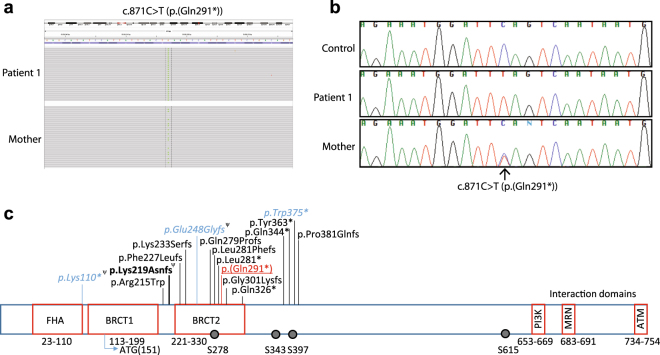
Patient 1 had 1821 variants detected by our DSD panel, of which 416 were predicted to affect the coding sequence or splicing. Forty of these were rare, only one of which was inherited in a recessive manner (Table 1), an apparently homozygous c.871C>T (p.(Gln291*)) nonsense variant in NBN (NM_002485.4, ClinVar SCV000692468). The variant had only a moderate quality score of 46 and depth of 26, but manual inspection of the variant and Sanger sequencing verified that it was a true variant (Fig. 1a, b). The patient’s mother was heterozygous for the variant. Paternal DNA was not available for testing. The variant was not found in the ~120,000 NBN alleles sequenced in the ExAC database. We did not perform Sanger sequencing of control women with normal menopause, as a large number are presumably sequenced within the publicly available population data, from which the variant is absent. Consanguinity was not reported. To determine whether one allele was a deletion that escaped detection by NGS, we performed a microarray of patient DNA. This analysis revealed an 8.4-Mb-long contiguous stretch of homozygosity on 8q21.3 containing the NBN gene, suggesting that the patient inherited a homozygous variant identical by descent. Using the criteria established by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology [9], this variant was considered function-altering (Table 2).
Table 1.
Variant filtering of NGS data for each patient
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| SNVs | 1821 | 1797 | 1732 | 1856 |
| Coding/splicinga | 416 | 403 | 381 | 417 |
| Rareb | 40 | 48 | 28 | 31 |
| Recessivec | 1 | 6 | 1 | 1 |
| Confidentd | 1 | 5 | 0 | 0 |
| POI-related | 1 | 1 | 0 | 0 |
aSNVs predicted to affect the coding sequence or splicing of protein products (snpeff)
b<0.01 in public databases and < 15 counts in previous sequencing runs
cSNVs fitting with recessive inheritance, taking into account parental DNA where available
dLikely errors were discounted by manual inspection using IGV
Fig. 1.
Function-altering variant detected in NBN. a IGV view of c.871C>T NBN (NM_002485.4) variant in Patient 1 (homozygous) and her mother (heterozygous). b Sanger sequencing confirmation of the c.871C>T NBN variant. c Schematic diagram showing patient variant with respect to previously reported variants in patients with Nijmegen breakage syndrome. Patient variant is underlined. Variants found in patients with mild NBS or isolated infertility are shown in italics. Ψ indicates variants that have been shown to also cause alternative protein products. The common founder variant, responsible for most cases of NBS, is shown in bold. Phosphorylation sites are shown as circles. Functional domains are indicated
Table 2.
Analysis of pathogenicity of variants using American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines [9]
| Patient 1 | Patient 2 | |
|---|---|---|
| NBN (NM_002485.4) c.871C>T (p.(Gln291*)) (hom) |
EIF2B2 (NM_014239.3) c.514C>T (p.(Arg172*)) (het) |
EIF2B2 (NM_014239.3) c.818A>G (p.(Lys273Arg)) (het) |
| PVS1: Truncating mutation in a gene where LOF is a known mechanism of disease | PVS1: Truncating mutation in a gene where LOF is a known mechanism of disease | PS1: Same amino acid change as a previously established pathogenic variant |
| PM1: Located in a critical and well-established functional domain | PM2: Extremely low frequency in Exome Sequencing Project, 1000 Genomes Project, and Exome Aggregation Consortium | PS3: Well-established in vitro and in vivo functional studies supportive of a damaging effect on the gene or gene product |
| PM2: Absent from controls in Exome Sequencing Project, 1000 Genomes Project, and Exome Aggregation Consortium | PM3: Detected in trans with a pathogenic variant | PM1: Located in a mutational hot spot and/or critical and well-established functional domain |
| PP3: Multiple lines of computational evidence support a deleterious effect on the gene or gene product | PP3: Multiple lines of computational evidence support a deleterious effect on the gene or gene product | PM2: Extremely low frequency in Exome Sequencing Project, 1000 Genomes Project, and Exome Aggregation Consortium |
| PP4: Patient’s phenotype is highly specific for a disease with a single genetic etiology | PP4: Patient’s phenotype is highly specific for a disease with a single genetic etiology | PM3: Detected in trans with a pathogenic variant |
| PP2: Missense variant in a gene that has a low rate of benign missense variation and in which missense variants are a common mechanism of disease | ||
| PP3: Multiple lines of computational evidence support a deleterious effect on the gene or gene product | ||
| PP4: Patient’s phenotype is highly specific for a disease with a single genetic etiology | ||
| Conclusiona: Pathogenic (1 very strong+2 moderate+2 supporting criteria) |
Conclusiona: Pathogenic (1 very strong + 2 moderate + 2 supporting criteria) |
Conclusiona: Pathogenic (2 strong + 3 moderate + 3 supporting criteria) |
Het heterozygous, hom homozygous, PVS very strong evidence, PS strong evidence, PM moderate evidence, PP supporting evidence
aBased on the scoring matrix outlined in American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines [9]
Patient 1 has a NBN variant affecting function but an atypical phenotype
There are at least 15 different clinically relevant NBN variants reported (Fig. 1c). These typically cause Nijmegen breakage syndrome (NBS), which is an autosomal recessive disorder characterized by microcephaly, growth retardation, dysmorphic faces, intellectual disability, combined immunodeficiency, radiosensitivity, and predisposition to malignancies. There is a high incidence of POI in affected females [10]. Heterozygous NBN variants are associated with increased cancer risk [11, 12].
Patient 1 was referred to the Gynaecology Department of the Royal Children’s Hospital (Melbourne, Australia) for investigation into primary amenorrhea at 16 years of age. The patient’s AMH level was <3.0 pmol/L (normal reference range > 14.0). FMR1 analysis was normal and karyotype analysis revealed a normal 46,XX chromosomal complement. She had no history of ovarian surgery or chemo- or radiotherapy. She had no history of recurrent infections or malignancy. Immunoglobulin investigations were normal. Anti-nuclear antibodies (ANA) were positive. A complete autoimmune profile revealed a slightly elevated anti-dsDNA of 28 IU/mL (normal reference range < 20 IU/mL). No microcephaly, growth retardation, or facial dysmorphia were apparent. The patient received a diagnosis of isolated POI, and NBS was never considered based on the clinical phenotype at presentation.
Patient 1 has chromosomal instability
NBN encodes nibrin which is one of the three subunits of the MRN complex (MRE11:RAD50:NBN). The MRN complex has important roles in repair of DNA double-strand breaks, cell-cycle control, telomere maintenance, and recombination [13–15]. One of the key roles of the MRN complex is to activate ATM which subsequently interacts with hundreds of effectors to trigger the DNA damage response [16]. NBN is responsible for the translocation of MRN to the nucleus [17], as well as its assembly at sites of DNA damage [18]. The important domains and residues of NBN are shown in Fig. 1c. Because NBN plays such a key role in the DNA damage response, NBN variants that impair function result in chromosomal instability. This chromosomal instability is responsible for the cancer predisposition and immunodeficiency of NBS patients.
To test whether our patient has cellular predisposition to chromosomal breakage, we performed a cytogenetic analysis of patient blood. Of 25 cells examined, four cells showed non-clonal abnormalities. One cell had a translocation of chromosomes 7 and 14: t(7;14)(q32;q11.2), whereas the other three cells had t(3;19)(p21;q13.3), dup(18)(q11.2q23), and t(3;10)(q21;q21.2), respectively. Spontaneous non-clonal rearrangements, particularly of chromosome 7 and 14, are consistent with NBS and suggest a high degree of chromosomal instability. Such aberrations are extremely rare in individuals without chromosomal breakage disorders.
The c.871C>T NBN variant is expressed in Patient 1 cDNA
In the mouse, deletion of Nbn causes embryonic lethality [19]. Consistent with this, human NBN variants found in NBS patients are likely hypomorphic rather than null variants. This has been confirmed for the most common causative NBN variant which results in the generation of partially functional truncated protein due to the use of a cryptic start site upstream of the variant [20].
Considering that Nbn null variants in mice are lethal and that alternative NBN protein products are common in patients with NBS, we hypothesized that the c.871C>T (p.(Gln291*)) variant may similarly cause an alternative protein product. Analysis using Human Splicing Finder (http://www.umd.be/HSF3/), revealed that the variant interrupted an exonic splicing enhancer and introduced an exonic splicing repressor, suggesting that it may cause nonsense-mediated alternative splicing (NAS).
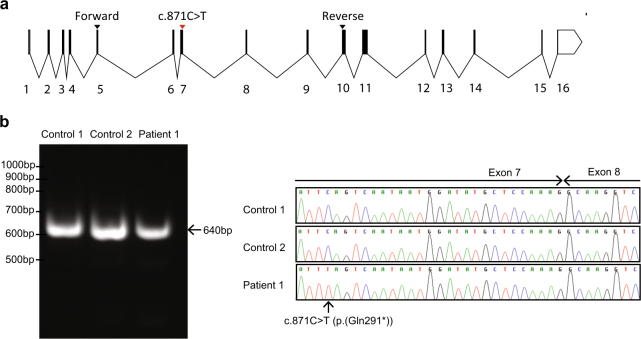
To test this theory, we sought to investigate NBN splicing in transformed patient lymphoblasts. Patient lymphoblasts grew poorly in culture and could not be propagated indefinitely. A poor proliferation capacity of NBN patient lymphocytes has been reported previously [21] and may be a consequence of chromosomal instability. We retrieved enough RNA to perform RT-PCR capturing exons 5–10 of the NBN gene (exons as per NG_008860.1) (Fig. 2a). This generated a single product of comparable size to controls (Fig. 2b). Sequencing revealed that the RT-PCR product was not different from the wild-type transcript, other than carrying the c.871C>T (p.(Gln291*)) nonsense variant and a benign c.553G>C (p.(Glu185Gln)) variant (Fig. 2c). The presence of normally spliced transcripts indicates that the nonsense variant does not cause complete nonsense-mediated decay, although any truncated protein would lack all the C-terminal domains important for the DNA damage response. Unfortunately, the poor proliferation and survival of patient lymphoblasts precluded protein analysis.
Fig. 2.
The NBN c.871C>T (p.(Gln291*)) (NM_002485.4) variant does not cause aberrant splicing or complete nonsense-mediated decay. a Schematic diagram showing numbered NBN exons and the location of the patient’s variant in the context of the NBN transcript (exons as per NG_008860.1). Primers for cDNA analysis are indicated. b RT-PCR demonstrates a single product comparable to wild-type cDNA from patient lymphoblasts. c Sequencing chromatograms demonstrate that patient lymphoblasts retain some normally spliced NBN transcripts
Patient 2 has compound heterozygous EIF2B2 variants
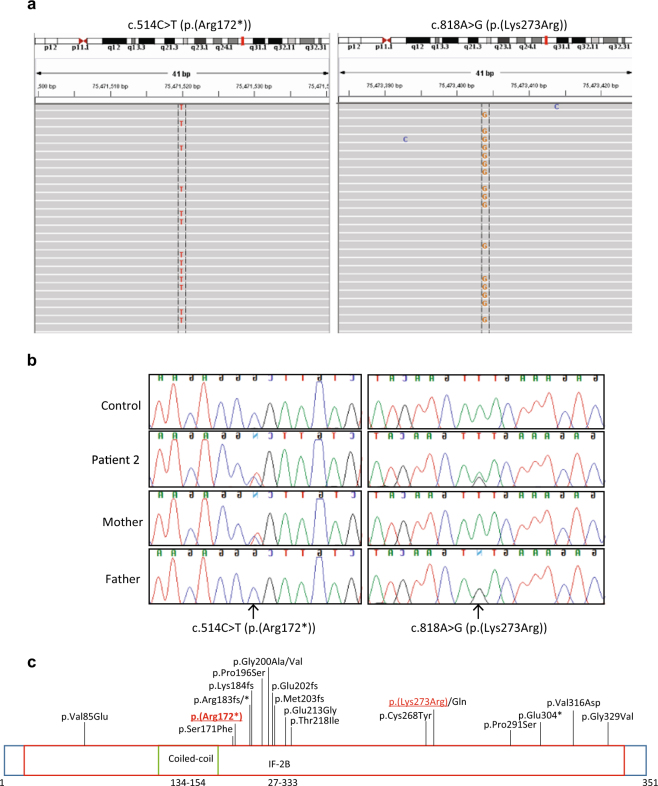
Patient 2 had 1797 variants identified by our sequencing panel. Of these, 403 were predicted to affect the coding sequence or splicing, and of these, 48 were considered rare. There were five genes fitting recessive inheritance, only one of which was a known POI-associated gene, EIF2B2 (Table 1). The patient harbored a c.514C>T (p.(Arg172*)) nonsense variant in exon 4, and a c.818A>G (p.(Lys273Arg)) missense variant in exon 6 (NM_014239.3, exons as per NG_013333.1, ClinVar SCV000692469, and SCV000692470, respectively). Both variants had a high quality score of 99 and a depth of >90 reads. Manual inspection of the NGS data confirmed the variants in a clearly heterozygous state (Fig. 3a). The variants were validated by Sanger sequencing (Fig. 3b). We did not sequence these variants in control women with normal menopause; however, they are very rare variants based on publicly available data. The c.514C>T (p.(Arg172*)) variant found in Patient 2, who was of Vietnamese descent, was found in 1 of 8578 alleles in the East Asian population but not in any other population according to the ExAC database [8]. The c.818A>G (p.(Lys273Arg)) variant was found in 1 of 16,500 alleles in the South Asian population but not in any other population [8]. The latter variant is, however, a previously reported variant affecting function, and associated with disease [22–24]. Sequencing parental DNA confirmed compound heterozygosity with the c.514C>T (p.(Arg172*)) variant inherited on the maternal allele, and the c.818A>G (p.(Lys273Arg)) variant inherited on the paternal allele (Fig. 3b). Using established guidelines [9], this variant is considered function-altering (Table 2).
Fig. 3.
Function-altering variants detected in EIF2B2. a IGV view of c.514C>T and c.818A>G (NM_014239.3) variants in Patient 2. b Sanger sequencing of c.514C>T and c.818A>G EIF2B2 variants in Patient 2 and her parents, confirming compound heterozygosity. c Schematic diagram showing patient variants with respect to previously reported variants. Patient variants are underlined. Variant not previously reported is indicated by bold text
Patient 2 has EIF2B2 variants affecting function but an atypical phenotype
EIF2B2 encodes one of the five subunits of the eukaryotic translation initiation factor 2B (EIF2B). EIF2B is a guanine nucleotide exchange factor and its role is to recharge EIF2 with GTP so that it can be available for future initiation of translation [25]. When EIF2 is phosphorylated as occurs under cellular stress, it preferentially binds EIF2B. EIF2B is less available to EIF2–GDP, thereby reducing the guanine nucleotide exchange and reducing protein synthesis [26]. EIF2B, therefore, controls the rate at which protein synthesis occurs, particularly under stress conditions.
Variants affecting the function of any of the five subunits of the EIF2B complex can cause autosomal recessive leukoencephalopathy with vanishing white matter (VWM) [27]. The classical presentation of VWM disease is onset of 2–5 years with cerebellar ataxia and spasticity, associated with cavitating cerebral white matter seen on MRI [28]. Other features that can be present include optic atrophy, epileptic seizures, psychiatric disturbances, and severe headache or migraine. The white-matter degeneration is progressive, and patients often experience episodic decline, particularly in the context of stress, such as infection/fever or minor head trauma. Disease presentation, however, can be highly variable ranging from fatal infantile forms to adult-onset forms with milder progression in cognitive and physical decline [29]. Due to the clinical variability, the term “eIF2B-related disorders” is now widely used. One eIF2B-related disorder is ovarioleukodystrophy [30]. Patients with ovarioleukodystrophy have POI, as well as neurodegeneration and evidence of VWM on MRI.
Patient 2, with EIF2B2 function-impairing variants, experienced primary amenorrhea and was diagnosed with POI when she was 19-years old. Her AMH level was <0.3 pmol/L and FSH was 54.3, 50.8, and 50.2 on three separate occasions with >1-month interval. Her karyotype was normal, as was FMR1 analysis. She had no relevant history of ovarian surgery or chemo- or radiotherapy. She is currently 29 years of age and does not report any difficulties with cognition or movement, and there were no apparent signs of neurological involvement. She received a diagnosis of isolated POI.
Because EIF2B2 variants are associated with VWM, Patient 2 underwent MRI investigation. MRI showed multiple abnormalities, including a focal region of gliosis and encephalomalacia in the medial right frontal lobe which was typical of a chronic right anterior cerebral artery territory infarct. Extensive confluent areas of white- matter signal abnormality were also observed, adjacent to the lateral ventricles, particularly the frontal horns and bodies bilaterally. Interestingly, the conclusion reached by radiology alone was that the infarct with cortical involvement was of vascular cause, such as perinatal insult or underlying vasculopathy/vasculitis, rather than a primary leukoencephalopathy. Further patient consultation revealed that she had experienced a parasitic infection following a trip to Vietnam. In addition to liver and lung lesions, she developed a brain abscess which required surgery. Several months later, she experienced high fever and clouded consciousness. These events were attributed to her prior infection, and resolved.
Considering the EIF2B2 variants carried by Patient 2, however, a likely explanation for the white-matter abnormality, and the clouded consciousness experienced after fever is ovarioleukodystrophy. Given that the patient is asymptomatic in terms of neurological functioning, and MRI alone did not indicate a primary leukodystrophy, her accurate clinical diagnosis relied on genetic analysis.
Discussion
We have used NGS to provide molecular diagnoses to patients with isolated POI. Surprisingly, two patients with a clinical diagnosis of isolated POI, had variants affecting function in genes classically associated with severe syndromic phenotypes.
NBN variants can cause “isolated” POI
Despite NBN function-altering variants typically causing NBS, there are two previous reports of variants that cause a milder phenotype. A sibling pair presented with isolated infertility but cellular features of a chromosomal instability disorder [31]. Another woman presented only with POI and mild microcephaly but, similarly, had the cellular features of NBS [4]. The mild phenotype is proposed to be due to partially functional NBN generated by alternative start site usage in the sibling pair and alternative splicing in the other patient. The latter patient appears resistant to cancer predisposition, because at the last report, she was 62-years old and had no history of malignancy. Interestingly, however, her sister died at an early age of lymphoma, a typical NBS cancer. Her sister also had POI and is suspected to have shared the NBN variant [4]. The discrepant phenotype between sisters, suggests that other factors may have protected the known living and healthy patient. Genetic or environmental modifiers may protect some patients from the full clinical phenotype. This remains a possible explanation for the mild phenotype in our patient, who is homozygous for a nonsense variant that is predicted to truncate the NBN protein, such that it lacks multiple functional domains.
In spite of a mild phenotype and the lack of malignancy in these previously reported patients [4, 31, 32], they had a high incidence of chromosomal aberrations. Similarly, our patient demonstrated a high degree of chromosomal instability. Presumably, the partially functional NBN protein in the previously reported patients allows some DNA damage response, so that such aberrations do not have the dire consequence of classical NBS variants. The lymphoma and subsequent death of one patient’s sister who is suspected of carrying an NBN variant [4], however, highlights the potential for a residual increased cancer risk. As more patients are identified, the prognosis of patients with NBN variants affecting function will become clearer.
NBN variants do not seem to be a common cause of isolated POI. Cytogenetic analysis of 96 patients with isolated POI and subsequent NBN sequencing in two patients who had evidence of chromosomal instability indicative of a repair defect, did not identify any variants [31]. Our patient is the second reported patient with isolated infertility, and the third reported patient with a mild phenotype associated with NBN variants, confirming that a broad clinical spectrum, including isolated POI, exists for patients with this genetic disruption.
Implications for Patient 1 management
Patient 1 enrolled in our study investigating the genetic basis of POI, with an established diagnosis of isolated POI. Her participation was preceded by thorough discussion at clinical consultation, and the information provided prior to NGS warned of incidental findings, but stated that variants unrelated to the condition in question would not be analyzed or reported.
However, when the molecular cause of POI was discovered in this patient, it raised several questions and harbors a potential burden. We cannot eliminate a possible cancer predisposition for this patient. It is reassuring that one previously reported patient has lived into late–middle age without any malignancy. As the previously reported patient’s sister died at a young age from lymphoma (a common type of cancer in NBS patients), it hints at a possible sensitivity to cancer. Indeed, our patient has evidence of chromosomal instability, which may put her at future risk of cancer development.
Because the NBN variant is directly related to the condition in question, for which the patient participated in our study, it is appropriate that our finding be communicated to her. In clinical consultation, it was emphasized that this is a research finding, and not diagnostic, and comes with some uncertainty. The patient was counseled about a possible predisposition to cancer. Clinical management of this patient may now be modified to include surveillance for cancer. Although the genetic diagnosis may have been unanticipated by the patient, both the counseling and management of this patient have changed, potentially improving her health and well-being.
EIF2B2 variants can cause “isolated” POI
All reported patients with homozygous or compound heterozygous-function-impairing variants in EIF2B, have neurological pathology. Patient 2 had no overt neurological impairment and her diagnosis at the outset of this study, was isolated POI.
In some cases of EIF2B-related ovarioleukodystrophy, ovarian failure can precede neurological decline, or neurological damage may be incidentally found on MRI [30]. Examples include a 16-year-old girl who experienced amenorrhea and had an MRI for recurrent headaches that detected leukodystrophy [30]; another woman experienced POI at the age of 24, and then progressive gait, cognitive, and behavioral changes from the age of 61 [33]; and a woman who experienced secondary amenorrhea and juvenile cataracts in her early 20s followed by clumsiness and gait disturbance at the age of 43 [34].
There is little genotype:phenotype correlation for EIF2B variants. There are over 100 different known variants responsible for EIF2B-related disorders [27], and at least 20 causative variants in EIF2B2 specifically (Fig. 3c). The same variant can have variable clinical consequences, even within a single family [24]. A small number of variants tend to be associated with more mild or more severe disease, such as the c.338G>A (p.(Arg113His)) variant in EIF2B5 which is associated with adult-onset milder disease [24], or the c.584G>A (p.(Arg195His)) variant in EIF2B5 which is associated with severe disease and death before the age of 2 years [24, 35], but there remain atypical cases, and genotype:phenotype correlations are not rigid.
The particular nature of the variants in Patient 2 is unlikely responsible for her milder disease. The spectrum of reported function-impairing EIF2B2 variants is shown in Fig. 3c. The majority of function-impairing variants cluster in the middle portion of the protein and cause childhood-onset VWM disease. The c.514C>T (p.(Arg172*)) variant identified in our patient falls within this cluster and is predicted to encode a truncated protein, like many disease-associated variants in its vicinity.
The second allele in Patient 2 harbors a previously reported c.818A>G (p.(Lys273Arg)) variant. This variant is causative in multiple unrelated patients with varying clinical severity [22–24], and the in vitro GEF activity of recombinant protein is impaired [36].
There is clear potential for environmental influence on the phenotype, with exposure to stress precipitating episodic decline. It is possible that our patient has experienced fewer stresses, leading to her preserved cognitive and physical function. It appears that female sex may be somewhat protective for disease severity with more females than males presenting with the adult-onset form of disease [37]. A correlation between the severity of POI and the age of onset of neurological decline has been demonstrated [30]. Therefore, it may be surprising that Patient 2 has not experienced any overt neurological decline, given her primary amenorrhea.
It is not yet established whether the EIF2B genes may have a role in isolated POI, or whether neurological decline is inevitable in patients identified with EIF2B function-impairing variants. Recognizing the potential role of EIF2B in isolated POI, Fogli et al. [38] screened 93 POI patients for variants that have previously been reported in ovarioleukodystrophy or mild VWM disease, however, no variants were identified. This study was limited in only screening for a subset of previously reported function-impairing variants, and other EIF2B variants may still have a role in isolated POI pathogenesis.
Implications for Patient 2 management
Patient 2, with causative EIF2B2 variants, is currently 29-years old and her clinical diagnosis at the study outset was isolated POI, unlike all reported cases of EIF2B2 variants which include neurological impairment, at least subclinically in the form of migraines, and abnormal MRI.
Similar to the aforementioned case, the diagnosis of causative EIF2B2 variants has potentially serious implications for her future health. It is likely, but not definite, that this patient will experience neurological decline. The discovery of EIF2B2 variants prompted neurological assessment by MRI, which showed extensive white-matter abnormality. The clinicians and counselors working with this patient had to be careful and sensitive, being aware that the diagnosis of a neurodegenerative disease might cause stress which could in itself trigger episodic decline.
Knowing about a risk of neurological decline may empower a patient to make practical or lifestyle changes to better accommodate future challenges. The patient can minimize the future risk of head trauma, fever, or fright, thereby reducing the threat of future neurological decline.
NGS for molecular diagnosis
NGS is clearly a powerful tool for the genetic diagnosis of disease, particularly for rare and heterogeneous diseases such as POI. Without NGS, the two cases described here would be unlikely to receive a genetic diagnosis. NBN and EIF2B2 would not be high priorities for candidate-gene sequencing in cases of isolated POI.
The two cases presented here are women with perceived isolated POI, who in fact harbored function-impairing variants in pleiotropic genes. This raises important issues in medical ethics in the current genomic era. There is much debate about the use of NGS and the potential for incidental finding; however, little attention is paid to the impact of pleiotropic results [39]. The two cases presented here, demonstrate scenarios that may increasingly be encountered. A full appreciation of the potential impact of genetic diagnosis must be appreciated by both clinicians and patients before commencing NGS.
Now that genetic studies can precede full clinical phenotype manifestation, it is also important that clinicians have an appreciation of the variable presentation of patients with variants in syndromic or pleiotropic genes, to ensure that appropriate information about potential health risks is communicated. It is important, for example, that future individuals found to have NBN variants are made aware that their clinical course may differ greatly from the severe and classical form of disease.
This study describes the successful identification of POI causative variants in two of four patients who underwent sequencing for a panel of ~1000 genes predicted to be involved in DSD. This is a small sample size of patients so we do not make any conclusions about the diagnostic yield of this panel for POI patients in general. This panel does not capture all known POI-related genes, so the two patients for whom we failed to identify causative variants, may have variants in genes not covered by our panel. Alternatively, they may have exonic deletions or duplications that are difficult to detect with NGS, or complex/multigenic inheritance that is difficult to identify and interpret. Future analysis of these patients using genomic tools such as whole-exome sequencing or copy-number variant analysis will hopefully yield genetic diagnoses.
Conclusion
Here, we have described the genetic diagnosis of two patients with apparent isolated POI. We have reported new function-altering variants in NBN and EIF2B2, encoding components of the DNA repair machinery and the translation initiation machinery, respectively. We have demonstrated the power, but also the challenges, of using NGS for molecular diagnosis. The patients described in this report raise questions about the management of pleiotropic gene variants when the full clinical manifestation of disease is unknown, and highlight the need for awareness of atypical presentation of genetic syndromes.
Acknowledgements
This work was supported by a Peter Doherty Early Career Fellowship (1054432 to EJT), a National Health and Medical Research Council program grant (1074258 to AHS), a fellowship (1062854 to AHS) from the Australian National Health and Medical Research Council, and the Victorian Government’s Operational Infrastructure Support Program.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Haller-Kikkatalo K, Uibo R, Kurg A, Salumets A. The prevalence and phenotypic characteristics of spontaneous premature ovarian failure: a general population registry-based study. Hum Reprod. 2015;30:1229–38. doi: 10.1093/humrep/dev021. [DOI] [PubMed] [Google Scholar]
- 2.Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr Rev. 2016;37:609–35. doi: 10.1210/er.2016-1047. [DOI] [PubMed] [Google Scholar]
- 3.Eggers S, Sadedin S, van den Bergen JA, et al. Disorders of sex development: insights from targeted gene sequencing of a large international patient cohort. Genome Biol. 2016;17:243. doi: 10.1186/s13059-016-1105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varon R, Dutrannoy V, Weikert G, et al. Mild Nijmegen breakage syndrome phenotype due to alternative splicing. Hum Mol Genet. 2006;15:679–89. doi: 10.1093/hmg/ddi482. [DOI] [PubMed] [Google Scholar]
- 5.Sadedin SP, Dashnow H, James PA, et al. Cpipe: a shared variant detection pipeline designed for diagnostic settings. Genome Med. 2015;7:68. doi: 10.1186/s13073-015-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ESP NGESP. Exome variant server. 2015. http://evs.gs.washington.edu/EVS/.
- 8.ExAC EAC. Exome aggregation consortium. 2015. http://exac.broadinstitute.org.
- 9.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chrzanowska KH, Szarras-Czapnik M, Gajdulewicz M, et al. High prevalence of primary ovarian insufficiency in girls and young women with Nijmegen breakage syndrome: evidence from a longitudinal study. J Clin Endocrinol Metab. 2010;95:3133–40. doi: 10.1210/jc.2009-2628. [DOI] [PubMed] [Google Scholar]
- 11.Steffen J, Varon R, Mosor M, et al. Increased cancer risk of heterozygotes with NBS1 germline mutations in Poland. Int J Cancer. 2004;111:67–71. doi: 10.1002/ijc.20239. [DOI] [PubMed] [Google Scholar]
- 12.Varon R, Reis A, Henze G, von Einsiedel HG, Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL) Cancer Res. 2001;61:3570–2. [PubMed] [Google Scholar]
- 13.Varon R, Vissinga C, Platzer M, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–76. doi: 10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 14.Carney JP, Maser RS, Olivares H, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–86. doi: 10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 15.Ranganathan V, Heine WF, Ciccone DN, et al. Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Curr Biol. 2001;11:962–6. doi: 10.1016/S0960-9822(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 16.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 17.Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–91. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res. 2004;45:473–8. doi: 10.1269/jrr.45.473. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–9. doi: 10.1016/S0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 20.Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–21. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 21.Ormerod E, Belengeanu V, Stoian M, et al. Nijmegen breakage syndrome: clinico-cytogenetic pattern. J Pediatr. 2009;7:19–24. [Google Scholar]
- 22.Zhang H, Dai L, Chen N, et al. Fifteen novel EIF2B1-5 mutations identified in Chinese children with leukoencephalopathy with vanishing white matter and a long term follow-up. PLoS ONE. 2015;10:e0118001. doi: 10.1371/journal.pone.0118001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leegwater PA, Vermeulen G, Konst AA, et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet. 2001;29:383–8. doi: 10.1038/ng764. [DOI] [PubMed] [Google Scholar]
- 24.Fogli A, Schiffmann R, Bertini E, et al. The effect of genotype on the natural history of eIF2B-related leukodystrophies. Neurology. 2004;62:1509–17. doi: 10.1212/01.WNL.0000123259.67815.DB. [DOI] [PubMed] [Google Scholar]
- 25.Pavitt GD. eIF2B, a mediator of general and gene-specific translational control. Biochem Soc Trans. 2005;33:1487–92. doi: 10.1042/BST0331487. [DOI] [PubMed] [Google Scholar]
- 26.Rowlands AG, Panniers R, Henshaw EC. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988;263:5526–33. [PubMed] [Google Scholar]
- 27.van der Knaap MS, Leegwater PA, Konst AA, et al. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol. 2002;51:264–70. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- 28.Pronk JC, van Kollenburg B, Scheper GC, van der Knaap MS. Vanishing white matter disease: a review with focus on its genetics. Ment Retard Dev Disabil Res Rev. 2006;12:123–8. doi: 10.1002/mrdd.20104. [DOI] [PubMed] [Google Scholar]
- 29.Fogli A, Boespflug-Tanguy O. The large spectrum of eIF2B-related diseases. Biochem Soc Trans. 2006;34:22–9. doi: 10.1042/BST0340022. [DOI] [PubMed] [Google Scholar]
- 30.Fogli A, Rodriguez D, Eymard-Pierre E, et al. Ovarian failure related to eukaryotic initiation factor 2B mutations. Am J Hum Genet. 2003;72:1544–50. doi: 10.1086/375404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warcoin M, Lespinasse J, Despouy G, et al. Fertility defects revealing germline biallelic nonsense NBN mutations. Hum Mutat. 2009;30:424–30. doi: 10.1002/humu.20904. [DOI] [PubMed] [Google Scholar]
- 32.Maraschio P, Peretti D, Lambiase S, et al. A new chromosome instability disorder. Clin Genet. 1986;30:353–65. doi: 10.1111/j.1399-0004.1986.tb01892.x. [DOI] [PubMed] [Google Scholar]
- 33.Ghezzi L, Scarpini E, Rango M, et al. A 66-year-old patient with vanishing white matter disease due to the p.Ala87Val EIF2B3 mutation. Neurology. 2012;79:2077–8. doi: 10.1212/WNL.0b013e3182749edc. [DOI] [PubMed] [Google Scholar]
- 34.Matsukawa T, Wang X, Liu R, et al. Adult-onset leukoencephalopathies with vanishing white matter with novel missense mutations in EIF2B2, EIF2B3, and EIF2B5. Neurogenetics. 2011;12:259–61. doi: 10.1007/s10048-011-0284-7. [DOI] [PubMed] [Google Scholar]
- 35.Fogli A, Wong K, Eymard-Pierre E, et al. Cree leukoencephalopathy and CACH/VWM disease are allelic at the EIF2B5 locus. Ann Neurol. 2002;52:506–10. doi: 10.1002/ana.10339. [DOI] [PubMed] [Google Scholar]
- 36.Liu R, van der Lei HD, Wang X, et al. Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum Mutat. 2011;32:1036–45. doi: 10.1002/humu.21535. [DOI] [PubMed] [Google Scholar]
- 37.Labauge P, Horzinski L, Ayrignac X, et al. Natural history of adult-onset eIF2B-related disorders: a multi-centric survey of 16 cases. Brain. 2009;132:2161–9. doi: 10.1093/brain/awp171. [DOI] [PubMed] [Google Scholar]
- 38.Fogli A, Gauthier-Barichard F, Schiffmann R, et al. Screening for known mutations in EIF2B genes in a large panel of patients with premature ovarian failure. BMC Women’s Health. 2004;4:8. doi: 10.1186/1472-6874-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kocarnik JM, Fullerton SM. Returning pleiotropic results from genetic testing to patients and research participants. JAMA. 2014;311:795–6. doi: 10.1001/jama.2014.369. [DOI] [PMC free article] [PubMed] [Google Scholar]