Abstract
The NAA10-NAA15 complex (NatA) is an N-terminal acetyltransferase that catalyzes N-terminal acetylation of ~40% of all human proteins. N-terminal acetylation has several different roles in the cell, including altering protein stability and degradation, protein localization and protein–protein interactions. In recent years several X-linked NAA10 variants have been associated with genetic disorders. We have identified a previously undescribed NAA10 c.215T>C p.(Ile72Thr) variant in three boys from two unrelated families with a milder phenotypic spectrum in comparison to most of the previously described patients with NAA10 variants. These boys have development delay, intellectual disability, and cardiac abnormalities as overlapping phenotypes. Functional studies reveal that NAA10 Ile72Thr is destabilized, while binding to NAA15 most likely is intact. Surprisingly, the NatA activity of NAA10 Ile72Thr appears normal while its monomeric activity is decreased. This study further broadens the phenotypic spectrum associated with NAA10 deficiency, and adds to the evidence that genotype–phenotype correlations for NAA10 variants are much more complex than initially anticipated.
Introduction
N-terminal (Nt-) acetylation is one of the most common protein modifications in eukaryotes [1, 2]. The functional consequences of Nt-acetylation are diverse, and include directing protein complex formation, subcellular localization, in addition to more general functions such as aiding in protein folding, and as a degradation signal marking proteins for degradation through the proteasome pathway [3–9]. N-terminal acetyltransferase A (NatA) consists of a catalytic subunit encoded by NAA10 (previously called Ard1, MIM 300013) and an auxiliary subunit encoded by NAA15 (previously called NAT1/NATH, MIM 608000) and is responsible for Nt-acetylation of ~40% of the human proteome [2,10–12]. Loss of NAA10 in D. melanogaster, T. brucei, and C. elegans results in lethality [13–15] and morpholino knockdown of NAA10 in zebrafish causes developmental defects and increased mortality rate of zebrafish embryos [16]. There are also numerous reports connecting human NAA10 to several different types of cancers, both as a tumor suppressor and as an oncogene (reviewed in [17]), and describing how loss of NAA10 expression induce cell cycle arrest and apoptosis [18–20].
In 2011, a NAA10 variant, c.109T>C p.(Ser37Pro), was identified as the cause of an X-linked recessive lethal disorder called Ogden syndrome (MIM 300855) [21]. Boys affected by Ogden syndrome had severe global development delays, craniofacial abnormalities, hypotonia, and cardiac anomalies or arrhythmia and all of the boys affected by this syndrome died between 8 and 16 months of age [21]. Molecular investigations revealed a reduced NatA complex formation and reduced Nt-acetylation of NatA substrates in cells derived from Ogden syndrome patients [22, 23]. Since then, several new NAA10 variants have been described and the phenotypic spectrum of patients with NAA10-related N-terminal acetylation deficiency has broadened [24–27]. Esmailpour and colleagues reported a NAA10 splice site variant, c.471+2T>A (NM_003491.3), that was identified in four males with the X-linked recessive disorder Lenz-micropthalmia syndrome (MIM 309800) [28, 29]. Popp and colleagues described two NAA10 variants, c.319G>T p.(Val107Phe) and c.346C>T p.(Arg116Trp), in a girl and a boy with severe intellectual disabilities, postnatal growth retardation, hypotonia, and behavioral anomalies [25, 30], and shortly later, Casey and colleagues reported another missense variant, c.128A>C p.(Tyr43Ser), in two brothers with developmental delay, facial dysmorphisms, scoliosis, and long QT [24]. Finally, three variants that affect NAA10 function, c.247C>T p.(Arg83Cys), c.384T>A p.(Phe128Ile), and c.382T>A p.(Phe128Leu), were identified in a total of 12 female and 1 male patients with development delay, severe intellectual disability, postnatal growth failure, and skeletal and cardiac anomalies [26, 27]. One of the female patients had inherited the variant from her mother, most likely through maternal germ line mosaicism, in all of the other female patients the NAA10 variants occurred de novo [25, 26].
We here report the identification of a novel NAA10 variant, c.215T>C p.(Ile72Thr), in two brothers and a third unrelated boy with global development delay and intellectual disability. The two brothers both have hypertrophic cardiomyopathy (HCM) and developmental delay with autistic features. The third patient has developmental delay, minor dysmorphic features, and cardiac abnormalities including hypertrophic cardiomyopathy and QT prolongation. He developed a malignant brain tumor at 2 years of age but remains in remission after treatment. N-terminal acetyltransferase assays reveal a clear reduction in catalytic activity of NAA10 Ile72Thr towards substrates representing in vitro monomeric NAA10 substrates, and a clearly increased protein turnover rate in cells, strongly suggesting that this variant is dysfunctional compared to NAA10 WT.
Materials and methods
Whole-exome sequencing
Whole-exome sequencing (WES) for family 1 (proband, affected sibling, and mother) was performed as previously described [31] in the Baylor College of Medicine Human Genome Sequencing Center using a custom VCRome 2.1 capture reagent. Sequence data were processed using the Mercury pipeline [32], with additional annotation performed in the Baylor Hopkins Center for Mendelian Genomics using the program Variant Analyzer under research protocol H-29697. The variant was submitted to ClinVar (SCV000494176). WES for family 2 was performed by GeneDX (USA).
Multiple sequence alignment and homology model
Multiple sequence alignments were created using ClustalX [33] and the illustration using ESPript3.0 [34]. The human NatA homology model was created and published previously [23].
Plasmid preparation and protein purification
pcDNA3.1-V5-NAA10 and pETM41-His/MPB-NAA10 plasmids coding for the p.Ile72Thr variant were created by site-directed mutagenesis (QuikChange® Multi Site-Directed Mutagenesis kit) according to the manufacturer’s protocol. Primer sequences are available on request. Protein purification was performed as described previously [25] with minor modifications.
Immunoprecipitation of NAA10-V5
HeLa cells were grown on Ø 10 cm plates and transiently transfected with 10 µg pcDNA3,1-V5-plasmids using X-tremeGENE 9 DNA transfection reagent (Roche). After 24 h medium was replaced. 48 h after transfection, cells were harvested and washed in cold PBS (pH 7.4), resuspended and lysed in 800 µl IPH lysis buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 5 mM EDTA; 0.5% NP-40; complete EDTA free protease inhibitor (Roche)), 20 min incubation on ice. Cell debris was removed by centrifugation (4 °C and 17,000×g for 10 min), the supernatant subjected to preclearing before 18 µg of V5 tag mouse monoclonal antibody (Invitrogen) was added to the supernatant. The mixture was incubated on a rotating wheel at 4 °C for 2 h before 225 µl of prewashed magnetic beads were added and incubated overnight. Beads were isolated using a magnet, washed three times in IPH lysis buffer, two times in acetylation buffer (50 mM Tris-HCl, 10% glycerol, 1 mM EDTA, pH 8.5) and equally divided into different tubes for Nt-acetylation assays and western blotting analysis. Anti-V5 (mouse, Invitrogen), anti-NAA15 and anti-NAA10 (rabbit, Biogenes [12]) antibodies were used to detect immunoprecipitated NAA10-V5 and NAA15. Protein bands of NAA10-V5 imaged by using ChemiDocTM XRS+ system of Bio-RAD were quantified by using the ImageLab 5.1 software from Bio-RAD.
Nt-acetylation assay
Equally divided magnetic beads containing NAA10-V5 were mixed with 247 µM Acetyl CoA, 66 µM Acetyl CoA [acetyl-1-14C] (Perkin Elmer), 300 µM substrate polypeptide, acetylation buffer, and incubated at 37 °C in a shaker for 1 h. As a negative control, water was added instead of substrate polypeptide. Product formation was detected as described [35]. For each condition, three measurements were performed. The measured product formation was correlated to the amount of NAA10-WT-V5 and NAA10-Ile72Thr-V5 present in each sample as determined by the V5-signal in Western blots. To account for the uncertainty and variance in the determination of NAA10-V5 in each sample, standard deviations were calculated by the equation , where A and dA represent the mean of measured radioactivity and standard deviation from the acetylation assay, and B and dB the mean of the quantification of the NAA10-V5 band intensities and the standard deviation from these measurements, respectively.
Protein turnover analysis
The protein turnover analysis of NAA10-Ile72Thr-V5 by Cycloheximide chase assay was performed as described previously [24]. In brief, HeLa cells were plated on six-well plates 16 h before transfection and transiently transfected with 2.6 µg NAA10-Ile72Thr-V5 and NAA10-WT-V5 and 50 µg/ml Cycloheximide was added 48 h after transfection Samples were analyzed by SDS-PAGE and Western blotting using anti-V5-tag antibody (Invitrogen, R960-25), anti-β-Tubulin antibody (Sigma Aldrich, T5293), anti-NAA10 antibody, and anti-NAA15 antibody [12]. Signals were detected and quantified as described above.
Results
Clinical report: family 1, patient II-1 and II-2
Individual II-1 is a boy who was first evaluated at 6 months of age by the pediatric cardiologist for suspected hypertrophic cardiomyopathy (HCM). The echocardiogram was suggestive of a small muscular ventricular septal defect, as well as severe left ventricular hypertrophy with evidence of mid-cavitary obliteration and some mitral insufficiency. Echo revealed moderate to severe concentric HCM with mild left ventricular outflow tract obstruction (LVOT). He was started and managed on β-blocker therapy (propanolol). At 1 year of age his echocardiogram revealed marked left ventricular hypertrophy with moderate outflow tract obstruction. By 2½ years his echocardiogram continued to reveal moderate left ventricular outflow tract obstruction with a peak velocity of 4.1 m/s. He had mild mitral regurgitation with a mildly dilated left atrium at that time. His propanolol dose was increased, and then changed to metoprolol, and verapamil was added, and treatment later changed to atenolol. His subsequent echocardiograms have shown mild to moderate LVOT. At the last evaluation by the pediatric cardiologist, when he was 8½ years of age, he was found to be relatively asymptomatic from a cardiac standpoint. His cardiomyopathy is described as severe concentric left ventricular HCM with moderate dynamic LVOTO, prolonged QTc on EKG. He has normal right ventricular size and function.
He has global developmental delay (DD) and intellectual disability (ID). MRI of the brain at 3 years of age showed a thin corpus callosum, with relative paucity of frontal lobe without focal parenchymal signal abnormality to explain his DD/ID. Gross motor milestones were delayed including holding head up (3–4 months), rolling over (4 months), crawling (12 months), standing (12 months), and walking (19 months). His speech has been particularly delayed, and he had no words at 4 years of age. He is presently 9 years old and attends special education class at school, defined as having a cognitive communication deficit and mixed receptive-expressive disorder, and he does not use signs. He has tactile aversion, and behavioral abnormalities: he may push, bite or pinch other children if he disagrees with their behavior. He copies others’ facial expression. He likes to clap his hands, jump, fake coughs, he has normal muscular tone, and normal hearing, and normal growth. At 4 years 4 months of age his height was 97.8 cm (5p for age according to CDC growth charts), weight 14.8 kg (10–25p for age), and 8 years 6 months of age his height was 124.7 cm (10–25p) and weight 24.2 kg (10–25p).
At 4 years of age he was hospitalized for severe symptomatic hypoglycemia (glucose of 29) and symptoms of fatigue, diaphoresis, and depressed responsiveness, had elevated medium chain acylcarnitines (C6, C8 and C10, but C10>C10:1). He was treated with dextrose and improved thereafter. The acylcarnitine profile was concluded to be consistent with diet, and not with medium chain acylcarnitine deficiency or another metabolic disorder. Metabolic screening tests were normal including plasma amino acids and urine organic acids. Sanger sequencing of the following cardiomyopathy related genes was performed prior to the WES: ACTC GLA, LAMP2, MYBPC3, MYH7, MYL2, MYL3, PRKG2, TNNT2, TNNI3, TNNC1, and TPM1. And chromosomal microarray (BCM oligoarray version 7.4) showed normal male karyotype without any pathological copy number variants. WES in the clinical laboratory did not reveal any suspected pathological variants, but detected variants of then unknown significance in two genes associated with cardiomyopathies, TTN & DSP. His mother was heterozygous for two of these (NM_133378.4(TTN): c.8525T>C p.(Leu2842Pro) and c.3002T>C p.(Met1001Thr)) and his father heterozygous for the third TTN variant (p.(Glu10731Lys)) and the two DSP variants (p.(Asn4Lys) and (p.Ser53Ala)). No variants were reported that could explain the developmental delay.
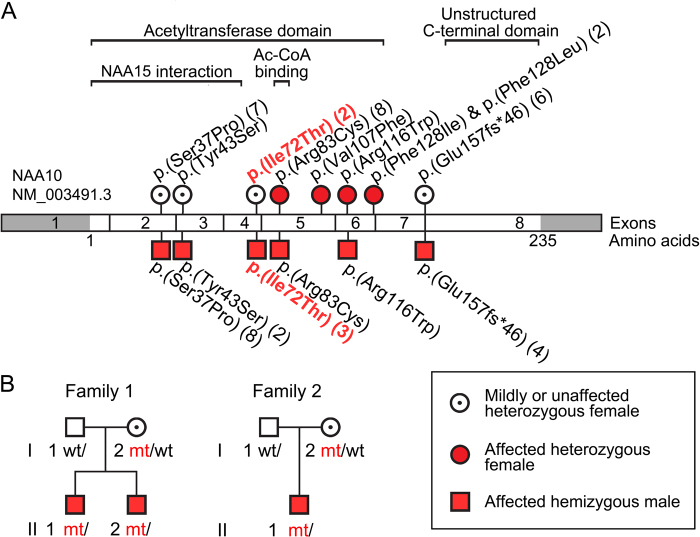
This proband in Family 1, individual II-1, was WES in the clinical lab, and further genetic studies including WES of his brother (individual II-2) and the mother were performed as part of the Baylor–Hopkins Center for Mendelian Genomics WGL2CMG study described previously by Eldomery and colleagues (cases BH5665_1 and BH5665_4) [31]. Both brothers were found to be hemizygous for the exon 4 variant in NAA10 c.215T>C (NM_003491.3, NG_031987.1), with the predicted protein effect p.(Ile72Thr) and the variant submitted to ClinVar (SCV000494176). The mother was found to be heterozygous for the c.215T>C variant. Both parents are healthy without any cardiac symptoms or abnormality. The mother is from El Salvador and the father from Mexico (Fig. 1).
Fig. 1.
NAA10 variants a NAA10 variants identified to date in patients with global development delays and/or intellectual disabilities. Variants found in female patients are shown as red circles; variants identified in male patients as red squares. Heterozygous females with no or very mild phenotypes reported are shown as white circles with a black dot. b Pedigrees for family 1 and family 2 presented in this study
His brother, Individual II-2 in Family 1, was born full term after a normal pregnancy and had birth weight 3.629 kg. He has had normal growth since birth, at 5 years 5 months of age his height was 103.7 cm (5–10p) and weight 17.5 kg (25–50p). He was already at 9 months of age referred to the pediatric cardiologist for the possibility of HCM due to the positive family history, and obstructive hypertrophic cardiomyopathy was confirmed in him as well. At 1 year of age the echocardiogram showed findings consistent with obstructive hypertrophic cardiomyopathy, severe asymmetric septal hypertrophy with mild dynamic obstruction, left ventricular end-diastolic dimension measures below normal for BSA and qualitatively hyperdynamic systolic function. He had moderate right ventricular hypertrophy, a small fenestrated secundum ASD, and small membranous VSD. He was started on propranolol, later transitioned from propranalol to atenolol, and his atenolol has been up-titrated at the follow-up visits to the hospital and pediatric cardiologist. At 4½ years of age he has a stable moderate asymmetric septal hypertrophy and mild dynamic LVOTO (~2 m/s). He had speech and gross motor delay, but less severe as compared his older brother. He has a mixed expressive/receptive language disorder, DD, and ‘behavioral arrests’ in the setting of normal EEG, thought to be behavioral rather than epileptic in etiology. Brain imaging has not been performed.
Clinical report: family 2, patient II-1
The third patient (family 2, II-1) with the NAA10 c.215T>C p.(Ile72Thr) variant was referred for genetic evaluation at 18 months of age for developmental delay in speech and language, and dysmorphic features. These features were thought to be mild and not warranting extensive evaluation so close clinical follow-up was planned. An MRI of the brain at that time showed mild periventricular leukomalacia of unclear significance. At 25 months of age, he was re-evaluated for sudden onset and rapidly worsening gait ataxia. Laboratory evaluation revealed no immediate cause for his abnormalities. While a repeat brain MRI was pending, concerns for a possible genetic etiology led to initiation of WES. The repeat brain MRI identified a new posterior fossa mass that developed in the interim. A gross total resection of the tumor was performed and identified as a medulloblastoma. He was treated according to COG protocol ACNS0334 using three courses of standard chemotherapy followed by three courses of high-dose chemotherapy with autologous stem cell rescue and no radiation therapy. A cardiac evaluation in preparation for high-dose chemotherapy demonstrated hypertrophic cardiomyopathy and QT interval prolongation. The NAA10 c.215T>C p.(Ile72Thr) variant was identified at this time and considered as the likely cause for his cardiac abnormalities. His treatment course was relatively uncomplicated with the exception of one episode of cardiac decompensation accompanying septic shock that was thought to be related to his underlying cardiac abnormalities. He completed that treatment course and there is no evidence of tumor recurrence 16 months later. The family is of Oklahoma decent, both parents are are healthy without any cardiac symptoms or abnormality. Phenotypic features of all three patients are summarized in Table 1.
Table 1.
Clinical features of the three patients
| General information | Patient II-1, Family 1 | Patient II-2, Family 1 | Patient II-1, Family 2 |
|---|---|---|---|
| Gender | M | M | M |
| Variant in NAA10 (coding, protein and genomic position) | NM_003491.3, c.215T>C,p.(Ile72Thr), chrX:153198002A>G (hg19) | NM_003491.3, c.215T>C,p.(Ile72Thr), chrX:153198002A>G (hg19) | NM_003491.3, c.215T>C,p.(Ile72Thr), chrX:153198002A>G (hg19) |
| Variant detected by | WES, confirmed Sanger | WES, confirmed Sanger | WES, confirmed Sanger |
| Inheritance | Maternal | Maternal | Maternal |
| Gestational week | NA | Full term | Full term |
| Age at last investigation | 8y 6mo | 5y 6mo | 3y 3mo |
| Birth and growth parameters | |||
| Weight (kg/SDS) | At birth: NA | At birth: 3.629 kg (28,3%) | At birth: 3.827 kg |
| 8y 6mo: 24.2 kg (17%) | 5y 6mo: 17.5 kg (36%) | 3y 3mo: 13.73 kg (25%) | |
| Length (cm/SDS) | At birth: NA | At birth: (25%) | At birth: 52.7 cm |
| 8y 6mo: 124.7 cm (24%) | 5y 6mo: 103.7 cm (15%) | 3y 3mo: 91.2 cm (7%) | |
| Head circumference (cm/SDS) | At birth: NA | At birth (37%) | At birth: NA |
| NA | NA | 3y: 49.4 cm | |
| Neurological | |||
| Development delay or ID | Yes | Yes | Yes |
| Behavioral anomalies | Yes | No | No |
| Delayed motor development | Yes | Yes | Yes |
| Age of walking | 19mo | NA | 18mo |
| Age of first words | At 4 years not 2 words together | NA | 15mo |
| Speech abilities | Delayed | Delayed | Delayed |
| Brain imaging | Thin corpus callosum, relative paucity of frontal lobe | Not performed | Mild periventricular leukomalacia, medulloblastoma |
| Organs | Hypertrophic cardiomyopathy, inguinal hernia | Hypertrophic cardiomyopathy | Hypertrophic cardiomyopathy |
| Skeletal | — | — | Delayed closure of anterior fontanelle, barrel chest, high-arched palate |
| Facial | — | — | Rather thick lips, large, wide-spaced teeth, very blond |
| Other | — | — | Dysphagiogram because of excessive drooling |
NA not available
Multiple sequence alignment and structural modeling
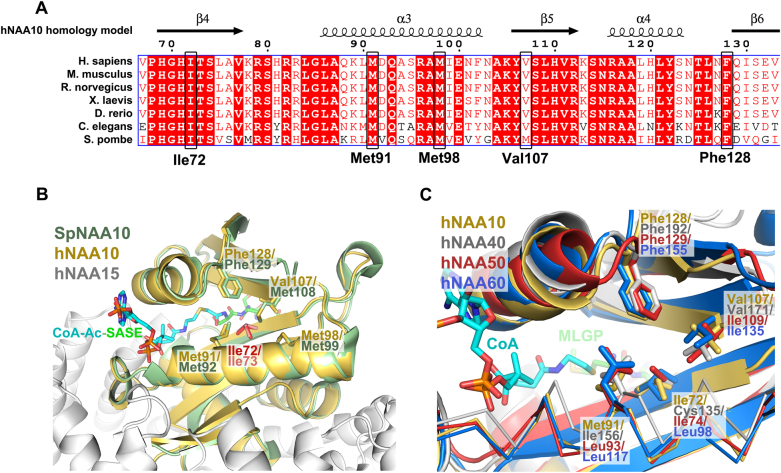
NAA10 multiple sequence alignment reveal that Ile72 is highly conserved through evolution from human down to yeast. Ile72 is located in the core of the protein with its side chain forming a hydrophobic pocket together with residues Met91, Met99, Val107 and Phe128 (of human NAA10), all of which also are highly conserved (Fig. 2a, b). Two of these residues (Val107 and Phe128) have previously been found altered by missense variants in four female patients with intellectual disabilities and developmental delay as part of the phenotype [25, 26]. These NAA10 variants (c.319G>T p.(Val107Phe), c.384T>A p.(Phe128Ile) and c.382T>A p.(Phe128Leu)) were shown to have reduced protein stability, and reduced catalytic activity. The hydrophobic pocket formed by these residues is also present in other NATs (NAA10 orthologues) for which X-ray crystal structures have been solved (NAA40, NAA50, NAA60) (Fig. 2c), in an archaea NAT (PDB ID: 4LX9, data not shown) and also in several lysine acetyltransferases that adopt the GCN5-related N-acetyltransferase (GNAT) fold (PDB ID: 1QST, 1BOB, 2P0W, data not shown). Altogether this suggests that introducing residues that disrupts or alters this hydrophobic pocket will have severe effects on NAA10 protein stability and/or catalytic function.
Fig. 2.
Multiple sequence alignment and structural alignment of NAA10. a Multiple sequence alignment of NAA10 orthologues from human, mouse, rat, frog, zebrafish, roundworm, and yeast. Numbering of amino acids correspond to the residue number in human NAA10. b Structural alignment of a hNatA homology model [23] with the crystal structure of NatA complexed S. pombe NAA10 [40] (SpNAA10) bound to a bisubstrate analog (PDB ID of SpNatA: 4KVM). SpNAA10 is shown in green, hNAA10 in yellow and hNAA15 in white. The bisubstrate analog bound to SpNAA10 is shown in cyan and green sticks. c Structural alignment of human NATs. The hNAA10 homology model is structurally aligned with crystal structures of hNAA40 [41] (gray cartoon), hNAA50 [42] (red cartoon), and hNAA60 [43] (blue cartoon) (PDB IDs: 4U9W, 3TFY, and 5ICV)
Functional testing of Naa10 Ile72Thr
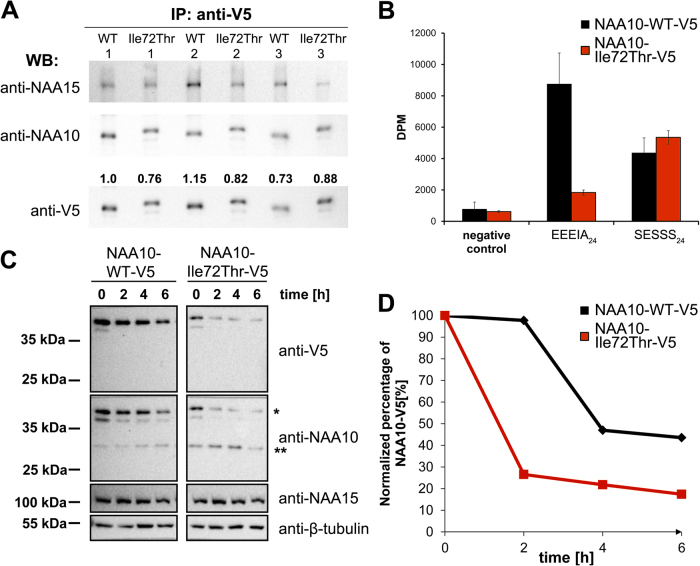
In order to study the catalytic activity of NAA10 Ile72Thr and NAA10 WT, we overexpressed and immunoprecipitated V5-tagged NAA10 from HeLa cells using antibodies towards the V5 fusion tag. Immunoprecipitated samples were subjected to in vitro NAT acetylation assays, and the amount of NAA10-WT-V5 or NAA10-Ile72Thr-V5 in each sample was determined by Western blotting (Fig. 3a). NAA15 co-immunoprecipitated with both NAA10-WT-V5 and NAA10-Ile72Thr-V5, suggesting that both NAA10 variants are able to form a NatA complex in our assay. The catalytic activity of the immunoprecipitated sample was then tested toward peptides EEEIA24 and SESSS24 which represent in vitro substrates for monomeric NAA10 and the NatA complex, respectively [2, 36]. Results from these experiments revealed an ~75% reduction in catalytic activity towards the in vitro monomeric NAA10 substrate EEEIA24, and surprisingly no reduction in catalytic activity when tested for the canonical NatA substrate SESSS24 (Fig. 3b). As NAA10 is dependent on the interaction with NAA15 for acetylation of canonical NatA substrates, our data thus suggest that NAA10 Ile72Thr is able to form a functional NatA complex, but has impaired function in a monomeric state.
Fig. 3.
Activity measurements and turnover rate of NAA10-Ile72Thr-V5 versus NAA10-WT-V5. a Quantification of immunoprecipitated NAA10-WT-V5 and NAA10-Ile72Thr-V5 used in acetylation assays. Quantification of V5-bands were performed using the ChemiDocTM XRS+ system, and the ImageLab 5.1 software from Bio-RAD. b Catalytic activity of immunoprecipitated NAA10 variants. The catalytic activity was measured and correlated to the amount of NAA10 present in each sample as determined by densitometry analysis in a. c Western blot of cycloheximide chase experiments. Samples were Western blotted and analyzed with anti-V5, anti-NAA10, anti-NAA15, and anti-β-tubulin as a loading control. Anti-NAA10 recognize two main bands in the western blot, denoted * and **, bands marked with * corresponds to the expected size of V5-tagged NAA10 while ** corresponds to the expected size of endogenous untagged NAA10. d Band intensities of V5-tagged NAA10 variants were quantified as in a, and the value for each time point were correlated to the measured density of t0
In order to further asses the stability of NAA10 Ile72Thr, we transfected HeLa cells with plasmids encoding V5-tagged NAA10 WT or NAA10 Ile72Thr and performed Cycloheximide chase experiments with antibodies against the V5 tag, NAA10, NAA15 and β-tubulin. These experiments revealed a clearly higher turnover rate for NAA10-Ile72Thr-V5 compared to NAA10-WT-V5 (Fig. 3c, d) suggesting that NAA10-Ile72Thr-V5 has a decreased stability and shorter half-life when expressed in HeLa cells compared to NAA10 WT.
DISCUSSION
We have identified a NAA10 missense variant, c.215T>C p.(Ile72Thr), in three male patients from two unrelated families with mild development delay, intellectual disabilities and HCM as overlapping phenotypes. Ile72 is highly conserved, and has an important structural role in NAA10, forming a hydrophobic pocket together with other amino acids (Val107, Phe128) that previously have been identified substituted in female patients with similar (although more severe) phenotypes [26]. The NAA10 p.(Ile72Thr) missense variant is not contained in EcAC or gnomAD (i.e., no occurrences of the variant, reviewed 19 December 2017). Functional studies of NAA10-Ile72Thr-V5 revealed an increased turnover rate when expressed in human cell lines (~60–70% of proteins being degraded in the first 2 h after Cycloheximide treatment, while the remaining 20–30% only slowly decayed in the following 4 h), and a reduced stability when recombinantly expressed in bacteria (we were not able to purify this protein as a monomer (Figure S1)). This together with our results from the IP NAA10-V5 NAT activity assays suggests that NAA10 Ile72Thr together with NAA15 is able to form a functional NatA complex, but at the same time that NAA10 in a monomeric form is destabilized and rapidly degraded. We speculate that while monomeric NAA10 is affected by the Ile72Thr substitution, a portion of the overexpressed NAA10 molecules in our assays are interacting with endogenous NAA15. This may stabilize and protect NAA10 Ile72Thr from being degraded and thus rescues the catalytic activity of the protein towards NatA substrates. Altogether, according to ACMG criteria, based on the functional studies presented here (PS3), the missense constraint metric of 2.59 in EcAC (PM2, PP1 and PP2), the damaging predictions in MutationTaster, SIFT, PolyPhen (data not shown) and a CADD score of 26.2 (PP3) we consider the variant as likely pathogenic.
Since the discovery of Ogden syndrome [21], several variants that affect NAA10 function have been identified in both boys and girls, greatly expanding the phenotypic spectrum associated with variants in NAA10 [24-27] (Table 2). The three patients described here with mild development delay, intellectual disabilities and HCM as overlapping phenotypes, have milder phenotypes than all of the previously described patients with variants affecting NAA10 function. Similar to previously described patients, patient II-1 from family 2 also had mild skeletal and facial anomalies [21,24–26], and MRI of patient II-1 from family 1 revealed thin corpus callosum which has been described previously in patients with NAA10 variants. MRI of patient II-1 from family 2 revealed periventricular leukomalacia and medulloblastoma. NAA10 has repeatedly been connected to the development of different types of cancers [17] and we cannot exclude the possibility that the p.(Ile72Thr) variant has played a role in the development of malignancy. That said, this has not been seen in any of the previously described patients with NAA10 variants affecting function, and NAA10 has to our knowledge not been previously connected to development of medulloblastoma.
Table 2.
Overview of all NAA10 variants with phenotype and functional data
| Variant no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| This study | Casey et al. [24] | Popp et al. [25], Saunier et al. [26] | Saunier et al. [26] | Saunier et al. [26] | Saunier et al. [26], Sidhu et al. [27] | Popp et al. [25], Saunier et al. [26] | Rope et al. [21], Myklebust et al. [23] | Esmailpour et al. [28], Forrester et al. [29] | |
| NAA10 variant (NM_003491.3) | c.215T>C p.(Ile72Thr) | c.128A>C p.(Tyr43Ser) | c.319G>T p.(Val107Phe) | c.384T>A p.(Phe128Ile) | c.382T>A p.(Phe128Leu) | c.247C>T p.(Arg83Cys) | c.346C>T p.(Arg116Trp) | c.109T>C p.(Ser37Pro) | c.471+2T>A (p.Glu157fs45*) |
| Patients | Male (3/3) | Male (2/2) | Female (1/1) | Female (1/1) | Female (2/2) | Female (8/9) Male (1/9) | Female (1/2) Male (1/2), | Male (8/8) | Male (4/4) |
| Inheritance | Inherited | Inherited | De novo | De novo | De novo | De novo/MGM | De novo | Inherited | Inherited |
| Age of last examination (age of death) | 3y 3mo–8y 6mo | 20y–25y | 8y 2mo | 6y 5mo | 17mo–8y 6mo | 2y 2mo–14y (male patient died 1 week old) | 5y 11mo–8y 1mo | 5–16mo (5–16 mo) | 14y–28y |
| X-inactivation | — | — | Random | Random | Random (1/2), NA (1/2) | Random (1/8), 82% (1/8), 100% (1/8), NA (5/8) | Random | — | — |
| Female carriers | No reported phenotypes | Mild ID, cardiac arrhythmias and long QT, down slanting PF, VT, premature CAD, dysmorphic facial features, random XCI | — | — | — | — | — | No reported phenotypes, >90% X-inactivation | Toe syndactyly, short terminal phalanges |
| Patient phenotype | |||||||||
| DD/ID | Yes | Mild (1/2) moderate (1/2) | Severe | Yes | Profound (1/2) severe (1/2) | Severe (6/9), moderate (1/9), yes (1/9), NA (1/9) | Severe (1/2) moderate (1/2) | Severe | Severe (3/4), mild (1/4) |
| Growth failure | No | Yes (1/2) | Yes | Yes | Yes (1/2) | Yes (6/9), No (2/9), NA (1/9) | Yes (1/2) | Yes | Yes |
| Neurological | Thin CC (1/3), relative paucity of frontal lobe (1/3), mild PVL (1/3), medulloblastoma (1/3) | Hypo, mild dilation of LV, mild CA, seizures (1/2), prominent SF (1/2) | Axial hypo/periph hyper, thin cc, borderline normal ventricles | Axial Hypo/periph hyper, superventricular cyst without hydrocephaly | Axial Hypo/ periph hyper (1/2), severe hypo (1/2), thin CC, parenchymal atrophy, seizures /infantile spasms | Hypo (7/9), hyper (2/9), WMVL (2/9), thin CC (1/9), VM (1/9), PVL and HIE (1/9) | Axial hypo/ periph hyper, enlarged ventricles, reduced periventricular volume, thin CC. | Truncal hypo (4/8), CA or immature CC (3/8), enlarged ventricles (2/8) | Hypo, bilateral anopthalmia/micropthalmia, seizures (2/4), ASCVD (1/4) |
| Organs | Hypertrophic cardiomyopathy, inguinal hernia (1/3) | Cardiac arrhythmias, long QT, innocent HM, inguinal hernia, VT (1/2) | PAS, ASD, long QT | IRBBB | Mild left ventricular diastolic dysfunction (1/2) | PAS (1/9), PFO vs. ASD (1/9), long QT (1/9), SVT (1/9), pulmonary hyper (1/9), VH (2/9), IRBBB (1/9) | IRBBB (1/2) | Cardiac anomalies (VSD, PAS) (6/8) cardiac arrhythmias (5/8), inguinal hernia (3/8) | Right VH (1/4) |
| Skeletal | Delayed closure of anterior fontanelle (1/3), barrel chest (1/3), high-arched palate (1/3) | Scoliosis, small hands/feet, BAD, Toe syndactyly (1/2) | Delayed closure of fontanels, large fontanels, delayed bone age, broad great toes, mild pectus carinatum | — | — | Large fontanels (4/9), small feet/hands (2/9), clinodactyly (1/9), pectus exavatum (3/9), broad great toes (1/9) | Small feet/hands (1/2), high-arched palate (1/2), wide interdental spaces (1/2) | Scoliosis, large fontanels (5/8), broad or widely spaced toes (2/8), delayed osseous development (1/8) | High-arched palate, clinodactyly or syndactyly, scoliosis (3/4), pectus exavatum (3/4), pes planus (2/4) |
| Other | Minor unspecific dysmorphic facial features (1/3) | Prominent dysmorphic facial features | Minor unspecific dysmorphic facial features | Minor unspecific dysmorphic facial features | Minor unspecific dysmorphic facial features | Minor unspecific dysmorphic facial features, autism spectrum disorder | Minor unspecific dysmorphic facial features, hypoplastic scrotum | Dysmorphic facial features, large ears (6/8), prominent eyes (4/8), cryptorchidism (5/8) | Dysmorphic facial features, large abnormally formed ears and abnormally developed eyes |
| Functional studies | |||||||||
| Catalytic activity | Reduced NAA10 activity, most likely intact NatA activity | Reduced NAA10 activity | Reduced NAA10 activity | NA | Reduced NAA10 activity | Reduced NAA10 activity, most likely reduced NatA activity | Reduced NAA10 activity | Reduced NAA10 activity, reduced NatA activity | NA |
| Protein stability | Reduced | Reduced | Most likely reduced | NA | Reduced | No reduction in protein stability | No reduction in protein stability | No reduction in protein stability | NA |
| Other features | — | — | — | — | — | — | — | Reduced NatA complex formation | Loss of NAA10 expression |
This table lists main phenotypic features and main findings from functional studies only. For a full overview of patient phenotypes and results from functional studies for each variant, please see respective references.
ASCVD, arteriosclerotic vascular disease; ASD, atrial septal defect; BAD, bilateral acetabular dysplasia; CA, cerebral atrophy; CAD; coronary artery disease; CC, corpus calossum; DD, development delay; HIE, hypoxic-ischemic encephalopathy; HM, heart murmur; hyper, hypertonia; hypo, hypotonia; ID, intellectual disabilities; IRBBB, Incomplete right bundle branch block; LV, lateral ventricles; MGM, maternal germ line mosaicism; mo, months; NA, not available; PAS, Pulmonary artery stenosis; periph, peripheral; PF, palpebral fissures; PFO, Patent Foramen Ovale; PVL, periventricular leukomalacia; SF, sylvian fissures; SVT, superventricular tachycardia; VH, ventricular hypertrophy; VM, ventriculomegaly; VT, ventricular tachycardia; WMVL, white matter volume loss; XCI, X-chromosome inactivation; y, years.
When the first NAA10 variants emerged, the observed phenotypes were directly linked to loss of Nt-acetylation, and it was speculated in correlations between the severity of phenotypes and remaining catalytic NAT activity from Nt-acetylation assays [21, 25]. However, as more NAA10 variants emerge, our understanding of how these variants are causing disease is becoming increasingly complex, and it is likely that different variants are causing disease through different mechanisms. In Table 2, we have summarized phenotypes and results of all the NAA10 variants that have been functionally assessed and published so far. Similar to the functional studies of the NAA10 Ile72Thr presented here (Table 2, variant 1), both NAA10 Tyr43Ser, Phe128Leu and Phe128Ile (Table 2, variants 2, 3, and 4) were found to greatly destabilize NAA10 [24, 26]. The variants Arg83Cys and Arg116Trp (Table 2, variants 6 and 7) however were shown to not destabilize, but rather directly reduce the catalytic activity of NAA10 due to hampered Acetyl CoA binding, and the Ser37Pro variant (Table 2, variant 8) causing Ogden syndrome did not destabilize NAA10 but rather caused a reduced catalytic activity in addition to loss of NatA complex formation [21, 25, 26]. Interestingly, proteomic studies of patient-derived cell lines expressing the NAA10 p.(Ser37Pro) variant showed that although both a reduced NAA10 catalytic activity and NatA catalytic activity was observed in NAT in vitro assays, mainly NatA substrates had a reduced level of Nt-acetylation, suggesting that NAA10 p.(Ser37Pro) affects NatA function rather than NAA10 function. In the case of Ile72Thr, our data show that NatA function is not affected and thus suggest that the patient phenotypes may be caused by loss of NAA15- and NatA-independent NAA10 activities such as NAT activities toward acidic N-termini [36], KAT activities [37, 38], or even non-catalytic functions of NAA10 [39]. Based on this, we speculate that loss of NatA function is more severe than loss of monomeric NAA10 function and that this can explain the large discrepancy in phenotype seen between patients with NAA10 p.(Ile72Thr) and p.(Ser37Pro) variants. Another NAA10 variant, c.332 T > G p.(Val111Gly), was published while this manuscript was in press [44]. Interestingly, as for NAA10 Ile72Thr, NAA10 Val111Gly is destabilized and displays an impaired monomeric NAA10 NAT activity, while the NatA-mediated NAT-activity appears to be intact.
NAA10 is an X-linked gene and not surprisingly, males that are hemizygous of NAA10 variants affecting function are more severely affected clinically compared to heterozygous females. For several of the NAA10 variants, heterozygous females only have mild phenotypes or no phenotypes at all, most likely due to skewed X-inactivation [21, 23, 24, 28]. X-inactivation might also explain why the two mothers heterozygous for NAA10 p.(Ile72Thr) have no phenotypes. There are two cases in which a NAA10 missense variant is causing disease in both females and males (p.(Arg83Cys) and p.(Arg116Trp), Table 2, variants 6 and 7), in both cases the male patient had more severe phenotypes compared to heterozygous females [25, 26]. Variants 1 and 3–5 in Table 2 (p.(Ile72Thr), p.(Val107Phe), p.(Phe128Ile), and p.(Phe128Leu)) are all affecting the same hydrophobic pocket in NAA10 and functional testing suggest that the severity of each of these variants are similar on a molecular level. It is therefore very interesting that the phenotype of the three boys described here clearly is less severe than the phenotypic spectrum of the four female patients with random XCI and missense variants affecting the same hydrophobic pocket of NAA10 (Fig. 2b, Table 2) [25, 26]. Altogether the phenotypes of the three patients described here together with our functional assay suggest that the NAA10 p.(Ile72Thr) variant is likely pathogenic, but that the variant is less severe compared to most of the previously described variants that affect NAA10 function. Our data also underscore that each of the identified NAA10 variants needs to be revisited in order to achieve a better understanding of the molecular mechanisms underlying clinical manifestations associated with NAA10 deficiency.
Ethics approval and consent to participate
The Baylor-Hopkins Center for Mendelian Genomics WGL2CMG research study has been approved by the Baylor College of Medicine Institutional Review Board (protocol H-29697). The Baylor College of Medicine IRB (IORG number 0000055) is recognized by the United States Office of Human Research Protections (OHRP) and Food and Drug Administration (FDA) under the federal wide assurance program. The Baylor College of Medicine IRB is also fully accredited by the Association for the Accreditation of Human Research Protection Programs (AAHRPP). Prior to transferring WES data for the proband from clinical lab to BHCMG research lab, and performing WES of the additional family members, written informed consent was obtained for each individual included in the study.
Electronic supplementary material
Acknowledgements
We are very grateful for the participation of the two families that are involved in this study. The work has been supported by the Research Council of Norway (Project 249843), the Norwegian Health Authorities of Western Norway (Project 912176), and Bergen Research Foundation (BFS). The Baylor-Hopkins Center for Mendelian Genomics (BHCMG) has been funded by the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) grant number UM1HG006542
Compliance with ethical standards
Conflict of interest
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical exome sequencing. JRL serves on the Scientific Advisory Board of BG. JRL has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., and is a coinventor of US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. All the remaining authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0136-0) contains supplementary material, which is available to authorized users.
References
- 1.Bienvenut WV, Sumpton D, Martinez A, et al. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol Cell Proteom. 2012;11:M111 015131. doi: 10.1074/mcp.M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnesen T, Van Damme P, Polevoda B, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106:8157–62. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–8. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behnia R, Panic B, Whyte JR, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 2004;6:405–13. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- 5.Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9:e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 2004;6:414–9. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- 7.Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun. 2014;5:4383. doi: 10.1038/ncomms5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–7. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–51. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen JR, Kayne PS, Moerschell RP, et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–75. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park EC, Szostak JW. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11:2087–93. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnesen T, Anderson D, Baldersheim C, Lanotte M, Varhaug JE, Lillehaug JR. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem J. 2005;386(Pt 3):433–43. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingram AK, Cross GAM, Horn D. Genetic manipulation indicates that ARD1 is an essential N-alpha-acetyltransferase in Trypanosoma brucei. Mol Biochem Parasit. 2000;111:309–17. doi: 10.1016/S0166-6851(00)00322-4. [DOI] [PubMed] [Google Scholar]
- 14.Sonnichsen B, Koski LB, Walsh A, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Mijares M, Gall MD, et al. Drosophila variable nurse cells Encodes Arrest Defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Dev Dyn. 2010;239:2813–27. doi: 10.1002/dvdy.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ree R., Myklebust L. M., Thiel P., Foyn H., Fladmark K. E., Arnesen T. The N-terminal acetyltransferase Naa10 is essential for zebrafish development. Bioscience Reports. 2015;35(5):e00249–e00249. doi: 10.1042/BSR20150168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalvik TV, Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32:269–76. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- 18.Arnesen T, Gromyko D, Pendino F, Ryningen A, Varhaug JE, Lillehaug JR. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006;25:4350–60. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- 19.Gromyko D, Arnesen T, Ryningen A, Varhaug JE, Lillehaug JR. Depletion of the human Nalpha-terminal acetyltransferase A induces p53-dependent apoptosis and p53-independent growth inhibition. Int J Cancer. 2010;127:2777–89. doi: 10.1002/ijc.25275. [DOI] [PubMed] [Google Scholar]
- 20.Lim JH, Park JW, Chun YS. Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677–82. doi: 10.1158/0008-5472.CAN-06-3171. [DOI] [PubMed] [Google Scholar]
- 21.Rope AF, Wang K, Evjenth R, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Damme P, Stove SI, Glomnes N, Gevaert K, Arnesen T. A Saccharomyces cerevisiae model reveals in vivo functional impairment of the Ogden syndrome N-terminal acetyltransferase NAA10 Ser37Pro mutant. Mol Cell Proteom. 2014;13:2031–41. doi: 10.1074/mcp.M113.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myklebust LM, Van Damme P, Stove SI, et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Human Mol Genet. 2015;24:1956–76. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casey JP, Stove SI, McGorrian C, et al. NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment. Sci Rep. 2015;5:16022. doi: 10.1038/srep16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popp B, Stove SI, Endele S, et al. De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. Eur J Human Genet. 2015;23:602–9. doi: 10.1038/ejhg.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunier C, Stove SI, Popp B, et al. Expanding the phenotype associated with NAA10-related N-terminal acetylation deficiency. Hum Mutat. 2016;37:755–64. doi: 10.1002/humu.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidhu Mandeep, Brady Lauren, Tarnopolsky Mark, Ronen Gabriel M. Clinical Manifestations Associated With the N-Terminal-Acetyltransferase NAA10 Gene Mutation in a Girl: Ogden Syndrome. Pediatric Neurology. 2017;76:82–85. doi: 10.1016/j.pediatrneurol.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Esmailpour T, Riazifar H, Liu LN, et al. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J Med Genet. 2014;51:185–96. doi: 10.1136/jmedgenet-2013-101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forrester S, Kovach MJ, Reynolds NM, Urban R, Kimonis V. Manifestations in four males with and an obligate carrier of the Lenz microphthalmia syndrome. Am J Med Genet. 2001;98:92–100. doi: 10.1002/1096-8628(20010101)98:1<92::AID-AJMG1009>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 30.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 31.Eldomery MK, Coban-Akdemir Z, Harel T, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid JG, Carroll A, Veeraraghavan N, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinform. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drazic A, Arnesen T. [14C]-Acetyl-coenzyme A-based in vitro N-terminal acetylation assay. Methods Mol Biol. 2017;1574:1–8. doi: 10.1007/978-1-4939-6850-3_1. [DOI] [PubMed] [Google Scholar]
- 36.Van Damme P, Evjenth R, Foyn H, et al. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N{alpha}-acetyltransferases and point to hNaa10p as the post-translational actin N{alpha}-acetyltransferase. Mol Cell Proteom. 2011;10:M110 004580. doi: 10.1074/mcp.M110.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian X, Li X, Cai Q, et al. Phosphoglycerate kinase 1 phosphorylates Beclin1 to induce autophagy. Mol Cell. 2017;65:917–31 e916. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon H, Kim HL, Chun YS, et al. NAA10 controls osteoblast differentiation and bone formation as a feedback regulator of Runx2. Nat Commun. 2014;5:5176. doi: 10.1038/ncomms6176. [DOI] [PubMed] [Google Scholar]
- 39.Lee CC, Peng SH, Shen L, et al. The Role of N-alpha-acetyltransferase 10 protein in DNA methylation and genomic imprinting. Mol Cell. 2017;68:89–103 e107. doi: 10.1016/j.molcel.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liszczak G, Goldberg JM, Foyn H, Petersson EJ, Arnesen T, Marmorstein R. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol. 2013;20:1098–105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magin RS, Liszczak GP, Marmorstein R. The molecular basis for histone H4- and H2A-specific amino-terminal acetylation by NatD. Structure. 2015;23:332–41. doi: 10.1016/j.str.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liszczak G, Arnesen T, Marmorstein R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J Biol Chem. 2011;286:37002–10. doi: 10.1074/jbc.M111.282863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stove SI, Magin RS, Foyn H, Haug BE, Marmorstein R, Arnesen T. Crystal structure of the Golgi-associated human Nalpha-acetyltransferase 60 reveals the molecular determinants for substrate-specific acetylation. Structure. 2016;24:1044–56. doi: 10.1016/j.str.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McTiernan N, Støve SI, Aukrust I, Mårli MT, Myklebust LM, Houge G, Arnesen T. NAA10 dysfunction with normal NatA-complex activity in a girl with non-syndromic ID and a de novo NAA10 p.(V111G) variant - a case report. BMC Med Genet. 2018;19:47. doi: 10.1186/s12881-018-0559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.