Abstract
Bortezomib alone and in combination with other anticancer agents are extensively used for chemotherapeutic treatment of multiple myeloma (MM) patients and are being developed for treating other cancers. Bortezomib acts through multiple pathways, and in this study with ANBL-6 and RPMI 8226 MM cells we show that bortezomib inhibited growth and induced apoptosis and that this was accompanied by downregulation of specificity protein (Sp) 1, Sp3, and Sp4 transcription factors that are overexpressed in these cells. Similar results were observed in pancreatic and colon cancer cells. The functional importance of this pathway was confirmed by showing that individual knockdown of Sp1, Sp3, and Sp4 in MM cells inhibited cell growth and induced apoptosis, and that this correlates with the results of previous studies in pancreatic, colon, and other cancer cell lines. The mechanism of bortezomib-mediated downregulation of Sp transcription factors in MM was due to the induction of caspase-8 and upstream factors, including Fas-associated death domain. These results demonstrate that an important underlying mechanism of action of bortezomib was due to the activation of caspase-8–dependent downregulation of Sp1, Sp3, Sp4, and pro-oncogenic Sp-regulated genes.
Introduction
Multiple myeloma (MM) is a B-cell malignancy associated with terminally differentiated plasma cells that proliferate in the bone marrow, and complications from MM include bone marrow failure, renal disease, and osteolytic bone disease (Cook, 2008; Dimopoulos et al., 2015; Paiva et al., 2015; Anderson, 2016). It is estimated that 30,770 new cases of MM will be diagnosed in 2018 and 12,770 patients will die of this disease in the United States (Siegel et al., 2018). Among all cancers, the number of Food and Drug Administration approvals for new therapies are among the highest for MM (Anderson, 2016; Bates, 2016; Neri et al., 2016; Orlowski and Lonial, 2016), and this has contributed to the improvements in overall survival from this disease from 3 years in the 1960s to 8–10 years (Anderson, 2016). Among the most recent and prominent therapies for treating patients with MM include the class of proteasome inhibitors such as bortezomib (Velcade; Takeda Pharmaceuticals International Corporation, Cambridge, MA) and immunotherapies that are being developed for many cancers (Anderson, 2016; Bates, 2016; Neri et al., 2016; Orlowski and Lonial, 2016).
One of the major advances in the treatment of MM was the development and clinical applications of bortezomib, a proteasome inhibitor that is used extensively in MM chemotherapy (Hideshima et al., 2001, 2003; LeBlanc et al., 2002; Mitsiades et al., 2002; Richardson et al., 2005; San Miguel et al., 2008) and in drug combination therapies. Bortezomib and related analogs target the ubiquitin-proteasome pathway by interacting with N-terminal threonine residues in the active site of the proteasome catalytic region, thereby blocking the function of the 26S proteasome (Pandit and Gartel, 2011). Although bortezomib was initially characterized as a proteasome inhibitor, there is evidence that this agent induces anticancer activities in MM and other cancer cell lines through many other pathways, including the induction of reactive oxygen species (ROS), intracellular stress, and apoptosis (Laussmann et al., 2011; Lipchick et al., 2016; Xian et al., 2017). Treatment of MM cells and other cancer cell lines with bortezomib has also been linked to the downregulation of specificity protein (Sp) 1 transcription factor. For example, bortezomib induces microRNA-29b (miR-29b) in MM cells and acute myeloid leukemia cells, and this is associated with the downregulation of Sp1 and Sp1-regulated genes/responses (Liu et al., 2008; Amodio et al., 2012; Blum et al., 2012); in MM cells, miR-29b and Sp1 are part of feedback loop where decreased expression of Sp1 induces miR-29b (Amodio et al., 2012). Another study (Bat-Erdene et al., 2016) showed that bortezomib decreased the expression of Sp1 in MM cells, and this was accompanied by the decreased expression of Sp1-regulated survival factors IRF4 and cMyc.
Research in our laboratory has focused on Sp transcription factors as drug targets, and it has been demonstrated that Sp1, Sp3, and Sp4 are overexpressed in multiple cancer cell lines (Mertens-Talcott et al., 2007; Chadalapaka et al., 2012; Nair et al., 2013; Jutooru et al., 2014; Hedrick et al., 2015, 2017; Kasiappan et al., 2016; Karki et al., 2017; Safe et al., 2018), whereas levels of Sp2 and Sp5 were either low or variable and were not further investigated. RNA interference studies show that Sp1, Sp3, and Sp4 individually regulate cell proliferation, survival, and migration of cancer cells (Hedrick et al., 2016). Bortezomib is a widely used anticancer agent, and it is essential to understand its mechanism of action, particularly in MM, since insights into its mechanisms are important for designing drug combination therapies. The results of this study demonstrate that not only Sp1, but also Sp3 and Sp4 are highly expressed and exhibit pro-oncogenic activities in MM cells, as previously observed in other cancer cell lines. Moreover, bortezomib induces caspase-8–dependent downregulation of Sp1, Sp3, and Sp4 in MM cells, suggesting that an important mechanism of action of this drug is due to targeting the downregulation of Sp1, Sp3, and Sp4.
Materials and Methods
Cell Lines, Antibodies, and Reagents.
The MM cell lines (ANBL-6 and RPMI 8226) were gifts from Robert Z. Orlowski (Department of Lymphoma and Myeloma, The University of Texas MD Anderson Cancer Center, Houston, TX). Panc1, L3.6pL, and SW480 cells were purchased from American Type Culture Collection (Manassas, VA). ANBL-6 and RPMI 8226 cells were maintained at 37°C in the presence of 5% CO2 in RPMI 1640 medium with 10% fetal bovine serum (FBS), and for ANBL-6 cells, interleukin-6 (1 ng/ml) was added. Similarly, Panc1, L3.6pL, MiaPaCa2, and SW480 cells were grown and maintained at 37°C in the presence of 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 medium supplemented with 10% FBS. DMEM, RPMI 1640 medium, FBS, formaldehyde, sodium orthovanadate, and trypsin were purchased from Sigma-Aldrich (St. Louis, MO). cMyc (cat. no. 9402s), survivin (cat. no. 2808s), cleaved poly(ADP-ribose) polymerase (cPARP; cat. no. 9541T), cyclin D1 (cat. no. 2978s), phospo (p)–Fas-associated death domain (FADD) (cat. no. 2781s), caspase-3 (cat. no. 9662s), cleaved caspase-3 (cat. no. 9661L), caspase-8 (cat. no. 9746T), FADD (cat. no. 2782T), and cleaved caspase-8 (cat. no. 9496T) antibodies were obtained from Cell Signaling Technology (Boston, MA). Sp1 (cat. no. ab13370) antibody was purchased from Abcam (Cambridge, MA); Sp3 (cat. no. sc-644), Sp4 (cat. no. sc-645), epidermal growth factor receptor (EGFR) (cat. no. sc-373746), and bcl2 (cat. no. sc-7382) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX); and β-actin (cat. no. A5316) antibody from Sigma-Aldrich. Immobilon Western chemiluminescence reagents for Western blot imaging were purchased from Millipore (Billerica, MA), and bortezomib was purchased from LC Laboratories (Woburn, MA). The Apoptotic, Necrotic, and Healthy Cells Quantification Kit was purchased from Biotium (Hayward, CA); the ROS Determination Kit was purchased from Invitrogen (Grand Island, NY); and the XTT Cell Viability Kit was obtained from Cell Signaling Technology. Caspase-8 inhibitor (Z-IETD-FMK; cat. no. 51-69401U) and interleukin-6 (cat. no. 10395-HNAE-25) recombinant human protein mixture were purchased from BD Biosciences (San Jose, CA) and Invitrogen (Carlsbad, CA), respectively. The caspase-3 inhibition was obtained from R & D Systems (Minneapolis, MN).
Cell Viability Assay.
Cells were plated in 96-well plates at a density of 10,000 per well with RPMI 1640 medium and DMEM containing 2.5% charcoal-stripped FBS. Cells were treated with dimethylsulfoxide (solvent control) and different concentrations of bortezomib with DMEM containing 2.5% charcoal-stripped FBS for 0–48 hours After treatment, 25 μl (XTT reagent with 1% of electron coupling solution) was added to each well and incubated for 4 hours as outlined in the manufacturer instructions (Cell Signaling Technology). Absorbance was measured at a wavelength of 450 nm in a 96-well plate reader after incubation for 4 hours in 5% CO2 at 37°C.
Measurement of ROS.
ROS levels were measured using the cell-permeable probe CM-H2DCFDA [5-(and 6)-chloromethyl-2-,7- dichlorodihydrofluorescein diacetate acetyl ester] as outlined in the manufacturer instructions (Life Technologies, Carlsbad, CA). Cells were seeded at a density of 1.5 × 105/ml in six-well plates; pretreated with glutathione (GSH) for 30 minutes; treated with vehicle (dimethylsulfoxide), bortezomib alone, or in combination with GSH, N-acetylcysteine (NAC), and catalase for 30 minutes; and incubated for up to 9 hours. ROS levels were measured by flow cytometry as previously described (Karki et al., 2017).
Measurement of Apoptosis (Annexin V Staining).
Cancer cells were seeded at density of 1.5 × 105/ml in six-well plates and pretreated with the caspase-8 inhibitor Z-IETD-FMK for 30 minutes and then treated with vehicle alone, bortezomib alone, or a combination with 15 μM caspase-8 inhibitor. Cells were stained and analyzed by flow cytometry using the Dead Cell Apoptosis Kit and Alexa Fluor 488 Assay Kit according to the manufacturer protocol (Invitrogen).
Western Blot Analysis.
ANBL-6, RPMI 8226, Panc1, L3.6pL, and SW480 cells were seeded at density of 1.5 × 105/ml in six-well plates and were treated with various concentrations of bortezomib alone or in combination with caspase-8 inhibitors, and whole-cell proteins were extracted using radioimmunoprecipitation assay lysis buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 (w/v), 0.5% sodium deoxycholate, and 0.1% SDS with protease and phosphatase inhibitor cocktail. Protein concentrations were measured using Lowry’s method, and equal amounts of protein were separated in 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Polyvinylidene difluoride membranes were incubated overnight at 4°C with primary antibodies in 5% skimmed milk and then were incubated for 2–3 hours with secondary antibodies conjugated with horseradish peroxidase. Membranes were then exposed to horseradish peroxidase substrate, and immune-reacted proteins were detected with a chemiluminescence reagent. The same Sp3 antibody detects both the high– and low–molecular weight forms of Sp3, as previously reported (Kasiappan et al., 2016; Karki et al., 2017).
Small Interfering RNA Interference Assay.
Small interfering RNA (siRNA) experiments were conducted using siRNA by electroporation (ECM-830; Harvard Apparatus, Holliston, MA). ANBL-6 cells (2 × 106) in 400 μl of serum-free media in a 4-mm-gap cuvette were electroporated using the following conditions: 100 V and three pulses for 30 milliseconds. RPMI 8226 cells (2 × 106) in 400 μl of serum-free media in a 4-mm-gap cuvette were also electroporated at a voltage of 250 V, in one pulse with a pulse length of 10 milliseconds. One microliter of the siRNAs (100 μM) was used during electroporation. The complexes used in the study are as follows:
iNS-5′: CGU ACG CGG AAU ACU UCG A (nonspecific)
siSp1: SASI_Hs02_00333289 [1] SASI_Hs01_ 00140198 [2] SASI_Hs01_00070994 [3]
siSp3: SASI_Hs01_00211941 [1] SASI_Hs01_ 00211942 [2] SASI_Hs01_00211943 [3]
siSp4: SASI_Hs01_00114420 [1] SASI_Hs01_ 00114421 [2] SASI_Hs01_00114420 [3]
siCaspase-8: SASI_Hs02_0031422 [1] SASI_Hs02_ 00314221_AS
The effects of knockdown were determined 72 hours after initial transfection.
Statistical Analysis.
One-way analysis of variance and Dunnett’s test were used to determine the statistical significance between two groups. To confirm the reproducibility of the data, the experiments were performed at least three independent times, and the results are expressed as the mean ± S.D. P values <0.05 were considered to be statistically significant.
Results
Bortezomib Decreases Expression of Sp Transcription Factors in MM Cells and Other Cancer Cell Lines.
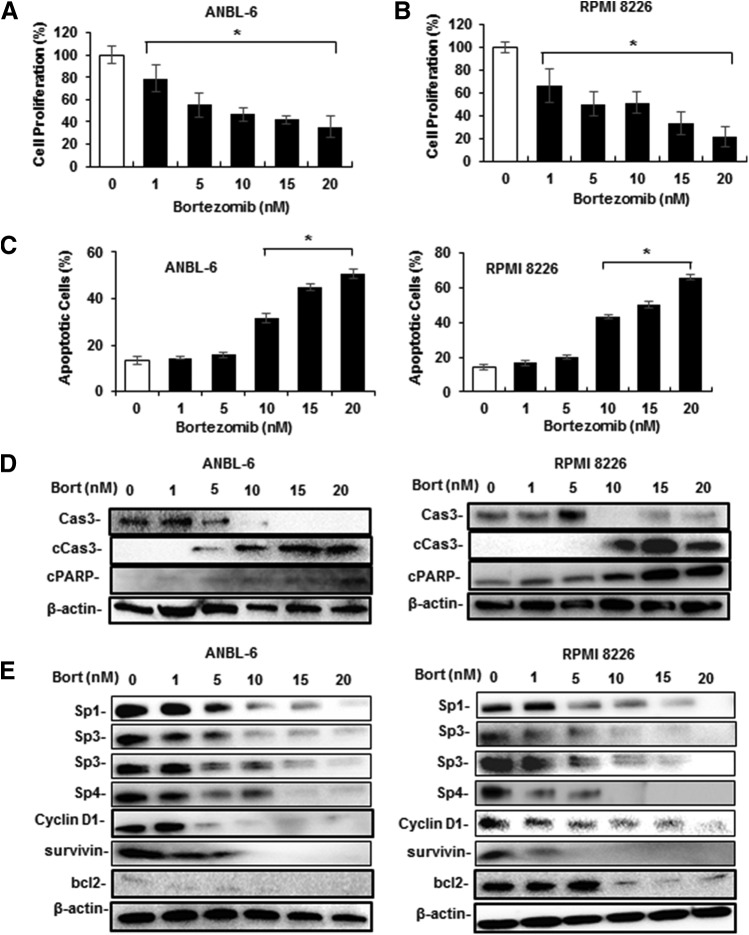
In this study, we used human ANBL-6 and RPMI 8226 MM cell lines, and treatment with 1–20 nM bortezomib decreased the growth of both cell lines with significant growth inhibition observed at 1 nM bortezomib after treatment for 24 and 48 hours (Fig. 1, A and B). Bortezomib also induced Annexin V staining (apoptosis marker) in both cell lines (Fig. 1C; Supplemental Fig. 1A), and statistically significant induction was observed after treatment with 5–10 nM bortezomib. This concentration range also induced caspase-3 cleavage in ANBL-6 and RPMI 8226 cells (Fig. 1D). Previous studies have shown that Sp1 is expressed in MM cells and bortezomib decreased Sp1 levels, and the results in Fig. 1E confirms that bortezomib downregulated Sp1 in ANBL-6 and RMPI 8226 cells. However, it is also evident that Sp3 and Sp4 are highly expressed in both cell lines, and that bortezomib decreased the levels of Sp3 and Sp4 proteins; the effects were observed in cells after treatment with 1–5 nM bortezomib. Cyclin D1, survivin, and bcl-2 are typical Sp-regulated genes (Safe et al., 2018), and bortezomib decreased the levels of these proteins in MM cells.
Fig. 1.
Bortezomib (Bort) inhibits MM cell growth and survival, and downregulates Sp1, Sp3, and Sp4. ANBL-6 cells (A), RPMI 8226 cells (B), and both cell lines (C) were treated with 1–20 nM bortezomib for 24 hours, and the effects on cell growth and Annexin V staining were determined as outlined in Materials and Methods. ANBL-6 and RPMI 8226 cells were treated with 1–20 nM bortezomib for 24 hours, and whole-cell lysates were analyzed for markers of apoptosis (D) and Sp transcription factors and Sp-regulated genes (E) by Western blots. (A–C) Results are expressed as the mean ± S.D. for at least three replicated experiments, and significant effects of treatment compared with solvent-treated controls are indicated (*P < 0.05). Both high–molecular weight (top) and low–molecular weight (bottom) forms of Sp3 are shown in all Western blots. Cas3, caspase-3.
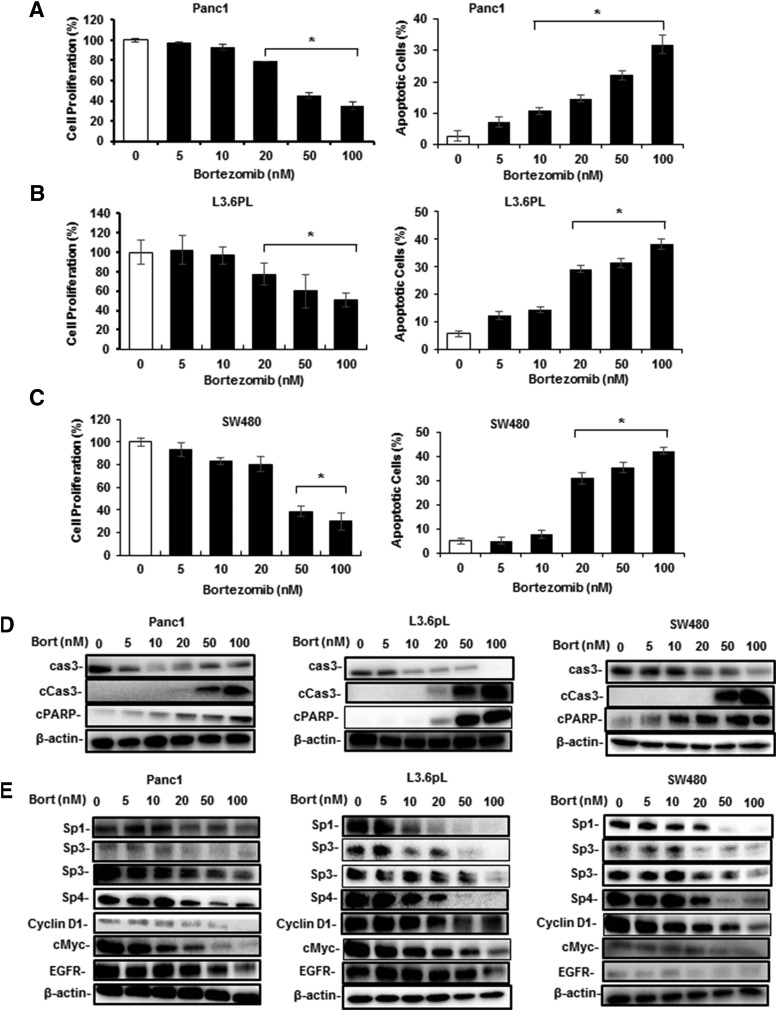
Since bortezomib is being developed to treat multiple cancers, we next investigated the effects of this compound in Panc1 and L3.6pL pancreatic and SW480 colon cancer cell lines. Figure 2, A–C demonstrates that 5–100 nM bortezomib treatment for 24–48 hours decreased the growth of all three cell lines. Western blot analysis also showed that bortezomib induced caspase-3 cleavage in these cell lines (Fig. 2D). In addition, bortezomib also decreased the expression of Sp1, Sp3, and Sp4 in Panc1, L3.6pL, and SW480 cells, and levels of Sp-regulated cMyc and EGFR were also decreased (Fig. 2E). Previous studies have reported several other anticancer agents induce similar responses in these cell lines (Nair et al., 2013; Jutooru et al., 2014; Kasiappan et al., 2016; Safe et al., 2018).
Fig. 2.
Bortezomib (Bort) inhibits pancreatic and colon cancer cell growth, induces apoptosis and downregulates Sp1, Sp3, and Sp4. Panc1 (A), L3.6pL (B), and SW480 (C) cancer cell lines were treated with 5–100 nM bortezomib, and effects on cell proliferation and Annexin V staining were determined. The cells were treated with 5–100 nM bortezomib for 24 hours, and whole-cell lysates were analyzed for markers of apoptosis (D) and Sp downregulation (E) by Western blots. (A–C) Results are expressed as the mean ± S.D. for at least three replicate experiments and significant effects of treatment (compared with solvent-treated controls) are indicated (*P < 0.05). Cas3, caspase-3.
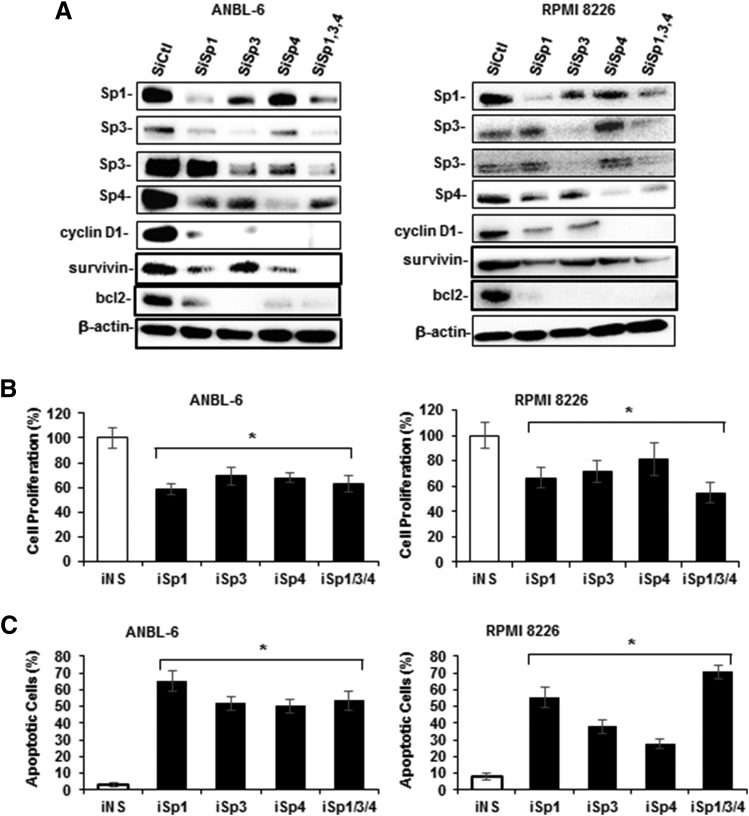
RNA interference studies in Panc1, L3.6pL, and SW480 cells and other cancer cell lines show that the knockdown of Sp1, Sp3, and Sp4 individually and in combination resulted in growth inhibition and the induction of apoptosis (Hedrick et al., 2016). Therefore, ANBL-6 and RPMI 8226 cells were transfected with oligonucleotides that target Sp1 (iSp1), Sp3 (iSp3), Sp4 (iSp4), and their combination (iSp1/3/4), and the effects on Sp knockdown were determined by Western blots of whole-cell lysates (Fig. 3A). Knockdown of Sp1 decreased the expression of Sp1 and also Sp4 in both cell lines; knockdown of Sp3 decreased Sp3 and also Sp1 (ANBL-6 and RPMI 8226) and Sp4 (ANBL-6); and knockdown of Sp4 primarily decreased only the target protein. The specificity of Sp knockdown is cell context dependent (Hedrick et al., 2016), and their self-regulation is due, in part, to common GC-rich cis-elements in the 5′-promoter regions of Sp1, Sp3, and Sp4 genes (Nicolás et al., 2001; Song et al., 2001; Lou et al., 2005). Transfection of these oligonucleotides individually also decreased proliferation (Fig. 3B) and induced Annex V staining (Fig. 3C) in ANBL-6 and RPMI 8226 cells. Thus, Sp1, Sp3, and Sp4 individually regulate MM cell growth and survival, and the knockdown of one Sp transcription factor is not compensated by the other two; this has previously been observed in other cancer cell lines, including Panc1, L3.6pL, and SW480 cells (Hedrick et al., 2016). Thus, bortezomib-mediated downregulation of not only Sp1, but also Sp3 and Sp4 contributes to the effects of this compound on growth inhibition and induction of apoptosis.
Fig. 3.
Functional effects of Sp knockdown in MM cells. (A) ANBL-6 and RPMI 8226 cells were transfected with oligonucleotides targeting Sp1 (iSp1), Sp3 (iSp3), Sp4 (iSp4), or their combination (iSp1/3/4), or a nonspecific control (iCtl); and whole-cell lysates were analyzed by Western blots. Cells were transfected with the same set of oligonucleotides, and after 72 hours the effects of Sp knockdown on cell proliferation (B) or Annexin V staining (C) were determined as outlined in the Materials and Methods. (B and C) Results are expressed as the mean ± S.D. for at least three replicate experiments, and significant (*P < 0.05) changes compared with iCtl (control) are indicated.
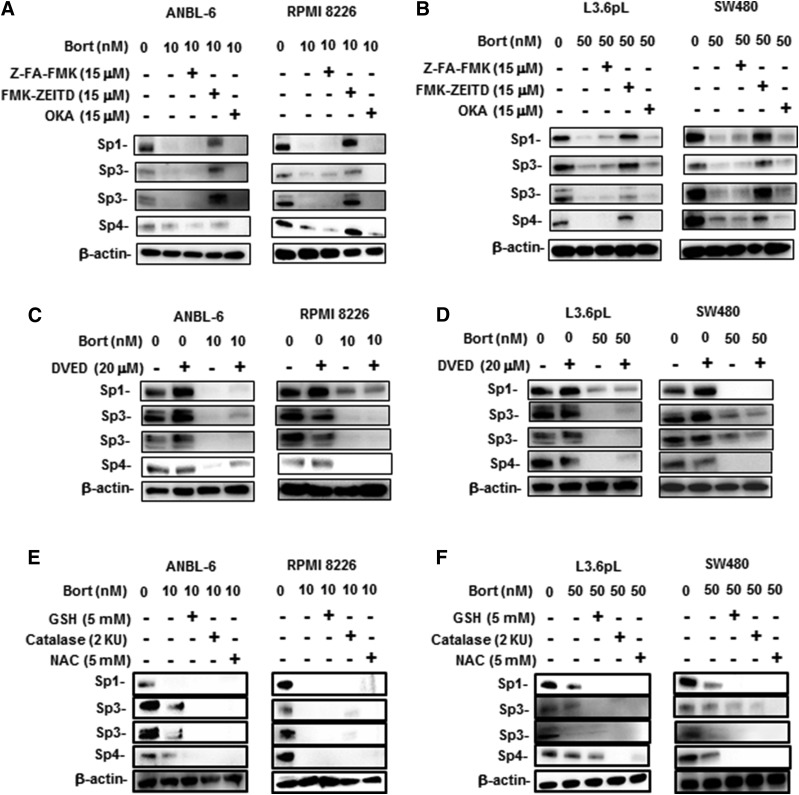
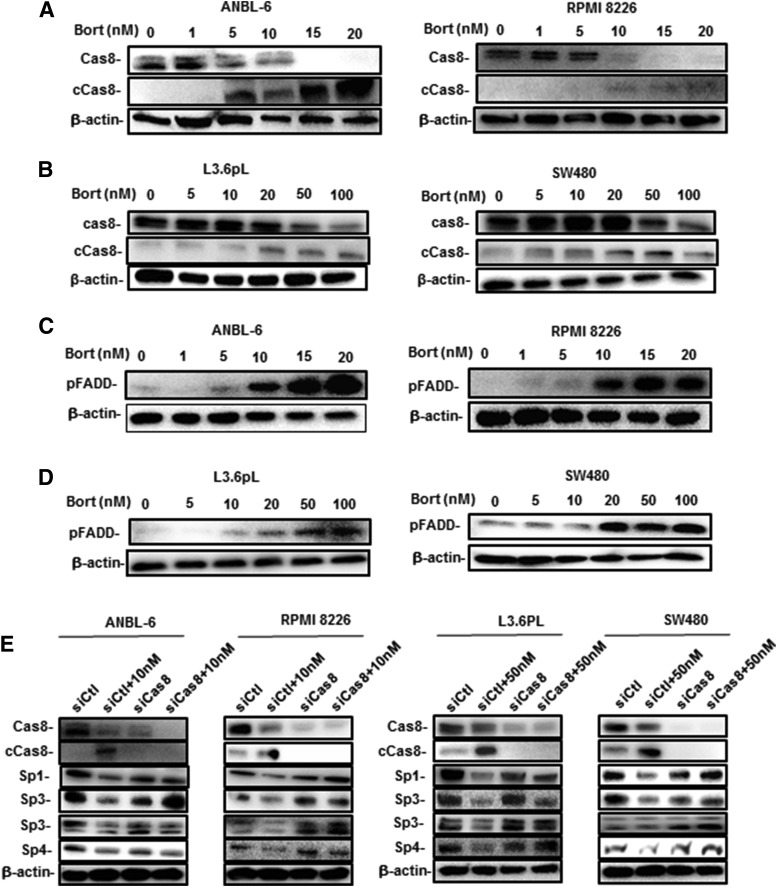
Previous studies show that drug-induced downregulation of Sp transcription factors is due to the induction of proteolytic enzymes or phosphatases, or through the induction of ROS (Safe et al., 2018). Results illustrated in Fig. 4A demonstrate that the phosphatase inhibitor okadaic acid (OKA) did not affect bortezomib-induced Sp downregulation, whereas the caspase-8 inhibitor FMK-ZEITD but not Z-FA-FMK (nonspecific) inhibited degradation of Sp1, Sp3, and Sp4 in ANBL-6 and RPMI 8226 cells. Similar results were observed in L3.6pL and SW480 cells (Fig. 4B), and we did not observe any inhibitory effects by the caspase-3 inhibitor DEVD (Fig. 4, C and D). Bortezomib-induced downregulation of Sp1, Sp3, and Sp4 was not inhibited by the antioxidants GSH or NAC in ANBL-6 and RPMI 8226 cells (Fig. 4E) and L3.6pL and SW480 cells (Fig. 4F), and therefore was not ROS dependent; this was consistent with the failure of bortezomib to induce ROS in ANBL-6 and RPMI 8226 cells (Supplemental Fig. 2). Interestingly, previous studies have reported that bortezomib induces the activation of caspase-8 (Liu et al., 2007; Laussmann et al., 2011; Amodio et al., 2012; Bullenkamp et al., 2014; Bat-Erdene et al., 2016) and bortezomib-mediated Sp1 degradation is blocked by FMK-ZEITD (Bat-Erdene et al., 2016). A role for caspase-3–dependent cleavage of Sp1 has also been reported (Torabi et al., 2018). The inhibition of bortezomib-induced effects by FMK-ZEITD is consistent with the observed induction of caspase-8 in MM cells (ANBL-6 and RPMI 8226) (Fig. 5A) and the L3.6pL pancreatic and SW480 colon cancer cells (Fig. 5B). We also investigated possible upstream activators of caspase-8 and the extrinsic apoptosis pathway and observed that FADD was induced by bortezomib in MM cells (Fig. 5C), as previously observed in Hela cervical cancer cells (Laussmann et al., 2011). FADD was also induced in pancreatic and colon cancer cells (Fig. 5D). The role of caspase-8 in mediating bortezomib-induced Sp downregulation was confirmed in MM, SW480, and L3.6pL cells since treatment of the cell lines with bortezomib plus an oligonucleotide-targeting caspase-8 blocked the downregulation of Sp1, Sp3, and Sp4 (Fig. 5E).
Fig. 4.
Potential inhibitors of bortezomib (Bort)-induced Sp downregulations in MM, colon, and pancreatic cancer cells. ANBL-6 and RPMI 8226 cells (A) and L3.6pL and SW480 cells (B) were treated with bortezomib alone or in combination with Z-FA-FMK, FMK-ZEITD, or OKA for 24 hours, and whole-cell lysates were analyzed by Western blots. Treatment with the inhibitors alone (Z-FA-FMK, FMK-ZEITD, or OKA) did not affect Sp expression (data not shown). ANBL-6 and RPMI 8226 (C) and L3.6pL and SW480 (D) cells were also treated with bortezomib alone or in combination with DVED, and whole-cell lysates were analyzed by Western blots. ANBL-6 and RPMI 8226 cells (E) and L3.6pL and SW480 cells (F) were treated with bortezomib alone or in combination with antioxidants, and whole-cell lysates were analyzed by Western blots.
Fig. 5.
Bortezomib (Bort) induces FADD and caspase-8 (Cas8) in cancer cells, and caspase-8 is required for Sp degradation. ANBL-6 and RPMI 8226 cells (A) and L3.6pL and SW480 cells (B) were treated with different concentrations of bortezomib for 24 hours, and whole-cell lysates were analyzed by Western blots for caspase-8 activation (cleavage). ANBL-6 and RPMI 8226 cells (C), and Panc1, L3.6pL, and SW480 cells (D) were treated as described in (A) and (B), and whole-cell lysates were analyzed by Western blots for the induction of FADD. (E) ANBL-6, RPMI 8226, L3.6pL, and SW480 cells were treated with bortezomib alone and in combination with an oligonucleotide targeted to caspase-8, and whole-cell lysates were isolated and analyzed by Western blots.
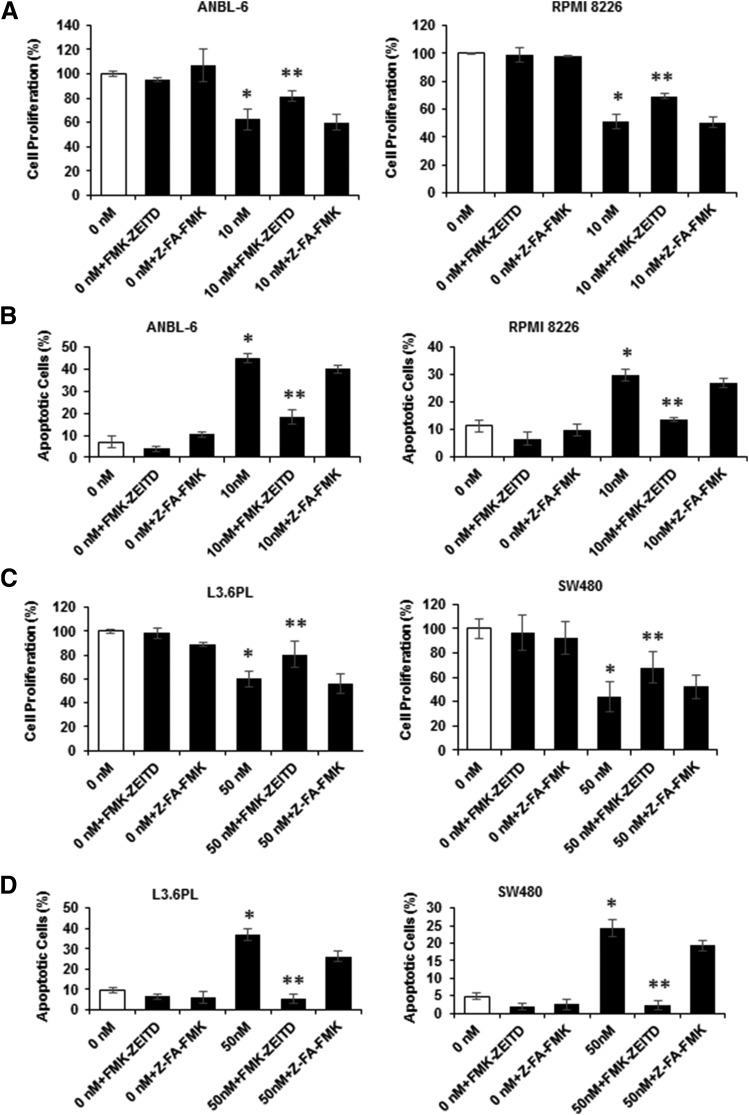
The role of bortezomib-induced caspase-8 on the effects of this compound on MM cell proliferation was determined in ANBL-6 and RPMI 8826 cells treated with FMK-ZEITD, Z-FA-FMK, and bortezomib alone and in combination. Bortezomib-mediated inhibition of MM cell proliferation was inhibited by FMK-ZEITD but not Z-FA-FMK (Fig. 6A), and similar results were observed for Annexin V staining (Fig. 6B; Supplemental Fig. 1B). Not surprisingly, the effects of FMK-ZEITD were more pronounced for the Annexin V assay. We also observed similar effects in L3.6pL and SW480 cells (Fig. 6, C and D), demonstrating that bortezomib-induced activation of caspase-8 plays an important role in mediating the growth inhibitory and proapoptotic effects of this compound in MM and other cancer cell lines.
Fig. 6.
Effects of caspase inhibitors on bortezomib-mediated inhibition of growth and induction of apoptosis. (A) ANBL-6 and RPMI 8226 cells were treated with bortezomib, Z-FA-FMK, and FMK-ZEITD alone or in combination for 24 hours, and the effects on cell proliferation were determined as outlined in Materials and Methods. (B) ANBL-6 and RPMI 8226 cells were treated as described in (A), and the effects on induction of Annexin V staining were determined. L3.6pL and SW480 cells were treated as outlined in (A), and the effects on cell proliferation (C) and Annexin V staining (D) were determined as outlined in Materials and Methods. (A–C) Results are expressed as the mean ± S.D. for at least three replicate determinations; significant (*P < 0.05) changes compared with control are indicated, and significant (**P < 0.05) reversal of the effects are also indicated.
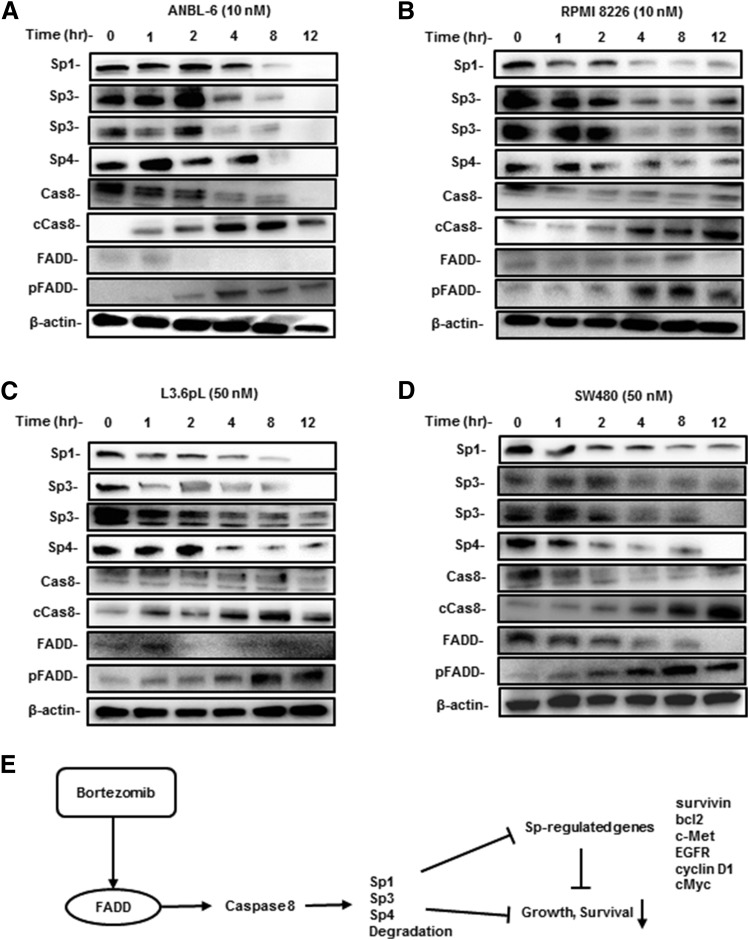
We also examined the time course–dependent effects of bortezomib on FADD, caspase-8, Sp1, Sp3, and Sp4 expression in the MM cell lines. In ANBL-6 and RPMI 8226 cells, the loss of Sp proteins was observed after 2 or 4 hours; cleaved caspase-8 was induced within 1–2 hours and pFADD was enhanced after 2 hours, suggesting that, in addition to FADD, other factors upstream from caspase-8 may also be activated by bortezomib (Fig. 7, A and B). Similar results were observed for L3.6pL and SW480 cells (Fig. 7, C and D). Figure 7E summarizes a possible mechanism of bortezomib-induced downregulation of Sp1, Sp3, and Sp4, which involves caspase-8–dependent proteolysis. Western blots at early time points show some cleavage products (data not shown), and the cleavage sites and rates of Sp1, Sp3, and Sp4 degradation will be investigated in future studies. Current studies are investigating the mechanisms associated with bortezomib-dependent induction of FADD and other upstream factors that activate caspase-8.
Fig. 7.
Time-dependent effects of bortezomib. ANBL-6 (A), RPMI 8226 (B), L3.6pL (C), and SW480 (D) cells were treated with bortezomib, and effects on the expression of various proteins at different treatment times were determined by Western blot analysis. (E) Proposed model for bortezomib-induced downregulation of Sp1, Sp3, Sp4, and Sp-regulated genes that play a role in cell proliferation and survival.
Discussion
Bortezomib and related proteasome inhibitors are used in drug combination therapies for the treatment of MM and are being evaluated for the treatment of other tumors (Anderson, 2016; Bates, 2016; Neri et al., 2016; Orlowski and Lonial, 2016). Therefore, it is essential that the mechanism of action of this compound be understood so that combination therapies can take advantage of mechanism-based drug-drug interactions. Bortezomib works through multiple pathways in MM and other cancer cells, and our initial hypothesis was that bortezomib induced ROS, which results in an ROS-dependent pathway resulting in the repression of Sp1, Sp3, and Sp4 mRNAs/proteins and pro-oncogenic Sp-regulated genes. This pathway has been worked out for several ROS-inducing anticancer agents including benzyl- and phenethylisothiocyanates, histone deacetylase inhibitors, piperlongumine, and penfluridol (Jutooru et al., 2014; Hedrick et al., 2015, 2017; Kasiappan et al., 2016; Karki et al., 2017). The treatment of MM cells with bortezomib decreased Sp1 protein expression, as previously reported; however, this response was not affected by cotreatment with bortezomib plus GSH or NAC (Fig. 4E), and the effects of bortezomib appeared to be ROS independent (Supplemental Fig. 2).
Previous studies have identified a miR-29b–Sp1 loop where miR-29b targets Sp1 (3′-untranslated region) to decrease gene expression; however, our results show that bortezomib not only decreased levels of Sp1 but also of Sp3 and Sp4 in MM cells and other cancer lines, suggesting that bortezomib may induce degradation pathways that simultaneously target all three Sp transcription factors. Individual knockdown of Sp1, Sp3, and Sp4 in lung, kidney, breast, pancreatic, colon, and rhabdomyosarcoma cancer cell lines results in decreased cell proliferation, survival, and migration/invasion and decreased expression of pro-oncogenic Sp-regulated genes, suggesting that Sp transcription factors are nononcogene addiction genes (Hedrick et al., 2016). Similar results were observed in MM cells (Fig. 3), and this indicates that not only Sp1, but also Sp3 and Sp4 play an important role in the growth and survival of MM cells. Moreover, among these three Sp transcription factors that target GC-rich cis-elements, the loss of one factor is not compensated by the other two in MM cells, and this is consistent with their differential regulation of gene expression previously demonstrated in pancreatic, colon, and other cancer cells (Hedrick et al., 2016).
Among those agents that inhibit drug-induced Sp downregulation including antioxidants, proteasome inhibitors, phosphatase inhibitors (OKA), zinc ions, and caspase inhibitors (reviewed in Safe et al., 2018), only the caspase-8 inhibitor FMK-ZEITD inhibited bortezomib-induced downregulation of Sp1, Sp3, and Sp4 (Fig. 4). This observation is consistent with previous reports showing that bortezomib activates caspase-8 and the extrinsic pathway of apoptosis (Liu et al., 2007; Laussmann et al., 2011; Bullenkamp et al., 2014; Bat-Erdene et al., 2016), and one of those reports also shows that the inhibition of caspase-8 blocked bortezomib-induced cell death (Bullenkamp et al., 2014), as observed in this study (Fig. 5).
Previous studies showed that the nonsteroidal anti-inflammatory drug tolfenamic acid also induced the degradation of Sp1, Sp3, and Sp4 in SW480 colon cancer cells (Pathi et al., 2014), and this response was blocked in cells cotreated with tolfenamic acid plus the caspase-8 inhibitor FMK-ZEITD. Thus, at least in SW480 cells, both bortezomib (Fig. 2 and 5) and tolfenamic acid exhibited a common mechanism of action involving caspase-8 (Fig. 7E), and this was also observed for bortezomib in pancreatic cancer and MM cell lines. Moreover, in these same cell lines, bortezomib induced FADD, which is upstream from caspase-8, and this was previously reported in Hela cells where bortezomib also induced FADD protein expression (Laussmann et al., 2011). Current studies in this laboratory are investigating the mechanisms associated with bortezomib-mediated induction of FADD and other genes upstream from caspase-8 and the mechanisms of caspase-8–dependent degradation of Sp1, Sp3, and Sp4.
In summary, this study shows for the first time that the bortezomib-dependent activation of caspase-8 results in the degradation of not only Sp1, but also Sp3 and Sp4, which are important nononcogene addiction genes (Hedrick et al., 2016) that are highly expressed in MM cells. Moreover, individual knockdown of Sp1, Sp3, and Sp4 induced the inhibition of MM cell growth and survival (Fig. 3), suggesting that Sp transcription factors are also pro-oncogenic in MM cells, as previously observed in colon, pancreatic, breast, lung, and kidney cancer cells (Hedrick et al., 2016). Thus, the contributions of bortezomib-induced downregulation of Sp1, Sp3, and Sp4 to the overall efficacy of this drug should be considered in the development of clinical applications of bortezomib since several Sp-regulated genes are associated with drug and radiation resistance (Safe et al., 2018).
Abbreviations
- cPARP
cleaved poly(ADP-ribose) polymerase
- DMEM
Dulbecco’s modified Eagle’s medium
- EGFR
epidermal growth factor receptor
- FADD
Fas-associated death domain
- FBS
fetal bovine serum
- GSH
glutathione
- miR-29b
microRNA-29b
- MM
multiple myeoloma
- NAC
N-acetylcysteine
- OKA
okadaic acid
- p
phospho
- ROS
reactive oxygen species
- siRNA
small interfering RNA
- Sp
specificity protein
Authorship Contributions
Participated in research design: Karki, Safe.
Conducted experiments: Karki, Harishchandra.
Contributed new reagents or analytic tools: Safe.
Performed data analysis: Karki, Harishchandra.
Wrote or contributed to the writing of the manuscript: Karki, Safe.
Footnotes
This work was supported by the National Institutes of Health [grant T32-ES-026568, K.K.], Texas AgriLife Research, and the Sid Kyle Endowment. No potential conflicts of interest relevant to this article are reported.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, Gullà AM, Pitari MR, Conforti F, Rossi M, et al. (2012) miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis 3:e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KC. (2016) Progress and paradigms in multiple myeloma. Clin Cancer Res 22:5419–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bat-Erdene A, Miki H, Oda A, Nakamura S, Teramachi J, Amachi R, Tenshin H, Hiasa M, Iwasa M, Harada T, et al. (2016) Synergistic targeting of Sp1, a critical transcription factor for myeloma cell growth and survival, by panobinostat and proteasome inhibitors. Oncotarget 7:79064–79075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates SE. (2016) Multiple myeloma: multiplying therapies. Clin Cancer Res 22:5418. [DOI] [PubMed] [Google Scholar]
- Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld AK, Whitman S, Walker A, Klisovic R, Byrd JC, Santhanam R, et al. (2012) Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood 119:6025–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullenkamp J, Raulf N, Ayaz B, Walczak H, Kulms D, Odell E, Thavaraj S, Tavassoli M. (2014) Bortezomib sensitises TRAIL-resistant HPV-positive head and neck cancer cells to TRAIL through a caspase-dependent, E6-independent mechanism. Cell Death Dis 5:e1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadalapaka G, Jutooru I, Safe S. (2012) Celastrol decreases specificity proteins (Sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis 33:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R. (2008) Economic and clinical impact of multiple myeloma to managed care. J Manag Care Pharm 14 (7 Suppl):19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos MA, Richardson PG, Moreau P, Anderson KC. (2015) Current treatment landscape for relapsed and/or refractory multiple myeloma. Nat Rev Clin Oncol 12:42–54. [DOI] [PubMed] [Google Scholar]
- Hedrick E, Cheng Y, Jin UH, Kim K, Safe S. (2016) Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget 7:22245–22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick E, Crose L, Linardic CM, Safe S. (2015) Histone deacetylase inhibitors inhibit rhabdomyosarcoma by reactive oxygen species-dependent targeting of specificity protein transcription factors. Mol Cancer Ther 14:2143–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick E, Li X, Safe S. (2017) Penfluridol represses integrin expression in breast cancer through induction of reactive oxygen species and downregulation of Sp transcription factors. Mol Cancer Ther 16:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, et al. (2003) Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 101:1530–1534. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. (2001) The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res 61:3071–3076. [PubMed] [Google Scholar]
- Jutooru I, Guthrie AS, Chadalapaka G, Pathi S, Kim K, Burghardt R, Jin UH, Safe S. (2014) Mechanism of action of phenethylisothiocyanate and other reactive oxygen species-inducing anticancer agents. Mol Cell Biol 34:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki K, Hedrick E, Kasiappan R, Jin UH, Safe S. (2017) Piperlongumine induces reactive oxygen species (ROS)-dependent downregulation of specificity protein transcription factors. Cancer Prev Res (Phila) 10:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasiappan R, Jutooru I, Karki K, Hedrick E, Safe SH. (2016) Benzyl isothiocyanate (BITC) induces reactive oxygen species-dependent repression of STAT3 by down-regulation of specificity proteins in pancreatic cancer. J Biol Chem 291:27122–27133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann MA, Passante E, Düssmann H, Rauen JA, Würstle ML, Delgado ME, Devocelle M, Prehn JH, Rehm M. (2011) Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell Death Differ 18:1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc R, Catley LP, Hideshima T, Lentzsch S, Mitsiades CS, Mitsiades N, Neuberg D, Goloubeva O, Pien CS, Adams J, et al. (2002) Proteasome inhibitor PS-341 inhibits human myeloma cell growth in vivo and prolongs survival in a murine model. Cancer Res 62:4996–5000. [PubMed] [Google Scholar]
- Lipchick BC, Fink EE, Nikiforov MA. (2016) Oxidative stress and proteasome inhibitors in multiple myeloma. Pharmacol Res 105:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann E, Huynh L, Vukosavljevic T, Takeki M, Klisovic RB, et al. (2008) Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood 111:2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue P, Chen S, Hu L, Lonial S, Khuri FR, Sun SY. (2007) The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res 67:4981–4988. [DOI] [PubMed] [Google Scholar]
- Lou Z, Maher VM, McCormick JJ. (2005) Identification of the promoter of human transcription factor Sp3 and evidence of the role of factors Sp1 and Sp3 in the expression of Sp3 protein. Gene 351:51–59. [DOI] [PubMed] [Google Scholar]
- Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. (2007) The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 67:11001–11011. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M, Libermann TA, Treon SP, et al. (2002) Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci USA 99:14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair V, Pathi S, Jutooru I, Sreevalsan S, Basha R, Abdelrahim M, Samudio I, Safe S. (2013) Metformin inhibits pancreatic cancer cell and tumor growth and downregulates Sp transcription factors. Carcinogenesis 34:2870–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri P, Bahlis NJ, Lonial S. (2016) New strategies in multiple myeloma: immunotherapy as a novel approach to treat patients with multiple myeloma. Clin Cancer Res 22:5959–5965. [DOI] [PubMed] [Google Scholar]
- Nicolás M, Noé V, Jensen KB, Ciudad CJ. (2001) Cloning and characterization of the 5′-flanking region of the human transcription factor Sp1 gene. J Biol Chem 276:22126–22132. [DOI] [PubMed] [Google Scholar]
- Orlowski RZ, Lonial S. (2016) Integration of novel agents into the care of patients with multiple myeloma. Clin Cancer Res 22:5443–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva B, van Dongen JJ, Orfao A. (2015) New criteria for response assessment: role of minimal residual disease in multiple myeloma. Blood 125:3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit B, Gartel AL. (2011) Thiazole antibiotic thiostrepton synergize with bortezomib to induce apoptosis in cancer cells. PLoS One 6:e17110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathi S, Li X, Safe S. (2014) Tolfenamic acid inhibits colon cancer cell and tumor growth and induces degradation of specificity protein (Sp) transcription factors. Mol Carcinog 53 (Suppl 1):E53–E61. [DOI] [PubMed] [Google Scholar]
- Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, et al. Assessment of Proteasome Inhibition for Extending Remissions (APEX) Investigators (2005) Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med 352:2487–2498. [DOI] [PubMed] [Google Scholar]
- Safe S, Abbruzzese JL, Abdelrahim M, Hedrick E. (2018) Specificity protein transcription factors and cancer: opportunities for drug development. Cancer Prev Res (Phila) 11:371–382. [DOI] [PubMed] [Google Scholar]
- San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, Kropff M, Spicka I, Petrucci MT, Palumbo A, Samoilova OS, et al. VISTA Trial Investigators (2008) Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med 359:906–917. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30. [DOI] [PubMed] [Google Scholar]
- Song J, Mangold M, Suske G, Geltinger C, Kanazawa I, Sun K, Yokoyama KK. (2001) Characterization and promoter analysis of the mouse gene for transcription factor Sp4. Gene 264:19–27. [DOI] [PubMed] [Google Scholar]
- Torabi B, Flashner S, Beishline K, Sowash A, Donovan K, Bassett G, Azizkhan-Clifford J. (2018) Caspase cleavage of transcription factor Sp1 enhances apoptosis. Apoptosis 23:65–78. [DOI] [PubMed] [Google Scholar]
- Xian M, Cao H, Cao J, Shao X, Zhu D, Zhang N, Huang P, Li W, Yang B, Ying M, et al. (2017) Bortezomib sensitizes human osteosarcoma cells to adriamycin-induced apoptosis through ROS-dependent activation of p-eIF2α/ATF4/CHOP axis. Int J Cancer 141:1029–1041. [DOI] [PubMed] [Google Scholar]