Abstract
Chronic hepatitis C virus infection often leads to liver cirrhosis and primary liver cancer. In 2015, an estimated 71 million people were living with chronic HCV. Although infection rates have decreased in many parts of the world over the last several decades, incidence of HCV infection doubled between 2010 and 2014 in the United States mainly due to increases in intravenous drug use. The approval of direct acting antiviral treatments is a necessary component in the elimination of HCV, but inherent barriers to treatment (e.g., cost, lack of access to healthcare, adherence to treatment, resistance, etc.) prevent dramatic improvements in infection rates. An effective HCV vaccine would significantly slow the spread of the disease. Difficulties in the development of an HCV culture model system and expression of properly folded- and natively modified-HCV envelope glycoproteins E1 and E2 have hindered vaccine development efforts. The recent structural and biophysical studies of these proteins have demonstrated that the binding sites for the cellular receptor CD-81 and neutralizing antibodies are highly flexible in nature, which complicate vaccine design. Furthermore, the interactions between E1 and E2 throughout HCV infection is poorly understood, and structural flexibility may play a role in shielding antigenic epitopes during infection. Here we discuss the structural complexities of HCV E1 and E2.
Keywords: hepatitis C virus, envelope glycoprotein, vaccine design, HCV, E2, E1
Introduction
Hepatitis C virus (HCV) presents with mild symptoms; as an acute illness that resolves within weeks; or a lifelong, chronic infection that can lead to cirrhosis, liver cancer, and, if left untreated, death. End-stage, liver disease caused by chronic HCV infection is the leading cause of liver transplantation in the United States, Europe, and Japan (1–3). According to the World Health Organization, there were 1.75 million new HCV infections and 71 million people living with chronic HCV infection worldwide in 2015. Intravenous drug use and unsafe healthcare practices are responsible for a majority of new infections, contributing heavily to the doubling of HCV incidence in the United States between 2010 and 2014 (4, 5).
Despite FDA approval of several direct acting antiviral (DAA) treatments for HCV with very high success rates (>90%) for all genotypes, many at-risk groups are still spreading infection faster than they are being cured (5–8). Chronic HCV prevalence is about 1% of the total world population, but is much higher in many areas where healthcare is not widely accessible. Mongolia, Uzbekistan, Egypt, and Gabon, for example, have HCV prevalence ranging from 4 to 7% (5) and specific populations in the Nile Delta and Upper Egypt can have infection rates as high as 28%, varying heavily based on socioeconomic status (9). The poorest and least educated in Egypt have the highest HCV infection rates and simply do not have the means to receive treatment. Furthermore, intravenous drug use accounts for about 23% of new HCV infections (10). Populations of intravenous drug users worldwide must overcome several barriers to treatment such as high cost, access to healthcare, compliance, and fear of being discovered as a drug user (8). After a successful course of treatment however, if the patient continues engaging in risky behaviors, they are still at risk to be re-infected. These factors prevent a dramatic improvement in HCV infection rates worldwide. Therefore, it seems unlikely that DAAs alone will eliminate HCV infection without an effective vaccine.
HCV is an enveloped virus containing a positive-sense, single stranded RNA genome. The lipid envelope, derived from the host membrane, is embedded with two type I transmembrane proteins, envelope glycoproteins E1 and E2, which form a heterodimer (11). HCV particles are uniquely associated with lipids and apolipoproteins, which play a role in proper formation and function of secreted virions (12–19). These associations give viral particles an overall low buoyant density (16). The E1/E2 heterodimer is responsible for viral entry from recognition of host cell receptors to membrane fusion. Initial host-virus attachment interactions are through glycosaminoglycans and low-density lipoprotein receptor (20). Several receptors have a necessary role in entry such as claudin-1, occludin, CD81, and scavenger receptor class B type 1, mainly through interaction with E2, although the role of E1 is not fully understood (21–24). E1 and E2 are on the surface of the virion, available for host immune recognition, and are ideal for studies in immunogenicity ultimately leading to vaccine design; however, the conformation of the E1/E2 heterodimer and its interactions have not been well characterized throughout the various stages of virus assembly, host cell attachment, and membrane fusion. High quality, fully glycosylated and disulfide-linked envelope glycoproteins have proven to be difficult to produce in large quantities for biophysical study until recently.
Envelope glycoprotein E1
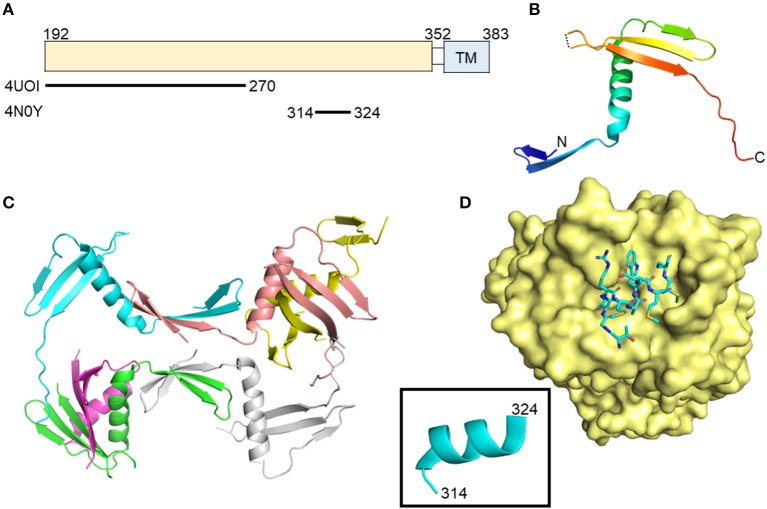
The exact role(s) of E1 during entry, egress, and immune escape is not fully understood (21–24). It has an N-terminal ectodomain of approximately 160 amino acids and exists as a trimer on the surface of cell culture-produced HCV particles, driven by interactions in the E1 C-terminal transmembrane region (25) (Figure 1). E1 may aid in recognition of hepatocytes through interactions with apolipoproteins, particularly ApoE, which further interacts with cell surface heparin sulfate during early attachment (26, 27). Structural data of N-terminal 79 amino acids of HCV E1 (nE1) was determined by X-ray crystallography (28). This structure showed a covalently linked, domain-swapped homodimer with nE1 forming 16 amino-acid α-helix flanked by a β-hairpin N-terminally and a three-stranded antiparallel β-sheet C-terminally (Figures 1A–C). The N-terminus of E1 does not resemble a class II fusion protein as hypothesized, or any other fusion protein conformation, despite having a fusion peptide-like domain (29); however, the published structure may be in a post-fusion conformation as crystals were obtained at a low pH. The cross-neutralizing, anti-E1 antibody IGH526 was shown to bind to an α-helical epitope (residues 314–324) predicted to be highly flexible in molecular dynamics simulations (Figure 1D) (30). This is the first E1 antigenic epitope structure described, and may assist in future vaccine design.
Figure 1.
Crystal structures of the N-terminal domain of E1 and an E1 peptide in complex with antibody. (A) Linear diagram of E1 glycoprotein. The crystallized E1 constructs and PDB IDs are shown below the diagram. (B) N-terminal domain structure of E1 monomer (PDB ID: 4UOI). (C) The six molecules of the asymmetric unit of the E1 N-terminal domain. (D) Structure of E1 peptide (aa314–324) in complex with antibody IGH526 (PDB ID: 4N0Y). The surface of antibody IGH526 is colored yellow, with the E1 peptide colored according to atom type (light blue, red, orange, and dark blue for carbon, oxygen, sulfur and nitrogen, respectively). The E1 peptide is further shown as ribbon structure in the box.
Envelope glycoprotein E2
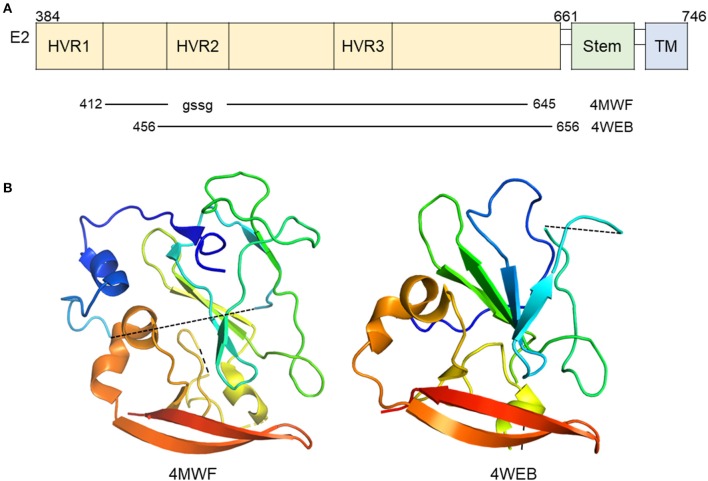
The functions of E2 have been more extensively studied relative to E1. E2 is responsible for mediating entry through interactions with several cellular receptors as mentioned above and is highly immunogenic (31–35). Two groups have published the structure of the core domain of E2 bound to Fabs (PDB ID: 4MWF and 4WEB) by X-ray crystallography (36, 37). The two studies employed a similar strategy with varying E2 expression constructs and antibodies for co-crystallization (Figure 2A). The 4MWF co-crystal was formed with E2 ectodomain (eE2) from HCV genotype 1a and a neutralizing, human Fab, AR3C that recognizing an N-terminal epitope in E2 and blocks E2-CD81 interaction. The eE2 in this structure does not contain hypervariable region 1 (HVR-1) and replaced HVR- 2 with a Gly-Ser-Ser-Gly linker. The 4WEB co-crystal was formed with eE2 from HCV genotype 2a, lacking the first 72 amino acids, and non-neutralizing Fab, 2A12, which binds a linear epitope at the C-terminus of eE2. Overall, both structures reveal a monomeric E2 with a globular nature (Figure 2B), unlike the class II fusion proteins that E2 was predicted to be similar to, and does not undergo major oligomeric or structural rearrangement upon exposure to low pH (37). Structural stability of the overall fold of the protein is provided by an extensive hydrophobic core and disulfide bonding. Follow-up alanine scanning studies mapped critically important E2 residues for neutralizing antibody recognition to core E2 stability elements and are in agreement with the published structures (38).
Figure 2.
Crystal structure of E2 in complex with antibody. (A) Linear diagram of E2 glycoprotein. The crystallized E2 core constructs and PDB IDs are shown below the diagram. (B) Ribbon representation of E2 (PDB ID: 4MWF and 4WEB).
The ordered portions of E2 are primarily arranged in β-sheets stabilized by disulfide bonds and hydrophobic interactions; however, a majority of eE2 (62% of it in the case of 4MWF) is in flexible loops or completely unstructured (36). Hydrogen-deuterium exchange and limited proteolysis experiments implicate the first 72 amino acids of eE2 containing HVR1 and region between HVR1 and HVR2 as highly flexible (37). In the 4MWF structure, the AR3C antibody binds this strand and provides stabilization for crystal formation (36). X-ray diffraction data for HVR2 could not be obtained (37). Therefore, in the absence of a stabilizing antibody, that leaves approximately the first 100 amino acids of eE2, containing several glycosylation sites, flexible and solvent exposed. This region is involved in epitope shielding, SR-BI binding, CD81 binding, and neutralizing antibody recognition (31–33, 39–45).
CD-81 binding site and neutralizing antibodies
Residues of E2 which form the CD81 binding site are found in clusters between aa412–446 and aa519–535 (termed the CD81 binding loop) of HCV genotype 1a strain H77 (36, 46, 47). Distant CD81 binding clusters are brought together by the overall fold of the protein. The two published eE2 structures, when compared, highlight the flexible nature of not only the CD81 binding loop, but the central immunoglobulin-like fold itself. In 4WEB, the CD81 binding loop is disordered, allowing hydrophobic residues to be solvent exposed. In the 4MWF structure, the CD81 binding loop is stabilized by a Fab fragment, bringing order to previously unstructured β-strand E and allowing residues such as F537 and L539 to be flipped into the hydrophobic core of the protein (Figure 3). In 2017, Vasiliauskaite et al. expanded on this observation by demonstrating that the hydrophobic residue positions and secondary structure in the CD81 binding loop of E2 were dependent on interactions with different neutralizing antibodies in both HCV pseudoparticles and cell culture-derived HCV particles (48). Given that the current evidence focus on binding to antibodies, the secondary structure of E2 bound to receptor CD81 may further reveal undescribed conformations.
Figure 3.
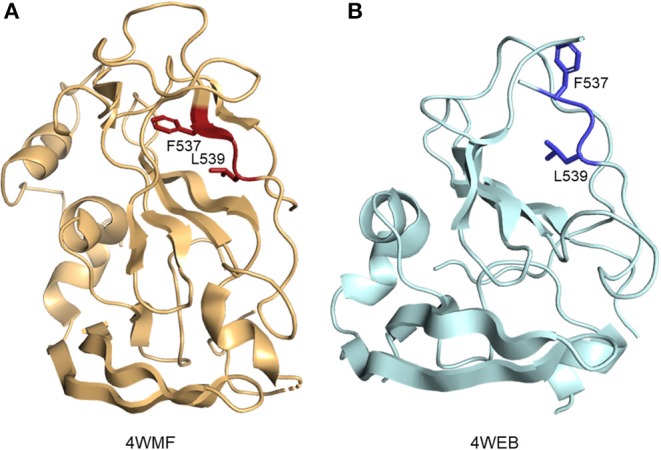
Ribbon representation of E2 hydrophobic residue positions in CD81 binding loop. (A) E2 residue F537 and L539 (side chains are shown as red sticks) in the presence of a stabilizing Fab fragment (not depicted) (PDB ID: 4MWF). (B) Solvent exposed positions of residue F537 and L539 (side chains are shown as blue sticks) (PDB ID: 4WEB).
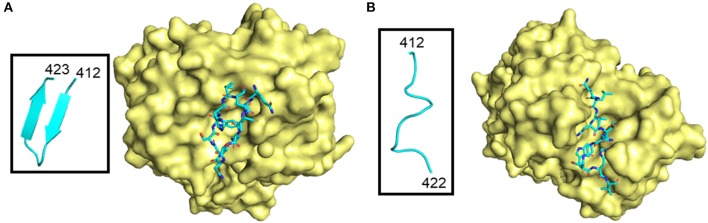
Many neutralizing antibody epitopes overlap CD81 binding residues of E2. For example, antibodies 3/11 and HCV1, as well as others, bind aa412–423, but recognize this flexible stretch of amino acids differently (32, 41, 49–52). The region adopts at least two different: an extended or “open” conformation or a β-hairpin (Figure 4) (50). Furthermore, the HCV1 antibody can bind from multiple angles, as visualized by electron microscopy, demonstrating not only local flexibility, but the long-reaching flexibility of the loop (53). Although the aa412–423 epitope is quite tempting for use in vaccine studies because of the cross-neutralizing antibody potential to functionally important residues, very few chronically infected HCV patients (<2.5%) produce a specific antibody response likely due to flexibility, and shielding by HVRs and glycans (45, 54–56). Recent studies seek to improve presentation of candidate epitopes and promote antigen recognition by the immune system using an engineered, cyclic immunogen. Initial data shows a designed E2 cyclic immunogen produced a strong antibody response in mice, whose serum was then used to successfully neutralize HCV infection in culture experiments (57). Further research will determine whether engineered derivations of this epitope will be useful in the pursuit of a viable HCV vaccine.
Figure 4.
Different conformations of E2 412–423 peptide in complex with antibodies. The antibody surface is colored yellow, and the peptide is shown as sticks colored according to atom type (light blue, red, orange, and dark blue for carbon, oxygen, sulfur and nitrogen, respectively). E2 peptides are further highlighted in boxes and shown as light blue ribbon structures. (A) β-hairpin peptide in complex with HCV1 antibody (PDB ID: 4DGY). (B) “Open” conformation peptide in complex with neutralizing antibody 3/11 (PDB ID: 4WHY).
HCV virion-associated E1 and E2
Structural information of the HCV virion is lacking, in sharp contrast to the closely related flaviviruses and alphaviruses. Obtaining a high-resolution, three dimensional structure of the HCV virion is difficult due to the low level of virus production in cell culture systems and the inherent heterogeneity of the particles owing to its association with apolipoproteins. Cryo-electron microscopy and tomography of HCV virions show spherical particles of highly heterogeneous size (40–100 nm in diameter). The particles displayed no obvious symmetry, no evidence of continuous membrane bilayer, and are covered by electron-dense material; although, the inherent low resolution of the electron micrographs may mask certain features (16). These findings perhaps call into question the hypothesis that HCV adopts a classical, icosahedral scaffold in which its two envelope glycoproteins anchor to the host cell-derived, double-layer lipid envelope. The lack of symmetry and membrane bilayer highlights the unique nature of the HCV virion relative to the other flaviviruses.
Higher-order aggregates of E1 and E2 on secreted virus particles appear to be covalently bonded, whereas non-covalently associated E1/E2 has been detected in the ER (58–60). At the moment, composition and structural information are unavailable for these higher order aggregates; however, one may glean insight from available structural information on E1 and E2 (28, 36, 37). The asymmetric unit of the nE1 structure contains six monomers stabilized by a series of intramolecular and intermolecular disulfide bonds (Figure 1C). It is possible that some or all of the intermolecular disulfides in the nE1 structure may be relevant to the higher order structures seen on the virion. The two eE2 structures are highly similar, with an RMSD of less than 0.8 Å for similar carbon-alpha positions with most of the differences located in loop regions. Interestingly, there are discrepancies in the disulfide bonding pattern in these regions. The overall fold of E2 core is unlikely to change in the virion, owing to its extensive hydrophobic core and three disulfide bonds formed between secondary structure elements.
The current structures available for E1 and E2 may reflect the immature forms of the proteins after initial synthesis and during virion assembly. Our hypothesis is that the folds found in these structures would be present on the virion, with the higher order aggregates formed via disulfide bonding through cysteines found in loop regions or within the core domains. During virion assembly and maturation, these core domains fold and higher order structures begin to form within the heterodimer or through interactions with other factors. The environment of the ER and Golgi apparatus during egress is oxidizing and compatible with disulfide bond formation and reshuffling, permitting the formation of the disulfide-linked aggregates. This maturation may contribute to the acid-resistance of extracellular HCV virions and have implications for the mechanisms of entry. Indeed, cell surface-bound HCV needs to be incubated for prolonged periods at 37°C for low-pH-mediated entry to occur (61). This suggests that post-binding events are required to prime the HCV envelope proteins for fusion.
Conclusion
Targeting a conserved epitope with known functional relevance is absolutely essential for production of a broadly neutralizing antibody. Structure-based vaccine design and innovative thinking with regard to stabilization of epitopes will be necessary to forward HCV vaccine efforts. Many of the vaccine studies in the past decade have been done with recombinant HCV E2 or E1E2; however, a majority of the human antibody responses were against E2 HVR1 and ultimately unsuccessful due to the high mutation rate in the region (62). The highly disordered and flexible nature of HCV E2 is a complicating factor to intelligent vaccine design. Not only is local flexibility seen between the same epitope partnered with different antibodies, but large portions of E2 are disordered and variably-sequenced between genotypes (i.e., the HVRs). The described structures of E1 and E2 are only representative of their respective serotypes and may or may not be representative of the many variable HCV isolates that exist. Within the HCV patient population, many circulating isolates are highly resistant to known broadly neutralizing antibodies, and many mutations that allow for resistance to neutralizing antibody recognition have been described (63, 64). Furthermore, inherently flexible, long-chain glycans are responsible for shielding targeted neutralizing antibody binding sites.
Available DAA treatments for HCV are undoubtedly necessary for infected patients; however, taking into account the rate at which high-risk groups are being infected with HCV, a vaccine is an imperative for preventative treatment. In order to achieve this goal, researchers must overcome the problem of HCV which uses an almost shapeshifting mechanism to evade immune detection: shrouding itself with a coat of apolipoproteins, flexibility, and hyper variability. HCV E2 has evolved to maintain a balance between the order of disulfide bonds and hydrophobic interactions necessary to form the overall protein fold, and the flexible chaos which allows the virus to replicate while evading the host immune response.
Author contributions
SY wrote the paper. YW and JM generated the figures. SY, YW, and JM participated in the critical review and revision of the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank C. Wang, J. Casiano, and A. Khan for helpful discussions.
Footnotes
Funding. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases and National Institutes of Health.
References
- 1.Tsoulfas G, Goulis I, Giakoustidis D, Akriviadis E, Agorastou P, Imvrios G, et al. Hepatitis C and liver transplantation. Hippokratia (2009) 13:211–5. [PMC free article] [PubMed] [Google Scholar]
- 2.Akamatsu N, Sugawara Y. Liver transplantation and hepatitis C. Int J Hepatol. (2012) 2012:686135. 10.1155/2012/686135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dultz G, Graubard BI, Martin P, Welker MW, Vermehren J, Zeuzem S, et al. Liver transplantation for chronic hepatitis C virus infection in the United States 2002-2014: an analysis of the UNOS/OPTN registry. PLoS ONE (2017) 12:e0186898. 10.1371/journal.pone.0186898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Academies of Sciences E, Medicine Eliminating the Public Health Problem of Hepatitis B and C in the United States: Phase One Report. Washington, DC: The National Academies Press; (2016). [PubMed] [Google Scholar]
- 5.Blach S, Zeuzem S, Manns M, Altraif I, Duberg A-S, Muljono DH, et al. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol. (2017) 2:161–76. 10.1016/s2468-1253(16)30181-9 [DOI] [PubMed] [Google Scholar]
- 6.Thokala P, Simpson EL, Tappenden P, Stevens JW, Dickinson K, Ryder S, et al. Ledipasvir-sofosbuvir for treating chronic hepatitis C: a NICE single technology appraisal-an evidence review group perspective. Pharmacoeconomics (2016) 34:741–50. 10.1007/s40273-016-0387-y [DOI] [PubMed] [Google Scholar]
- 7.Pradat P, Huleux T, Raffi F, Delobel P, Valantin MA, Poizot-Martin I, et al. Incidence of new hepatitis C virus infection is still increasing in French MSM living with HIV. AIDS (2018) 32:1077–82. 10.1097/QAD.000000000000178 [DOI] [PubMed] [Google Scholar]
- 8.Shiffman ML. The next wave of hepatitis C virus: the epidemic of intravenous drug use. Liver Int. (2018) 38(Suppl. 1):34–9. 10.1111/liv.13647 [DOI] [PubMed] [Google Scholar]
- 9.Wanis H. Hepatitis C Treatment in Egypt: Why Cost Remains a Challenge. Cairo: Egyptian Initiative for Personal Rights: Economic and Social Justice Unit; (2014). [Google Scholar]
- 10.WHO Global Hepatitis Report 2017. (2017). Available online at: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
- 11.Lavie M, Goffard A, Dubuisson J. HCV glycoproteins: assembly of a functional E1-E2 heterodimer. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk, VA: Horizon Bioscience; (2006). [PubMed] [Google Scholar]
- 12.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. (2002) 76:6919–28. 10.1128/JVI.76.14.6919-6928.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. (2006) 80:2418–28. 10.1128/JVI.80.5.2418-2428.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felmlee DJ, Sheridan DA, Bridge SH, Nielsen SU, Milne RW, Packard CJ, et al. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology (2010) 139:1774–83, 1783 e1771–6. 10.1053/j.gastro.2010.07.047 [DOI] [PubMed] [Google Scholar]
- 15.Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. (2011) 286:3018–32. 10.1074/jbc.M110.175018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, et al. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci USA. (2013) 110:9505–10. 10.1073/pnas.1307527110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JY, Acosta EG, Stoeck IK, Long G, Hiet MS, Mueller B, et al. Apolipoprotein E likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J Virol. (2014) 88:12422–37. 10.1128/JVI.01660-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefevre M, et al. Apolipoprotein E mediates evasion from hepatitis C virus neutralizing antibodies. Gastroenterology (2016) 150:206–17. e204. 10.1053/j.gastro.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Wang X, Chi X, Zhao F, Guo J, Ma P, et al. Neglected but important role of apolipoprotein E exchange in hepatitis C virus infection. J Virol. (2016) 90:9632–43. 10.1128/JVI.01353-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helle F, Dubuisson J. Hepatitis C virus entry into host cells. Cell Mol Life Sci. (2008) 65:100–12. 10.1007/s00018-007-7291-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of hepatitis C virus to CD81. Science (1998) 282:938–41. [DOI] [PubMed] [Google Scholar]
- 22.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. (2002) 21:5017–25. 10.1093/emboj/cdf529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature (2007) 446:801–5. 10.1038/nature05654 [DOI] [PubMed] [Google Scholar]
- 24.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature (2009) 457:882–6. 10.1038/nature07684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falson P, Bartosch B, Alsaleh K, Tews BA, Loquet A, Ciczora Y, et al. Hepatitis C virus envelope glycoprotein E1 forms trimers at the surface of the virion. J Virol. (2015) 89:10333–46. 10.1128/JVI.00991-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazumdar B, Banerjee A, Meyer K, Ray R. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology (2011) 54:1149–56. 10.1002/hep.24523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol. (2012) 86:7256–67. 10.1128/JVI.07222-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, et al. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun. (2014) 5:4874 10.1038/ncomms5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li HF, Huang CH, Ai LS, Chuang CK, Chen SS. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: involvement in cell fusion and virus entry. J Biomed Sci. (2009) 16:89. 10.1186/1423-0127-16-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong L, Kadam RU, Giang E, Ruwona TB, Nieusma T, Culhane JC, et al. Structure of hepatitis C virus envelope glycoprotein E1 antigenic site 314-324 in complex with antibody IGH526. J Mol Biol. (2015) 427:2617–28. 10.1016/j.jmb.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. (2012) 8:e1002653. 10.1371/journal.ppat.1002653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong L, Giang E, Robbins JB, Stanfield RL, Burton DR, Wilson IA, et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci USA. (2012) 109:9499–504. 10.1073/pnas.1202924109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahid A, Dubuisson J. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat. (2013) 20:369–76. 10.1111/jvh.12094 [DOI] [PubMed] [Google Scholar]
- 34.Li D, von Schaewen M, Wang X, Tao W, Zhang Y, Li L, et al. Altered glycosylation patterns increase immunogenicity of a subunit hepatitis C virus vaccine, inducing neutralizing antibodies which confer protection in mice. J Virol. (2016) 90:10486–98. 10.1128/JVI.01462-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vietheer PT, Boo I, Gu J, McCaffrey K, Edwards S, Owczarek C, et al. The core domain of hepatitis C virus glycoprotein E2 generates potent cross-neutralizing antibodies in guinea pigs. Hepatology (2017) 65:1117–31. 10.1002/hep.28989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science (2013) 342:1090–4. 10.1126/science.1243876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature (2014) 509:381–4. 10.1038/nature13117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce BG, Keck ZY, Lau P, Fauvelle C, Gowthaman R, Baumert TF, et al. Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design. Proc Natl Acad Sci USA. (2016) 113:E6946–54. 10.1073/pnas.1614942113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. (2003) 278:41624–30. 10.1074/jbc.M305289200 [DOI] [PubMed] [Google Scholar]
- 40.Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, et al. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. (2010) 84:34–43. 10.1128/JVI.02199-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong L, Giang E, Nieusma T, Robbins JB, Deller MC, Stanfield RL, et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J Virol. (2012) 86:13085–8. 10.1128/JVI.01939-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L, Zhong L, Struble E, Duan H, Ma L, Harman C, et al. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci USA. (2013) 110:7418–22. 10.1073/pnas.1305306110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alhammad Y, Gu J, Boo I, Harrison D, McCaffrey K, Vietheer PT, et al. Monoclonal antibodies directed toward the hepatitis C virus glycoprotein E2 detect antigenic differences modulated by the N-terminal hypervariable region 1 (HVR1), HVR2, and intergenotypic variable region. J Virol. (2015) 89:12245–61. 10.1128/JVI.02070-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cowton VM, Angus AG, Cole SJ, Markopoulou CK, Owsianka A, Dunlop JI, et al. Role of conserved E2 residue W420 in receptor binding and hepatitis C virus infection. J Virol. (2016) 90:7456–68. 10.1128/JVI.00685-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prentoe J, Velazquez-Moctezuma R, Foung SK, Law M, Bukh J. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology (2016) 64:1881–92. 10.1002/hep.28705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, et al. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol. (2006) 80:8695–704. 10.1128/JVI.00271-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothwangl KB, Manicassamy B, Uprichard SL, Rong L. Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding. Virol J. (2008) 5:46 10.1186/1743-422X-5-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasiliauskaite I, Owsianka A, England P, Khan AG, Cole S, Bankwitz D, et al. Conformational flexibility in the immunoglobulin-like domain of the hepatitis C virus glycoprotein E2. MBio (2017) 8:17. 10.1128/mBio.00382-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck ZY, Lau P, et al. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol. (2012) 86:12923–32. 10.1128/JVI.02052-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meola A, Tarr AW, England P, Meredith LW, McClure CP, Foung SK, et al. Structural flexibility of a conserved antigenic region in hepatitis C virus glycoprotein E2 recognized by broadly neutralizing antibodies. J Virol. (2015) 89:2170–81. 10.1128/JVI.02190-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aleman F, Tzarum N, Kong L, Nagy K, Zhu J, Wilson IA, et al. Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors. Proc Natl Acad Sci USA. (2018) 115:7569–74. 10.1073/pnas.1802378115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu J, Hardy JM, Boo I, Vietheer P, McCaffrey K, Pombourios P, et al. The antigen-binding fragment of MAb24 in complex with a peptide from Hepatitis C Virus E2 epitope I (412-423). (2018). Available online at: http://www.rcsb.org/structure/5VXR
- 53.Kong L, Lee DE, Kadam RU, Liu T, Giang E, Nieusma T, et al. Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen. Proc Natl Acad Sci USA. (2016) 113:12768–73. 10.1073/pnas.1609780113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarr AW, Owsianka AM, Jayaraj D, Brown RJ, Hickling TP, Irving WL, et al. Determination of the human antibody response to the epitope defined by the hepatitis C virus-neutralizing monoclonal antibody AP33. J Gen Virol. (2007) 88(Pt 11):2991–3001. 10.1099/vir.0.83065-0 [DOI] [PubMed] [Google Scholar]
- 55.Tarr AW, Urbanowicz RA, Jayaraj D, Brown RJ, McKeating JA, Irving WL, et al. Naturally occurring antibodies that recognize linear epitopes in the amino terminus of the hepatitis C virus E2 protein confer noninterfering, additive neutralization. J Virol. (2012) 86:2739–49. 10.1128/JVI.06492-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keck ZY, Girard-Blanc C, Wang W, Lau P, Zuiani A, Rey FA, et al. Antibody response to hypervariable region 1 interferes with broadly neutralizing antibodies to hepatitis C virus. J Virol. (2016) 90:3112–22. 10.1128/JVI.02458-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pierce BG, Boucher EN, Piepenbrink KH, Ejemel M, Rapp CA, Thomas WDJr, et al. Structure-based design of hepatitis C virus vaccines that elicit neutralizing antibody responses to a conserved epitope. J Virol. (2017) 91:e01032–17. 10.1128/JVI.01032-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drummer HE, Maerz A, Poumbourios P. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett (2003) 546:385–90. 10.1016/S0014-5793(03)00635-5 [DOI] [PubMed] [Google Scholar]
- 59.Flint M, Logvinoff C, Rice CM, McKeating JA. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J Virol. (2004) 78:6875–82. 10.1128/JVI.78.13.6875-6882.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol. (2010) 84:10159–68. 10.1128/JVI.01180-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. (2006) 80:1734–41. 10.1128/JVI.80.4.1734-1741.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ray R, Meyer K, Banerjee A, Basu A, Coates S, Abrignani S, et al. Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J Infect Dis. (2010) 202:862–6. 10.1086/655902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey JR, Wasilewski LN, Snider AE, El-Diwany R, Osburn WO, Keck Z, et al. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest. (2015) 125:437–47. 10.1172/JCI78794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urbanowicz RA, McClure CP, Brown RJ, Tsoleridis T, Persson MA, Krey T, et al. A diverse panel of hepatitis C virus glycoproteins for use in vaccine research reveals extremes of monoclonal antibody neutralization resistance. J Virol. (2015) 90:3288–301. 10.1128/JVI.02700-15 [DOI] [PMC free article] [PubMed] [Google Scholar]