Abstract
Cocaine is one of the most used psychostimulant drugs worldwide. MicroRNAs are post-transcriptional regulators of gene expression that are highly expressed in brain, and several studies have shown that cocaine can alter their expression. In a previous study, we identified several protein-coding genes that are differentially expressed in a dopaminergic neuron-like model after an acute exposure to cocaine. Now, we used the prediction tool WebGestalt to identify miRNA molecules potentially involved in the regulation of these genes. Using the same cellular model, we found that seven of these miRNAs are down-regulated by cocaine: miR-124-3p, miR-124-5p, miR-137, miR-101-3p, miR-9-5p, miR-369-3p and miR-153-3p, the last three not previously related to cocaine. Furthermore, we found that three of the miRNA genes that are differentially expressed in our model (hsa-miR-9-1, hsa-miR-153-1 and hsa-miR-124-3) are nominally associated with cocaine dependence in a case–control study (2,085 cases and 4,293 controls). In summary, we highlighted novel miRNAs that may be involved in those cocaine-induced changes of gene expression that underlie addiction. Moreover, we identified genetic variants that contribute to cocaine dependence in three of these miRNA genes, supporting the idea that genes differentially expressed under cocaine may play an important role in the susceptibility to cocaine dependence.
Introduction
Cocaine is one of the most used illicit drugs and its use is one of the major public health problems worldwide1. However, only a subset of individuals exposed to cocaine (around 15–16% of cocaine users) develop an addiction2.
Transcriptomic studies performed in animal models and human brain post-mortem samples have revealed that both acute and chronic exposure to cocaine produce changes in the expression of genes related to diverse functional categories, such as cell adhesion, extracellular matrix, synaptic communication and neuroplasticity, receptors, ion channels and transporters, oligodendrocytes and myelin, mitochondrial function, apoptosis and cell death, transcription factors and signal transduction3,4. Cocaine also produces epigenetic adaptations like changes in DNA methylation, chromatin remodeling or alterations in miRNA regulation5–7. Both epigenetic adaptations and changes in gene expression induced by cocaine trigger molecular and cellular adaptations in the central nervous system that may explain the persistence of drug-seeking behavior, even after extended periods of abstinence4,8–10.
The interaction between the individual’s genetic background, epigenetic factors and environment11 determine how neuronal circuits adapt to chronic cocaine exposure, establishing the development of addiction in some individuals but not in others10. Cocaine dependence is one of the most heritable psychiatric disorders (around 65–79%12–15), and some of those genetic risk factors may lie in genes that mediate cocaine’s effects, conferring initial vulnerability to the establishment of drug-induced neuroadaptations.
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression that bind to target mRNAs to inhibit translation or promote mRNA degradation. Each miRNA can regulate the expression of hundreds of different mRNAs, and each mRNA species can be targeted by several miRNAs. MiRNA regulation generates a very complex and dynamic system that allows the cells to fine-tune gene expression16–18. In addition to the canonical cytoplasmic function, there is evidence that miRNAs located in the nucleus can regulate mRNA stability in the nucleolus and modulate alternative splicing, as well as activate or inhibit the transcription of target genes19.
MiRNAs are very abundant in the central nervous system, especially in the synapto-dendritic compartment, where they control local mRNA translation in response to neuronal activity20, which is essential for synapses development, neuronal plasticity, memory and learning21,22. It is known that cocaine alters miRNA expression profiles in brain23. Using RNAseq, Eipper-Mains and collaborators inspected cocaine-responsive miRNAs in nucleus accumbens (NAc) and striatal post-synaptic densities (PSDs) in chronically cocaine-treated mice. They found several differentially expressed miRNAs, most of them belonging to four miRNA families (miR-8, miR-7, miR-142 and Let-7), which suggests that cocaine modulates expression of miRNAs that have similar target genes24. In a subsequent study, they used a list of mRNAs25 found enriched in PSD to identify predicted targets of these miRNAs26, using bioinformatic tools.
In a previous study we evaluated changes in gene expression induced by acute cocaine exposure in a dopaminergic neuron-like model (differentiated SH-SY5Y cells). We found differences in 756 protein-coding genes involved in several processes with potential relevance to central nervous system function, including regulation of transcription, chromatin modification, focal adhesion and cell projection, and neurotrophin and MAPK signaling pathways27. Furthermore, these genes showed an enrichment of predicted binding sites for several miRNAs, miR-124 among them. Based on these results, we hypothesized that the observed changes in gene expression can be produced, in part, by changes in miRNA expression. Here, we aimed at validating predicted changes in miRNA expression altered by cocaine in differentiated SH-SY5Y cells. Furthermore, we aimed to evaluate their possible contribution to the susceptibility to cocaine dependence through a case–control association study with common genetic variants.
Materials and methods
MiRNA selection
In a previous study27 we assessed transcriptomic changes induced by cocaine in a human dopaminergic neuron-like model (differentiated SH-SY5Y neuroblastoma cells) at 6 and 24 h after an acute 30-min exposure to cocaine at 1 or 5 μM. We identified 756 protein-coding genes showing differential expression only under 5 μM cocaine after 6 h (10% FDR), 337 of them down-regulated and 419 up-regulated27. In the present study we further evaluated the enrichment of miRNA-binding sites among the differentially expressed genes. We performed a “miRNAs target analysis” with the online tool WEB-based GEne SeT AnaLysis Toolkit (WebGestalt, http://www.webgestalt.org/webgestalt_2013)28,29 using the default settings and considering up-regulated and down-regulated subsets of genes separately (Supplementary Table 1). We filtered the results obtained using the following criteria: (i) miRNAs expressed in brain, according to the miRIAD database (http://bmi.ana.med.uni-muenchen.de/miriad/), (ii) every single miRNA has predicted binding sites in at least 10 target genes, (iii) an enrichment P-value <0.01. We also discarded predictions of miRNA families due to the difficulty to validate all the different miRNAs within them. We then prioritized miRNAs which predicted target genes were enriched in any biological function according to GO or KEGG, or miRNAs with a reported relation with cocaine.
MiRNA expression
Changes in the expression of miRNA genes were tested in new cultures of dopaminergic neuron-like cells exposed to cocaine. SH-SY5Y cells (ATCC, LGC Standards) were differentiated and validated as previously described27. After an exposure to 5 µM cocaine for 30 min, the medium was replaced and cells were retrieved at 3 or 6 h, performing 10 biological replicates per condition. Total RNA including miRNAs was isolated using the miRNeasy Micro Kit (Qiagen, Hilden, Germany). RNA concentration was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, TermoFisher Scientific Inc., Wilmington, DE, USA) and integrity was evaluated with the Bioanalyzer 2100 platform (Agilent Technologies, Santa Clara, CA, USA). The averaged RIN values (RNA Integrity Number) was 9.98, being 9.8 the lowest one.
Mature miRNAs were retrotranscribed using the miScript II RT Kit (Qiagen) and real-time quantitative PCR (qRT-PCR) experiments were performed with the miScript SYBR Green PCR Kit (Qiagen) in the LightCycler 480 II system (Roche Life Sciences, Branford, CT, USA). For primers sequences, see Supplementary Table 2. To select a reference gene we tested several miRNA genes, and compared and ranked them using the online tool RefFinder (http://150.216.56.64/referencegene.php?type=reference) that integrates different computational tools (geNorm, Normfinder, BestKeeper and the comparative ΔCt method). Relative quantification was performed for each miRNA by normalizing the level of expression to that of the selected reference miR-3911, which is stable across samples.
A one-way ANOVA with Dunnett’s post hoc test was performed to examine the gene expression changes among groups. Previously, normality and homogeneity of variance were ascertained in our samples using the Shapiro–Wilk and Bartlett tests, respectively. A value of P < 0.05 was considered to be statistically significant.
Gene networks
The differentially expressed miRNAs were subjected to analysis of gene networks using the Ingenuity Pathway Analysis 8.8 software (http://www.ingenuity.com/products/ipa; Ingenuity Systems, Redwood city, CA, USA).
Gene-based association analysis
We used MAGMA 1.05b30 to perform a gene-based association analysis to test the contribution to cocaine dependence susceptibility of the miRNAs which expression is altered by cocaine. We used the summary statistics from a GWAS meta-analysis of cocaine dependence (Ncases = 2,085; Ncontrols = 4,293)31, considering the SNP-wise mean model, in which the sum of −log(SNP P-value) for SNPs located within a gene and its regulatory regions was used as the statistic test. We selected all miRNA genes encoding each differentially expressed mature miRNA and the regulatory regions, including both that of the miRNA and that of the host protein-coding gene, identified according to histone marks H3K4me1, H3K4me3, H3K27me3 and H3K27Ac in UCSC Genome Browser (assembly GRCh37/hg19)32. The gene P-value was calculated using a known approximation of the sampling distribution33. MAGMA accounts for gene size, number of SNPs in a gene and linkage disequilibrium between markers (estimated using data from 1000Genomes Phase 3, European ancestry samples34).
Summary statistics used in this analysis derive from a case–control GWAS meta-analysis of cocaine dependence performed using four datasets from the dbGaP repository (https://www.ncbi.nlm.nih.gov/gap) under the project 10608. All cases used met DSM-IV criteria for cocaine dependence, although most of them are also dependent to other drugs of abuse. Control individuals were taken from the general population, except for those of the SAGE sample, where drug abuse or dependence were formally discarded31.
Results
In a previous study we identified 756 protein-coding genes showing differential expression in a dopaminergic neuron-like model after an acute exposure to cocaine27. Here we evaluated whether these changes in gene expression might be produced by changes in miRNA expression induced by cocaine.
Using the online prediction tool WebGestalt we tested enrichment for miRNA-binding sites in the differentially expressed mRNAs. We found enrichment for 6 miRNAs in the subset of down-regulated target genes, and 84 miRNAs for the up-regulated ones (Supplementary Table 1). After filtering them according to the criteria described above (see Materials and methods), we selected 11 miRNAs (hsa-miR-9-3p, hsa-miR-9-5p, hsa-miR-101-3p, hsa-miR-105-5p, hsa-miR-124-3p, hsa-miR-124-5p, hsa-miR-137, hsa-miR-153-3p, hsa-miR-181-5p, hsa-miR-186-5p and hsa-miR-369-3p), all of them predicted to bind up-regulated genes in our previous study. Thus, we expected to find these miRNAs down-regulated, as they are negative regulators of gene expression.
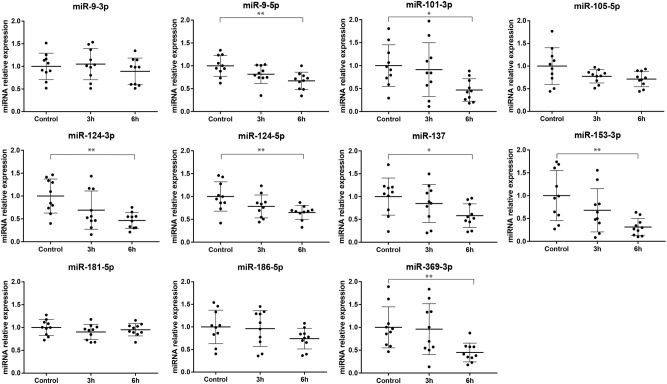
Since the RNA samples used in the previous work did not contain miRNAs, we performed new experiments of cocaine exposure in differentiated SH-SY5Y cells, to experimentally validate changes in miRNA expression, at 3 and 6 h after 30 min of cocaine treatment at 5 µM. The results showed that 6 h after the 30-min cocaine treatment, seven out of the 11 tested miRNAs were down-regulated, as predicted, when compared to untreated control cells: miR-9-5p (FC = −1.49, SE = 0.19, P = 1.3e-03), miR-101-3p (FC = −2.00, SE = 0.26, P = 0.012), miR-124-3p (FC = −2.17, SE = 0.17, P = 1.4e-03), miR-124-5p (FC = −1.54, SE = 0.15, P = 4e-03), miR-137 (FC = −1.69, SE = 0.26, P = 0.015), miR-153-3p (FC = −2.94, SE = 0.2, P = 1.3e-03) and miR-369-3p (FC = −2.22, SE = 0.2, P = 7.6e-03) (Fig. 1). None of them showed significant changes at 3 h after treatment, although in some cases a decreasing trend was observed (Fig. 1). The other four miRNAs did not change their expression significantly (Fig. 1).
Fig. 1.
Transcription levels of all tested miRNAs determined by qRT-PCR at 3 and 6 h after a 30-min exposure to 5 µM cocaine in dopaminergic neuron-like cells. Significant differences compared to control cells (not exposed to cocaine) normalized to miR-3911 are indicated (*P-value <0.05, **P-value <0.01). Mean and standard deviation are shown
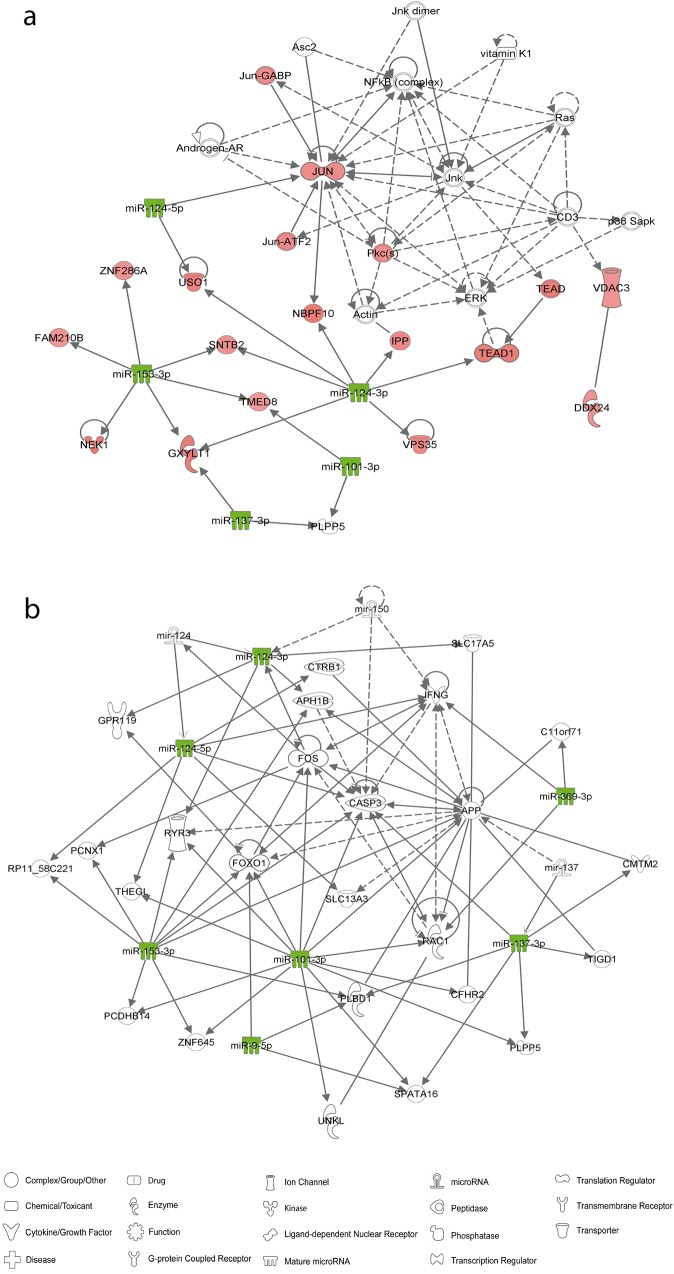
Then, we evaluated the relationship between these miRNAs and other genes by gene network analysis. First, we inspected the connections between the identified down-regulated miRNAs and their up-regulated target mRNA (predicted to contain miRNA-binding sites). We found five out of the seven differentially expressed miRNAs connected with 17 up-regulated genes in a network related to “Developmental Disorder, Ophthalmic Disease, Organismal Injury and Abnormalities” (score = 41; Fig. 2a). Second, we inspected how these differentially expressed miRNAs are connected with other genes in the genome and found that all seven miRNAs were present and highly connected in a network involved in “Cell-To-Cell Signaling and Interaction, Nervous System Development and Function, Inflammatory Disease” (score = 21; Fig. 2b), including target genes such as FOS, CASP3 and RYR3.
Fig. 2.
Gene networks involved in a) 'developmental disorder, ophthalmic disease, organismal injury and abnormalities' (score = 41) and b) 'cell-to-cell signaling and interaction, nervous system development and function, inflammatory disease' (score = 21). The green and red nodes in the pathway indicate the down- and up-regulated genes, respectively
Finally, we evaluated the possible contribution of these miRNAs to cocaine dependence susceptibility by assessing the presence of common genetic risk variants for cocaine dependence within the genes encoding these miRNAs. Thus, we performed a gene-based association analysis in a sample of 2,085 cocaine-dependent individuals and 4,293 controls. Interestingly, three miRNA genes were found nominally associated with cocaine dependence: hsa-miR-9-1 (P = 0.011), hsa-miR-153-1 (P = 0.036) and hsa-miR-124-3 (P = 0.042) (Table 1). These genes encode mature miRNAs that are highly connected in the identified networks (Fig. 2a, b).
Table 1.
Results of miRNA gene-based association analysis of common genetic variants in a GWAS meta-analysis of cocaine dependence in individuals of European ancestry
| Gene ID | Coordinates a | N SNPs | ZSTAT | P- value | SNP ID | Risk allele | SNP P- value |
|---|---|---|---|---|---|---|---|
| hsa-miR-101-1 | chr1:65523406-65534997 | 19 | 0.2260 | 0.4105 | rs486378 | G | 0.0420 |
| hsa-miR-101-2 | chr9:4828718-4857394 | 146 | 0.4890 | 0.3123 | rs7870037 | A | 0.0113 |
| hsa-miR-124-1 | chr8:9760113-9764675 | 6 | −0.8845 | 0.8117 | rs62489494 | C | 0.7111 |
| hsa-miR-124-2 | chr8:65283014-65291854 | 17 | 0.3783 | 0.3525 | rs190938 | G | 0.0518 |
| hsa-miR-124-3 | chr20:61804035-61811005 | 18 | 1.7294 | 0.0418 | rs76137972 | G | 0.0342 |
| hsa-miR-137 | chr1:98509277-98521215 | 30 | −0.1075 | 0.5428 | rs12744323 | C | 0.1661 |
| hsa-miR-153-1 | chr2:220158748-220161492 | 8 | 1.7939 | 0.0364 | rs6436132 | T | 0.0178 |
| hsa-miR-153-2 | chr7:157362028-157377114 | 38 | −0.7436 | 0.7714 | rs221296 | A | 0.2219 |
| hsa-miR-153-2_HGb | chr7:158377426-158412766 | 123 | 0.6251 | 0.2659 | rs12670306 | A | 0.0221 |
| hsa-miR-369 | chr14:101522238-101541542 | 78 | 0.8836 | 0.1884 | rs12431682 | T | 5.3E-03 |
| hsa-miR-9-1 | chr1:156389579-156394148 | 5 | 2.2778 | 0.0113 | rs78605853 | G | 6.0E-03 |
| hsa-miR-9-2 | chr5:87957415-87991755 | 57 | 0.6955 | 0.2433 | rs201864123 | G | 0.0484 |
| hsa-miR-9-3 | chr15:89899916-89911697 | 27 | −0.6557 | 0.7440 | rs961288 | C | 0.4393 |
Analysis performed using 2,085 cases and 4,293 controls
aGene coordinates based on GRCh37/hg19. NSNPs: number of SNPs per region included in the gene-based analysis; ZSTAT: the Z-value for the gene; SNP ID: SNP with lowest P-value
bPromoter region of miR-153-2 host gene (PTPRN2). In bold, nominally associated genes
Discussion
In this study we explored the possibility that cocaine-induced changes in gene expression found in a previous study of our group may be triggered by changes in the expression of miRNA molecules.
We used prediction tools to identify miRNAs that potentially regulate protein-coding genes which expression is altered by cocaine according to a previous study by our group in a dopaminergic cell model27. Seven of the 11 miRNAs selected were significantly down-regulated after an acute exposure to cocaine (miR-9-5p, miR-101-3p, miR-124-3p, miR-124-5p, miR-137, miR-153-3p and miR-369-3p), consistent with the predictions and with the up-regulation of their target genes. Moreover, through gene network analyses, we correlated down-regulated miRNAs with the up-regulated target genes identified in our previous study. This network includes several genes previously associated with cocaine dependence, such as PKC, encoding an enzyme involved in cocaine-induced neuroplasticity35–37, or JUN, an early immediate gene (EIG) that activates transcription through heterodimerization at AP-1 sites38–41. Furthermore, we found interaction between miR-124-3p and its validated target TEAD142,43, encoding a transcription factor regulated by the Hippo pathway that controls proliferation44. On the other hand, we performed a network analysis to test the interaction of the seven down-regulated miRNAs identified with all the genes in the genome. We identified an interesting network involved in “Cell-To-Cell Signaling and Interaction, Nervous System Development and Function, Inflammatory Disease” in which the seven differentially expressed miRNAs are present and highly interconnected. In this network we highlight three genes: FOS, CASP3 and RYR3. FOS is a very important EIG highly associated with cocaine dependence, as recently reviewed elsewhere45. CASP3 encodes a pro-apoptotic protein involved in cocaine-mediated cell death that is very important for both adult and fetal cocaine neurotoxicity46,47. And RYR3, a member of the RyR family involved in cocaine-induced place preference regulated by dopamine D1 receptor48. RYR3 is a highly confident predicted target of miR-124-3p and miR-153-3p according to different online miRNA:mRNA prediction tools like miRBD, DianaT, TargetScan, and PicTar. Furthermore, RYR3 was found up-regulated in dopaminergic-enriched regions from substantia nigra and ventral tegmental area in post-mortem brain samples from cocaine abusers38, as well as in our previous study on dopaminergic neuron-like cells treated with cocaine27.
Some of the miRNAs reported in the present study had also been found down-regulated by cocaine in previous works. For example, miR-124 is down-regulated in the NAc of mice chronically exposed to cocaine49 and in neuronal cell cultures (Be(2)-M17 and SH-SY5Y) treated with cocaine50. Moreover, overexpression of this gene in NAc attenuates cocaine-induced place preference in mice51. On the other hand, miR-137 and miR-101 are down-regulated in the NAc of cocaine self-administrated rats compared to controls52. This is the first time that miR-9-5p, miR-369-3p and miR-153-3p are found down-regulated after cocaine treatment. MiR-9 is one of the most highly expressed miRNAs in the adult vertebrate brain and it plays an important role in brain development, neurogenesis, synaptic plasticity and memory53–57. Furthermore, a recent study associated fetal alcohol exposure with an increase of miR-9 in the pituitary, which represses the D2r gene and its spliced variant D2s58. The miR-369 is located in the miR379–410 cluster, which contains 38 miRNAs involved in neuronal development and function59. MiR-369-3p regulates the expression of Ncad, Adam19, and TrappC8, very important in neurogenesis and neuronal migration60. Finally, miR-153 inhibits Cacna1c to suppress neuroendocrine secretion (insulin and dopamine)61.
Interestingly, three of the genes encoding differentially expressed mature miRNAs (hsa-miR-9-1, hsa-miR-153-1 and hsa-miR-124-3) were found to be nominally associated with cocaine dependence in this study. Previous studies had reported genes which expression is altered by cocaine and that also carry risk variants for cocaine dependence, or viceversa (NFAT5, PLCB1 and NTNG1)27,62,63. The findings in the present study, together with the previous ones, support the hypothesis that genes up-regulated or down-regulated by cocaine may also contribute to the susceptibility to cocaine dependence.
Strengths and limitations of the present study should be discussed. Our study was performed in a dopaminergic neuron-like model, from a tumor cell line, which performance may differ from the events that take place in the brain. However, we were able to detect previously described miRNAs that had shown down-regulation by cocaine in mice, and three new miRNAs. On the other hand, three genes encoding these altered miRNAs were found to carry genetic variants nominally associated with cocaine dependence in human samples, despite the relatively small sample size of the meta-analysis used. Increasing the number of individuals of this study may increase the strength of the associations. Finally, it is important to note that, although many tools/databases are available to predict mRNA:miRNA interactions, the overlap between the outputs is often very low17, so it is important to validate them. Here, we demonstrated that WebGestalt is a very useful tool to identify enrichment of miRNA-binding sites in a list of potential target genes, since we could validate changes in the expression of 7 out of 11 predicted miRNAs. This strategy was previously used by others with a similar performance64,65. Further studies are needed to validate the mRNA:miRNA interactions identified.
In conclusion, we found seven miRNAs down-regulated in dopaminergic neuron-like cells after an acute cocaine treatment, which might regulate several protein-coding genes previously reported to be up-regulated in this model. Here we highlighted new miRNAs that may be involved in cocaine-induced changes of gene expression. Furthermore, using a GWAS meta-analysis of clinical samples, we found association between cocaine dependence and three genes encoding these differentially expressed miRNAs. All these results support the idea that genes which expression is altered by cocaine might play an important role in the susceptibility to cocaine dependence, and also that miRNAs may be relevant players in modulating cocaine effects and dependence.
Electronic supplementary material
Acknowledgements
Major financial support for this research was received by B.C. from the Spanish ‘Ministerio de Economía y Competitividad’ (SAF2015-68341-R) and AGAUR, ‘Generalitat de Catalunya’ (2017-SGR-738). The research leading to these results has also received funding from the European Union Seventh Framework Program [FP7/2007-2013] under grant agreement no. 602805 (Aggressotype) and from the European Union H2020 Program [H2020/2014-2020] under grant agreement no. 667302 (CoCA). N.F-C. was supported by a contract from the ‘Centro de Investigación Biomédica en Red de Enfermedades Raras’ (CIBERER) and by an EMBO short-term fellowship (ASTF 573-2016). J.C-D. was supported by ‘Generalitat de Catalunya’ (2015 FI_B 00448). We thank Laura Pineda, Noèlia Benetó, and Bàrbara Torrico for their support with the cell differentiation experiments. The authors acknowledge the contribution of data from dbGAP accessed through project number 10608. The Study of Addiction: Genetics and Environment (SAGE), one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI (supported by U01 HG004422, U01 HG004446, U01HG004438, HHSN268200782096C; and support for collection of datasets and samples was provided by the COGA; U10 AA008401, COGEND; P01 CA089392 and FSCD; R01 DA013423). GWAS of Heroin Dependence (supported by R01DA17305); GWAS of Cocaine Dependence in Two Populations (supported by RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535); GWAS of Heroin Dependence (supported by R01DA17305). CRIC Study (supported by U01DK060990, U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK60980, U01DK060963, and U01DK060902). CIDR-Gelernter Study, a genome-wide association study funded as part of the Genetics of Alcohol Dependence in American Populations (supported by U01HG004438 and HHSN268200782096C). The COPDGene study (supported by U01HL089856 and U01HL089897, and the COPD Foundation through contributions made by an Industry Advisory Board comprised of Pfizer, AstraZeneca, Boehringer Ingelheim, Novartis, and Sunovion). Personalized Medicine Research Project (PMRP) (supported by U01HG004608, U01HG004438, and U01HG004603).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Noèlia Fernàndez-Castillo, Bru Cormand
Contributor Information
Bru Cormand, Email: bcormand@ub.edu.
Noèlia Fernàndez-Castillo, Email: noefernandez@ub.edu.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41398-018-0224-5).
References
- 1.United Nations Office on Drugs and Crime. World Drug Report 2017 (United Nations, Sales No. E.17.XI.6).
- 2.O’Brien MS, Anthony JC. Risk of becoming cocaine dependent: epidemiological estimates for the United States, 2000–2001. Neuropsychopharmacology. 2005;30:1006–1018. doi: 10.1038/sj.npp.1300681. [DOI] [PubMed] [Google Scholar]
- 3.Lull ME, Freeman WM, Vrana KE, Mash DC. Correlating human and animal studies of cocaine abuse and gene expression. Ann. NY Acad. Sci. 2008;1141:58–75. doi: 10.1196/annals.1441.013. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Z, Enoch MA, Goldman D. Gene expression in the addicted brain. Int. Rev. Neurobiol. 2014;116:251–273. doi: 10.1016/B978-0-12-801105-8.00010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaillancourt K, Ernst C, Mash D, Turecki G. DNA methylation dynamics and cocaine in the brain:progress and prospects. Genes (Basel) 2017;8:E138. doi: 10.3390/genes8050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76:259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenny PJ. Epigenetics, microRNA, and addiction. Dialog. Clin. Neurosci. 2014;16:335–344. doi: 10.31887/DCNS.2014.16.3/pkenny. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farris SP, Harris RA, Ponomarev I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Front. Neurosci. 2015;9:176. doi: 10.3389/fnins.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt HD, McGinty JF, West AE, Sadri-Vakili G. Epigenetics and psychostimulant addiction. Cold Spring Harb. Perspect. Med. 2013;3:a012047. doi: 10.1101/cshperspect.a012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 2012;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajonijebu DC, Abboussi O, Russell VA, Mabandla MV, Daniels WMU. Epigenetics: a link between addiction and social environment. Cell Mol. Life Sci. 2017;74:2735–2747. doi: 10.1007/s00018-017-2493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bevilacqua L, Goldman D. Genes and addictions. Clin. Pharmacol. Ther. 2009;85:359–361. doi: 10.1038/clpt.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat. Rev. Genet. 2005;6:521–32. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 14.Bühler KM, et al. Common single nucleotide variants underlying drug addiction: more than a decade of research. Addict. Biol. 2015;20:845–871. doi: 10.1111/adb.12204. [DOI] [PubMed] [Google Scholar]
- 15.Hall FS, Drgonova J, Jain S, Uhl GR. Implications of genome wide association studies for addiction: are our a priori assumptions all wrong? Pharmacol. Ther. 2013;140:267–279. doi: 10.1016/j.pharmthera.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 17.Afonso-Grunz F, Müller S. Principles of miRNA–mRNA interactions: beyond sequence complementarity. Cell Mol. Life Sci. 2015;72:3127–3141. doi: 10.1007/s00018-015-1922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J. Transl. Med. 2016;14:143. doi: 10.1186/s12967-016-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J. Mol. Sci. 2016;17:1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Most D, Workman E, Harris RA. Synaptic adaptations by alcohol and drugs of abuse: changes in microRNA expression and mRNA regulation. Front. Mol. Neurosci. 2014;7:85. doi: 10.3389/fnmol.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Kwon EJ, Tsai LH. MicroRNAs in learning, memory, and neurological diseases. Learn Mem. 2012;19:359–368. doi: 10.1101/lm.026492.112. [DOI] [PubMed] [Google Scholar]
- 22.Kolshus E, Dalton VS, Ryan KM, McLoughlin DM. When less is more—microRNAs and psychiatric disorders. Acta Psychiatr. Scand. 2013;129:241–56. doi: 10.1111/acps.12191. [DOI] [PubMed] [Google Scholar]
- 23.Jonkman S, Kenny PJ. Molecular, cellular, and structural mechanisms of cocaine addiction: a key role for microRNAs. Neuropsychopharmacology. 2013;38:198–211. doi: 10.1038/npp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eipper-Mains JE, et al. microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA. 2011;17:1529–1543. doi: 10.1261/rna.2775511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, Tian QB, Kuromitsu J, Kawai T, Endo S. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci. Res. 2007;57:61–85. doi: 10.1016/j.neures.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Eipper-Mains JE, Eipper BA, Mains RE. Global approaches to the role of miRNAs in drug-induced changes in gene expression. Front. Genet. 2012;3:109. doi: 10.3389/fgene.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernàndez-Castillo N, et al. Transcriptomic and genetic studies identify NFAT5 as a candidate gene for cocaine dependence. Transl. Psychiatry. 2015;5:e667. doi: 10.1038/tp.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabana-Dominguez, J., Shivalikanjli, A., Fernandez-Castillo, N. & Cormand, B. Genome-wide association meta-analysis of cocaine dependence: shared genetics with comorbid conditions. bioRxiv, 10.1101/374553 (2018). [DOI] [PubMed]
- 32.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou CD. A simple approximation for the distribution of the weighted combination of non-independent or independent probabilities. Stat. Probab. Lett. 2005;73:179–187. doi: 10.1016/j.spl.2004.11.028. [DOI] [Google Scholar]
- 34.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortinski PI, Briand LA, Pierce RC, Schmidt HD. Cocaine-seeking is associated with PKC-dependent reduction of excitatory signaling in accumbens shell D2 dopamine receptor-expressing neurons. Neuropharmacology. 2015;92:80–89. doi: 10.1016/j.neuropharm.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue B, Guo ML, Jin DZ, Mao LM, Wang JQ. Cocaine facilitates PKC maturation by upregulating its phosphorylation at the activation loop in rat striatal neurons in vivo. Brain Res. 2012;1435:146–153. doi: 10.1016/j.brainres.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller BW, et al. Cocaine craving during protracted withdrawal requires PKCε priming within vmPFC. Addict. Biol. 2017;22:629–639. doi: 10.1111/adb.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bannon MJ, et al. A molecular profile of cocaine abuse includes the differential expression of genes that regulate transcription, chromatin, and dopamine cell phenotype. Neuropsychopharmacology. 2014;39:2191–2199. doi: 10.1038/npp.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moratalla R, Elibol B, Vallejo M, Graybiel AM. Network-level changes in expression of inducible Fos-Jun proteins in the striatum during chronic cocaine treatment and withdrawal. Neuron. 1996;17:147–156. doi: 10.1016/S0896-6273(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 40.Malaplate-Armand C, et al. Effect of acute and chronic psychostimulant drugs on redox status, AP-1 activation and pro-enkephalin mRNA in the human astrocyte-like U373 MG cells. Neuropharmacology. 2005;48:673–684. doi: 10.1016/j.neuropharm.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Imam SZ, Duhart HM, Skinner JT, Ali SF. Cocaine induces a differential dose-dependent alteration in the expression profile of immediate early genes, transcription factors and caspases in PC12 cells: a possible mechanism of neurotoxic damage in cocaine addiction. Ann. NY Acad. Sci. 2005;1053:482–490. doi: 10.1196/annals.1344.042. [DOI] [PubMed] [Google Scholar]
- 42.Mucaj V, et al. MicroRNA-124 expression counteracts pro-survival stress responses in glioblastoma. Oncogene. 2015;34:2204–2214. doi: 10.1038/onc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 44.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandra R, Lobo MK. Beyond neuronal activity markers: select immediate early genes in striatal neuron subtypes functionally mediate psychostimulant addiction. Front. Behav. Neurosci. 2017;11:112. doi: 10.3389/fnbeh.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lepsch LB, Planeta CS, Scavone C. Cocaine causes apoptotic death in rat mesencephalon and striatum primary cultures. Biomed. Res. Int. 2015;2015:1–7. doi: 10.1155/2015/750752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dey S, Mactutus CF, Booze RM, Snow DM. Cocaine exposure in vitro inducesapoptosis in fetal locus coeruleus neurons by altering the Bax/Bcl-2 ratio and through caspase-3 apoptotic signaling. Neuroscience. 2007;144:509–521. doi: 10.1016/j.neuroscience.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurokawa K, et al. Cocaine increases ryanodine receptors via dopamine D1 receptors. Synapse. 2011;65:1106–1112. doi: 10.1002/syn.20935. [DOI] [PubMed] [Google Scholar]
- 49.Chandrasekar V, Dreyer JL. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell Neurosci. 2009;42:350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Rodrigues AC, et al. MicroRNA expression is differentially altered by xenobiotic drugs in different human cell lines. Biopharm. Drug Dispos. 2011;32:355–367. doi: 10.1002/bdd.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandrasekar V, Dreyer JL. Regulation of miR-124, let-7d, and miR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology. 2011;36:1149–1164. doi: 10.1038/npp.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn RK, et al. Temporally specific miRNA expression patterns in the dorsal and ventral striatum of addiction-prone rats. Addict. Biol. 2017;23:631–642. doi: 10.1111/adb.12520. [DOI] [PubMed] [Google Scholar]
- 53.Radhakrishnan B, Alwin Prem Anand A. Role of miRNA-9 in brain development. J. Exp. Neurosci. 2016;2016:101–120. doi: 10.4137/JEN.S32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sim SE, et al. The brain-enriched microRNA miR-9-3p regulates synaptic plasticity and memory. J. Neurosci. 2016;36:8641–8652. doi: 10.1523/JNEUROSCI.0630-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madelaine R, et al. MicroRNA-9 couples brain neurogenesis and angiogenesis. Cell Rep. 2017;20:1533–1542. doi: 10.1016/j.celrep.2017.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu X, et al. miR-124 and miR-9 mediated downregulation of HDAC5 promotes neurite development through activating MEF2C-GPM6A pathway. J. Cell Physiol. 2018;233:673–687. doi: 10.1002/jcp.25927. [DOI] [PubMed] [Google Scholar]
- 57.Roese-Koerner B, Stappert L, Brüstle O. Notch/Hes signaling and miR-9 engage in complex feedback interactions controlling neural progenitor cell proliferation and differentiation. Neurogenesis. 2017;4:e1313647. doi: 10.1080/23262133.2017.1313647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gangisetty O, Jabbar S, Wynne O, Sarkar DK. MicroRNA-9 regulates fetal alcohol-induced changes in D2 receptor to promote prolactin production. J. Endocrinol. 2017;235:1–14. doi: 10.1530/JOE-17-0135. [DOI] [PubMed] [Google Scholar]
- 59.Winter J. MicroRNAs of the miR379-410 cluster: new players in embryonic neurogenesis and regulators of neuronal function. Neurogenesis. 2015;2:e1004970. doi: 10.1080/23262133.2015.1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rago L, Beattie R, Taylor V, Winter J. miR379-410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin. EMBO J. 2014;33:906–20. doi: 10.1002/embj.201386591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu H, et al. The Ia-2β intronic miRNA, miR-153, is a negative regulator of insulin and dopamine secretion through its effect on the Cacna1c gene in mice. Diabetologia. 2015;58:2298–2306. doi: 10.1007/s00125-015-3683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelaï S, et al. Netrin g1: its downregulation in the nucleus accumbens of cocaine-conditioned mice and genetic association in human cocaine dependence. Addict. Biol. 2017;23:1–13. doi: 10.1111/adb.12485. [DOI] [PubMed] [Google Scholar]
- 63.Cabana-Domínguez J, et al. Association of the PLCB1 gene with drug dependence. Sci. Rep. 2017;7:1–8. doi: 10.1038/s41598-017-10207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng W, et al. Identification of key genes in glioma CpG island methylator phenotype via network analysis of gene expression data. Mol. Med. Rep. 2017;15:2795–2801. doi: 10.3892/mmr.2017.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng YE, JXIA Liu, ZHIP Yan, Yao XH, Liu XH. Potential microRNA biomarkers for acute ischemic stroke. Int. J. Mol. Med. 2015;36:1639–1647. doi: 10.3892/ijmm.2015.2367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.