Abstract
Key points
Classifying different subtypes of neurons in deep brain structures is a challenge and is crucial to better understand brain function.
Understanding the diversity of neurons in the globus pallidus (GP), a brain region positioned to influence afferent and efferent information processing within basal ganglia, could help to explain a variety of brain functions.
We present a classification of neurons from the GP using electrophysiological data from wild‐type mice and confirmation using transgenic mice.
This work will help researchers to identify specific neuronal subsets in the GP of wild‐type mice when transgenic mice with labelled neurons are lacking.
Abstract
Classification of the extensive neuronal diversity in the brain is fundamental for neuroscience. The globus pallidus external segment (GPe), also referred to as the globus pallidus in rodents, is a large nucleus located in the core of the basal ganglia whose circuitry is implicated in action control, decision‐making and reward. Although considerable progress has been made in characterizing different GPe neuronal subtypes, no work has directly attempted to characterize these neurons in non‐transgenic mice. Here, we provide data showing the degree of overlap in expression of neuronal PAS domain protein (Npas1), LIM homeobox 6 (Lhx6), parvalbumin (PV) and transcription factor FoxP2 biomarkers in mouse GPe neurons. We used an unbiased statistical method to classify neurons based on electrophysiological properties from nearly 200 neurons from C57BL/6J mice. In addition, we examined the subregion distribution of the neuronal subtypes. Cluster analysis using firing rate and hyperpolarization‐induced membrane potential sag variables revealed three distinct neuronal clusters: type 1, characterized by low firing rate and small sag potential; type 2, with low firing rate and larger sag potential; and type 3, with high firing rate and small sag potential. We used other electrophysiological variables and data from marker‐expressing neurons to evaluate the clusters. We propose that the GPe GABAergic neurons should be classified into three subgroups: arkypallidal, low‐firing prototypical and high‐firing prototypical neurons. This work will help researchers identify GPe neuron subtypes when transgenic mice with labelled neurons cannot be used.
Keywords: electrophysiology signature, cluster analysis, molecular signature
Key points
Classifying different subtypes of neurons in deep brain structures is a challenge and is crucial to better understand brain function.
Understanding the diversity of neurons in the globus pallidus (GP), a brain region positioned to influence afferent and efferent information processing within basal ganglia, could help to explain a variety of brain functions.
We present a classification of neurons from the GP using electrophysiological data from wild‐type mice and confirmation using transgenic mice.
This work will help researchers to identify specific neuronal subsets in the GP of wild‐type mice when transgenic mice with labelled neurons are lacking.
Introduction
The external part of the globus pallidus (GPe) (also referred to simply as globus pallidus in rodents) has been classically viewed as a relay station along the striatal indirect pathway (Albin et al. 1989). For many years, the GPe was considered to have uniform cellular constituents (Albin et al. 1989; DeLong, 1990; Smith et al. 1998). However, recent work has shown that the GPe is a widely connected nucleus influencing several parts of the cortico‐basal ganglia circuitry, and that it contains multiple subtypes of GABAergic neurons and a small percentage of cholinergic neurons (Hoover & Marshall, 1999; Mallet et al. 2012; Mastro et al. 2014; Dodson et al. 2015; Hernandez et al. 2015; for review see Hegeman et al. 2016).
The cellular heterogeneity in the GPe stems from early stages of brain development (Flandin et al. 2010; Nobrega‐Pereira et al. 2010). This raises the possibility of identifying GPe neuronal subtypes by specific molecular markers expressed during development. The vast majority of GPe neurons originate from the ganglionic eminence (GE), some from the lateral GE (LGE) and some from the medial GE (MGE) (Dodson et al. 2015). To date, GABAergic neurons in the GPe are classified into two subtypes: prototypical and arkypallidal neurons. The ‘prototypical’ neurons mostly express the transcription factor Nkx2‐1, exhibit fast firing rate and project strongly to the subthalamic nucleus (STN). The ‘arkypallidal’ neurons mostly express the opioid precursor preproenkephalin and the transcription factor FoxP2, exhibit a low firing rate and strongly project back to the striatum (for review see Hegeman et al. 2016). However, other biochemical markers label GPe neuronal subpopulations. A subset of the prototypical neurons (∼45% of Nkx2‐1‐positive neurons) also express parvalbumin (PV). In addition, about 34% of neurons in the GPe express the LIM homeobox 6 (Lhx6) protein, but not PV (Mastro et al. 2014). Dodson et al. (2015) and Hernández et al. (2015) showed that the Lhx6‐positive neurons are mostly Nkx2‐1‐positive and the neurons that show strong Lhx6 immunostaining tend to be PV‐negative. However, some PV‐positive neurons show weak Lhx6 immunostaining. The reported percentage of overlap between Lhx6 and PV neurons in GPe ranges from almost none to 50% in mice (Mastro et al. 2014; Dodson et al. 2015; Hernandez et al. 2015), or almost complete overlap in rats (Abdi et al. 2015; for review see Hegeman et al. 2016). In addition, a mouse expressing the fluorescent protein tdTomato (tdTm) in neuronal PAS domain protein (Npas1)‐positive neurons (the Npas1‐Cre‐2A‐tdTm BAC mouse) was developed and characterized and ∼25% of GPe neurons are labelled in this animal (Hernandez et al. 2015). The FoxP2 transcription factor is expressed in about 60% of these NPas1‐Cre‐2A‐tdTm‐positive GPe neurons, while PV is not expressed in these cells (Dodson et al. 2015; Hernandez et al. 2015).
Although molecular markers have been used as one of the main criteria to identify different neuronal subpopulations in a given brain region (Fishell & Heintz, 2013), more attention should be given to other properties of the neurons, such as morphology and physiological properties. Different studies have shown that the electrophysiological properties of GPe neurons are highly variable, indicating that different GPe neuron populations may have differing electrophysiological signatures. Although some attempts have been made to classify GPe neurons based on electrophysiology, the classification schemes were qualitative in nature, based on visual inspections of neuronal electrophysiological characteristics by the experimenter which does not allow an unbiased classification based on quantification of several electrophysiological properties and the separation of characteristics that are not clearly dichotomous (Kita & Kitai, 1991; Cooper & Stanford, 2000; Chuhma et al. 2011).
The present work characterizes the molecular profile of GPe neurons using two recently described transgenic mice, the Tg(Lhx6‐EGFP)BP221Gsat BAC (Mastro et al. 2014) and Npas1‐Cre‐2A‐ tdTm BAC mice (Hernandez et al. 2015). FoxP2‐Cre mice infected with a green fluorescent protein (GFP) reporter and biomarker staining for PV‐positive neurons from wild‐type C57BL/6J mice are also used to provide information on molecular signatures of GPe neurons. We combine this molecular phenotypic analysis with classification of neurons based on electrophysiological characteristics to further identify and separate subtypes of neurons in the GPe. We present a classification method of the GPe neurons from wild‐type C57BL/6J mice based upon cluster analysis of the neuronal electrophysiological properties and compare these findings to characteristics of neurons in the molecular marker‐expressing mice. We propose that the GPe GABAergic neurons should be classified into at least three subtypes: the arkypallidal neurons, the high firing rate prototypical neurons and the low firing rate prototypical neurons. This work will help researchers to identify specific subtypes of GPe neurons and to study environmental and drug effects in these GPe neuronal subtypes in wild‐type mice, and perhaps other species.
Methods
Ethical approval
All procedures used in this study were performed in accordance with the National Institutes of Health Guide to the Care and Use of Laboratory Animals and with approval of the National Institute on Alcohol Abuse and Alcoholism Animal Care and Use Committee.
Animals
Male C57BL/6J mice (IMSR cat. no. JAX:000664, RRID:IMSR_JAX:000664, The Jackson Laboratory, Bar Harbor, ME, USA) were used as ‘wild‐type’ animals and also for maintenance of the transgenic mouse colony. FoxP2‐Cre (B6.Cg‐Foxp2tm1.1(cre/GFP)Rpa/J, IMSR cat. no. JAX:030541, RRID:IMSR_JAX:030541) mice were used to target FoxP2‐expressing neurons with injection of a AAV9.CAG.Flex.tdTomato.WPRE virus (Addgene, Cambridge, MA, USA) into the GPe 3–4 weeks before electrophysiological recordings. Npas1‐ and Lhx6‐positive neurons were targeted using male and female Npas1‐Cre‐2A‐tdTm BAC mice (Npas1‐tdTm, provided by Dr Savio Chan, Northwestern Medicine, Chicago, IL, USA) (Hernandez et al. 2015) or Tg(Lhx6‐EGFP)BP221Gsat BAC (Lhx6‐EGFP, provided by Dr Aryn Gittis, Carnegie Mellon University (Pittsburgh, PA, USA) derived from the GENSAT line, MMRRC cat. no. 000246‐MU, RRID:MMRRC_000246‐MU), respectively. All mice were housed in groups of two to four on a 12 h light–dark cycle (lights on at 06.30 h) with ad libitum access to food and water.
Immunohistochemistry
We achieved dual transgenic expression of Tdtm in Npas1 neurons and GFP in Lhx6 neurons by breeding the Npas1‐Tdtm with the Lhx6‐EGFP mice. Mice (P30–45) positive for both fluorophores were anaesthetized with pentobarbital (50 mg kg−1, intraperitoneally) and transcardially perfused with PBS followed by 4% formaldehyde, and brains were removed and kept in 4% formaldehyde overnight. The brains were maintained in PBS until sliced with a Pelco easiSlicer vibratome (Ted Pella Inc., Redding, CA, USA) in 50 μm sections. An antigen retrieval protocol was performed in slices stained for FoxP2. Slices were incubated in 10 mM sodium citrate, pH 6.0, for 10 min at 95°C and quickly washed twice with PBST (PBS, 0.2% Triton X‐100). Slices were then blocked with 4% BSA for 4 h at room temperature and incubated overnight at 4°C with primary antibodies: chicken anti‐GFP (1:2000 v/v, Abcam, Cambridge, MA, USA, cat. no. ab13970, RRID:AB_300798), rabbit anti‐DS‐Red (1:500 v/v, Clontech Laboratories, Mountain View, CA, USA, Inc. cat. no. 632496, RRID:AB_10013483) and mouse anti‐PV (1:1000 v/v, Sigma‐Aldrich, St Louis, MO, USA, cat. no. P3088, RRID:AB_477329); or chicken anti‐GFP (1:2000 v/v, Abcam cat. no. ab13970, RRID:AB_300798), rat anti‐Tdtm (1:250 v/v, Kerafast, Boston, MA, USA, cat. no. EST203) and rabbit anti‐FoxP2 (1:500 v/v, Abcam cat. no. 16046, RRID:AB_2314424). Next, slices were washed four times with PBST for 1 h each. Slices were than incubated overnight at 4°C in secondary antibodies: goat anti‐chicken 488 (1:1000 v/v, Thermo Fisher Scientific, Waltham, MA, USA, cat. no. A‐11039, RRID:AB_142924), goat anti‐rabbit 568 (1:2500 v/v, Thermo Fisher Scientific, cat. no. A‐11010, RRID:AB_143156) or goat anti‐rat 568 (1:500 v/v, Thermo Fisher Scientific, cat. no. A‐11077, RRID:AB_141874) and goat anti‐mouse 350 (1:1000 v/v, Thermo Fisher Scientific, cat. no. A‐21049, RRID:AB_141456) or goat anti‐rabbit 350 (1:500 v/v, Thermo Fisher Scientific, cat. no. A‐21068, RRID:AB_141378). Slices were washed four times with PBST for 1 h each, and then kept in PBS before mounting.
In vitro patch‐clamp recording
Brain slices were harvested from 3‐ to 8‐week‐old mice. Mouse coronal GPe slices (250–300 μm) were prepared using a Leica VT1200S vibrating blade microtome (Leica Microsystems, Buffalo Grove, IL, USA). Male C57BL/6J and female or male FoxP2‐Cre, Npas1‐tdTm and Lhx6‐EGFP mice were anaesthetized with isoflurane, decapitated and the brain was quickly removed and immersed in ice‐cold ‘cutting’ artificial cerebrospinal fluid (aCSF) solution containing the following (in mm): 194 sucrose, 30 NaCl, 4.5 KCl, 26 NaHCO3, 1.2 NaH2PO4, 10 d‐glucose, 1 MgCl2, saturated with 95% O2–5% CO2. Slices were equilibrated for 30–40 min at 32°C in carbogen‐bubbled aCSF containing (in mm): 124 NaCl, 4.5 KCl, 26 NaHCO3, 1.2 NaH2PO4, 10 d‐glucose, 1 MgCl2 and 2 CaCl2. Slices were then incubated in aCSF at room temperature until transferred to the recording chamber.
Recordings from GPe neurons were performed in slices fully submerged in a superfusion chamber at 30–32°C with a ∼2 ml min−1 aCSF flow rate, using micropipettes (2–4 MΩ) made from 1.5 mm outer diameter borosilicate glass with a filament (World Precision Instruments, Sarasota, FL, USA) pulled on a Sutter Instruments (Novato, CA, USA) puller. Neurons were visualized using an upright microscope (Scientifica, Uckfield, UK) with a LUMPlanFL N 40×/0.80 W objective (Olympus, Waltham, MA, USA). Recording pipettes were filled with an internal solution containing (in mm): 140 potassium gluconate, 10 Hepes, 0.1 CaCl2, 2 MgCl2, 1 EGTA, 2 ATP‐Mg, 0.2 GTP‐Na, pH 7.25 (290–295 mOsm). When recording in slices from wild‐type C57BL/6J mice, 1% NeurobiotinTM Tracer (Vector Laboratories, Burlingame, CA, USA) was added to the internal solution for post hoc immunohistochemistry to examine PV expression with confocal microscope imaging. Recordings were obtained using a Multiclamp 700A amplifier, Digidata 1322A digitizer and analysed using pClamp 10.3 software (Molecular Devices, Sunnyvale, CA, USA, RRID:SCR_011323). A low‐pass filter of 2 kHz and sampling frequency of 10 kHz were used.
To compare the autonomous spontaneous activity of GPe neurons in cell‐attached and whole‐cell recording modes, a 1 min tight‐seal recording was made in the cell‐attached mode for most of the neurons before transitioning to the whole‐cell mode. Neurons were then recorded for 5 min in whole‐cell current clamp (I = 0) gap‐free mode and the last minute was used to measure the following electrophysiological properties: Firing rate (Hz), firing rate coefficient of variation (CoVar), interspike interval (ISI, ms), ISI CoVar, action potential (AP) threshold (mV) and AP width (ms). To further characterize the electrophysiological properties, a negative current injection (1 s duration) was applied and the following properties were calculated based on the –200 pA current step: input resistance (MΩ) and hyperpolarization‐induced membrane potential sag (mV). We also calculated the sag ratio. Figure 1 describes the definition and how each variable was calculated. We also constructed phase‐plot graphs of the membrane voltage against its rate of change (dV/dt versus V) during the action potential for different neuronal subtypes.
Figure 1. Definitions and calculation descriptions of the electrophysiological variables.
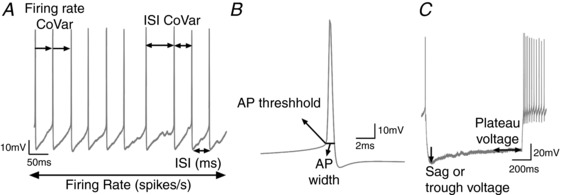
A, firing rate, firing rate coefficient of variation (CoVar), interspike interval (ISI) and ISI CoVar were calculated using positive threshold event detection in ClampFit 10.3. B, after averaging action potentials (AP) for 1 min during the fifth minute of recording, a single vector was exported to Excel and the first derivative was calculated. The AP threshold voltage was measured at 10 mV ms−1 change in the initial rising phase of the action potential. Another linear regression was calculated to determine the AP threshold on the descending phase of the AP to allow measurement of the AP width. C, input resistance was calculated in Clampfit from voltage responses to −200 pA hyperpolarizing current steps, by measuring the plateau voltage at 800–900 ms after the step onset relative to the baseline current injection. The hyperpolarization‐induced sag (Sag) (or trough voltage) was calculated in Clampfit from voltage responses to −200 pA hyperpolarizing current steps, by calculating the peak negative amplitude between 0 and 400 ms after step onset. sag ratio = trough voltage/plateau voltage. Capacitance was calculated by Clampex and shows the change in charge/change in potential.
BLAQ‐assisted immunostaining
Thick slices (250–300 μm) with Neurobiotin‐filled neurons were fixed overnight in 4% formaldehyde in PBS. Fixation was initially at room temperature and, after up to 2 h, sections were transferred to 4°C for 12–24 h. To reduce autofluorescence in thick brain sections and enable efficient imaging of Neurobiotin‐filled neurons we used the Brain BLAQ (Block Lipids and Aldehyde Quench) method (Kupferschmidt et al. 2015). Briefly, sections were washed for 1 h in PBST, and rinsed twice for 1 min in deionized water (diH2O). Slices were then incubated twice for 10 min in freshly prepared sodium borohydride (NaBH4; 5 mg ml−1 in diH2O). Following two 1 min rinses in diH2O, slices were incubated twice for 15 min at room temperature in filtered Sudan Black B solution (0.2% in 70% ethanol). Slices were washed twice for 30 min in room temperature PBS, before immunocytochemistry procedures. Slices were blocked using 4% BSA in PBST for 4 h and incubated for 72 h at 4°C with rabbit anti‐PV antibody (1:1000 v/v dilution; Swant, Marly FR, Switzerland, cat. no. PV27, RRID: AB_2631173). Following four washes in PBST over a total of 16–24 h, slices were incubated for 48 h at 4°C in the following secondary antibodies: 568 goat anti‐rabbit (1:1000 v/v dilution; Thermo Fisher Scientific, cat. no. A‐11011, RRID: AB_143157) and Streptavidin AlexaFluor‐488 conjugate (1:1000 v/v dilution; Thermo Fisher Scientific cat. no. S11223, RRID: AB_2336881). Slices were then washed four times in PBST over a total of 16–24 h and washed for 1 h in PBS before storage.
Confocal imaging
Coronal sections were mounted in Vectashield (Vector Laboratories) and imaged on a Zeiss 510 Meta confocal or a Zeiss LSM 880 laser confocal scan head mounted on a Zeiss Z1Axio Observer inverted microscope frame (Carl Zeiss, Oberkochen, German). Appropriate sets of filter cubes were used to image the fluorescence channels: a fluorescein isothiocyanate filter for the AlexaFluor‐488 (excitation 450–490 nm, dichroic 495, emission 500–550 nm), a Rhodamine filter for the 568 goat anti‐rabbit antibody (excitation 532–558 nm, dichroic 565, emission 570–640 nm) and a Chameleon 2 photon laser for the AlexaFluor‐350 (2P excitation 720nm, emission 390–470nm). Images were taken using CApochromat 40×/1.2 W DICIII or CApochromat 40×/1.2 W Korr FCS M27 objectives (water‐immersion solution: ImmersolTM W, Zeiss). Z‐stack images (10 stacks, each 2 μm thick) were acquired to analyse overlap among Lhx6 × Npas1 × PV (94 z‐projection images from 3 mice) or Lhx6 × Npas1 × FoxP2 (70 z‐projection images from 2 mice). The GPe was defined based on the cytoarchitecture and coordinates used in a previous study (Dodson et al. 2015).
Data analysis
Confocal Z‐stack images were exported to Fiji 3 (ImageJ, RRID:SCR_003070) (Schneider et al. 2012; Schindelin et al. 2015) where automatic macros were developed to: (1) split channels; (2) create Z‐projections of the stacks with maximal intensity; (3) run a minimum filter of 1–2 pixels; (4) run a Gaussian blur filter of sigma 200 scaled to be subtracted out of the original image; (5) run another Gaussian blur filter of sigma 5; (6) perform a auto local threshold using the Phansalkar method with radius = 5–15 pixels; (7) erode 3–4 times; (8) dilate 3–4 times; (9)perform a binary image watershed; and (10) analyse particles of sizes from 500 pixels to infinity with circularity between 0.10 and 1.00 (parameters were adjusted to size of 200 pixels to infinity and 0.40–1.00 circularity for images containing nuclear FoxP2 staining). Manual inspection of the masks created by Fiji 3 were performed and manual segmentation was performed when masks did not represent cell bodies from the naive images. Masks were then saved and transferred to CellProfiler (CellProfiler image analysis software, Broad Institute, Cambridge, MA, USA, RRID: SCR_007358) (Carpenter et al. 2006; Kamentsky et al. 2011), where the number of neurons and all possible colocalizations were automatically determined. Random samples of five images per animal were selected for manual counting. No significant difference between the automatic counting and the sample manual counting was observed in the Lhx6 × Npas1 × PV experiment (data not shown). Manual counting was only necessary to examine the overlap between Npas1 and FoxP2 biomarkers.
Electrophysiological data were analysed using ClampFit 10.3. Statistical analysis was performed in Clamp‐Fit 10.3, STATISTICA 12 (Dell software, Round Rock, TX, RRID: SCR_014213), Microsoft Office Excel software (Microsoft, Redmond, WA, USA) and Prism 7 (GraphPad Software, La Jolla, CA, USA, RRID:SCR_002798). GPe neurons are spontaneously active, and consistent action potential discharge can be seen as soon as the pipette forms a giga‐seal in the cell‐attached configuration. Most of our electrophysiological analysis was based on whole‐cell recording, and thus we wanted to determine the effect of the pipette internal solution on cell physiology. We thus correlated GPe neuronal firing rates in cell‐attached and whole‐cell mode using the Spearman rank order statistic. We detected a strong and significant correlation between the firing rates in tight‐seal cell‐attached and whole‐cell recording modes among 80 neurons from 36 male C57BL/6J mice used for this analysis (r = 0.91; P < 0.05; data not shown). The slope of the curve is 0.7, indicating that the firing rate in whole‐cell recordings is slightly lower than the firing rate in cell‐attached recordings. For all electrophysiological measures described in this study we used the whole‐cell patch‐clamp recording configuration.
Electrophysiological parameters from GPe neurons were collected from C57BL/6J, FoxP2, Npas1 and Lhx6 mice. Only neurons in which all parameters could be measured were considered for the following analysis. The analysis was performed on 177 GPe neurons from 62 male C57BL/6J mice. A proper Cluster Analysis should be run with non‐normally distributed and weakly correlated variables. Thus, a Shapiro–Wilk normality test was run to exclude any variable that had a normal distribution. Considering the non‐normally distributed variables, correlations were calculated by a Spearman rank order test. After standardization of data, an unsupervised hierarchical cluster analysis was performed using Euclidean distances and Ward's method of linkage rule (Ward, 1963). The final number of clusters was determined based on the Thorndike procedure (Thorndike, 1953). In this method, the average within‐group distance is plotted at each stage of clustering, resulting in a drop in the average within‐group distance as the number of clusters increases. The final number of clusters is then decided at the stage where the maximal drop is reached in this plot.
To evaluate the cluster analysis, we used electrophysiological measures of the GPe neurons from male C57BL/6J mice not included in the unsupervised hierarchical cluster analysis. In addition, we used data from neurons identified by markers for expression of FoxP2, Npas1, Lhx6 or PV, as well as the colocalization data we obtained from our immunohistochemical experiments. Fourteen neurons were recorded from three FoxP2‐Cre mice injected with a GFP reporter virus. Thirty‐nine neurons were recorded from 15 NPas1‐Cre‐TdTm mice. Forty‐three neurons were recorded from 13 Lhx6‐EGFP mice. Eleven PV‐positive immunohistochemically stained neurons from 11 C57BL/6J mice and Neurobiotin‐filled neurons were also used to evaluate the cluster analysis.
Results
Biomarker‐based identification of GPe neurons
Previous studies have shown that GPe neurons are mostly GABAergic (Nobrega‐Pereira et al. 2010). In addition, different groups have proposed select biomarkers to identify subtypes of GABAergic neurons in the GPe. Probably the best segregation observed by different groups is between PV‐ and FoxP2‐positive neurons that do not appear to overlap (Dodson et al. 2015; Hernandez et al. 2015). However, there is some controversy about the amount of overlap among other biomarkers in the GPe (Mastro et al. 2014; Dodson et al. 2015; Hernandez et al. 2015). These studies used different ages and strains of mice as well as different antibodies. In some cases, it is difficult to determine the specific antibodies used for each experiment and it is unclear if the co‐localization was done with double, triple or quadruple immunostaining approaches. Here we performed two separate experiments to elucidate the overlap among Npas1, Lhx6 and PV neurons and among Npas1, Lhx6 and FoxP2 neurons.
First, we used double hemizygous transgenic mice expressing Lhx6–EGFP and NPas1–Tdtm to perform a triple immunostaining experiment for GFP (in green, 488 nm), Tdtm (in red, 568 nm) and PV (in blue, 350 nm). We were able to count 1375 Npas1‐positive neurons (∼458 neurons per mouse), 1508 Lhx6‐positive neurons (∼502 neurons per mouse) and 1925 PV‐positive neurons (∼642 neurons per mouse) (Fig. 2 A–D). Out of the NPas1‐positive neurons, 45.8 ± 4.9% (mean ± standard deviation) expressed Npas1 alone, 36 ± 6.6% co‐expressed Lhx6, 12.6 ± 3.7% co‐expressed PV and 5.6 ± 1.05% co‐expressed Lhx6+PV (Fig. 2 E). Out of the Lhx6‐positive neurons: 46.5 ± 10.0% expressed Lhx6 alone, 32.8 ± 7.9% co‐expressed Npas1, 15.6 ± 2.4% co‐expressed PV and 5.1 ± 0.4% co‐expressed Npas1+PV (Fig. 2 F). Out of the PV‐positive neurons, 74.8 ± 4.5% expressed PV alone, 12.2 ± 4.6% co‐expressed Lhx6, 9 ± 2.9% co‐expressed Npas1 and 4.0 ± 2.7% co‐expressed Npas1+Lhx6 (Fig. 2 G). The number of neurons counted for each of the colocalization percentages mentioned above is shown in Fig. 2 E–G. Figure 2 H shows a proportional Euler diagram for the percentages of all GPe GABAergic neurons showing expression profiles in this experiment. We observed a higher number of PV‐positive neurons in the dorsolateral subregion of the GPe, while Npas1 and Lhx6 neurons are more regularly distributed across GPe subregions (Fig. 2 I). The relative subregional distribution of neurons and their overlap is shown in Fig. 2 J–O.
Figure 2. Molecular signature and overlap among Npas1, Lhx6 and PV biomarkers in neurons of the GPe.
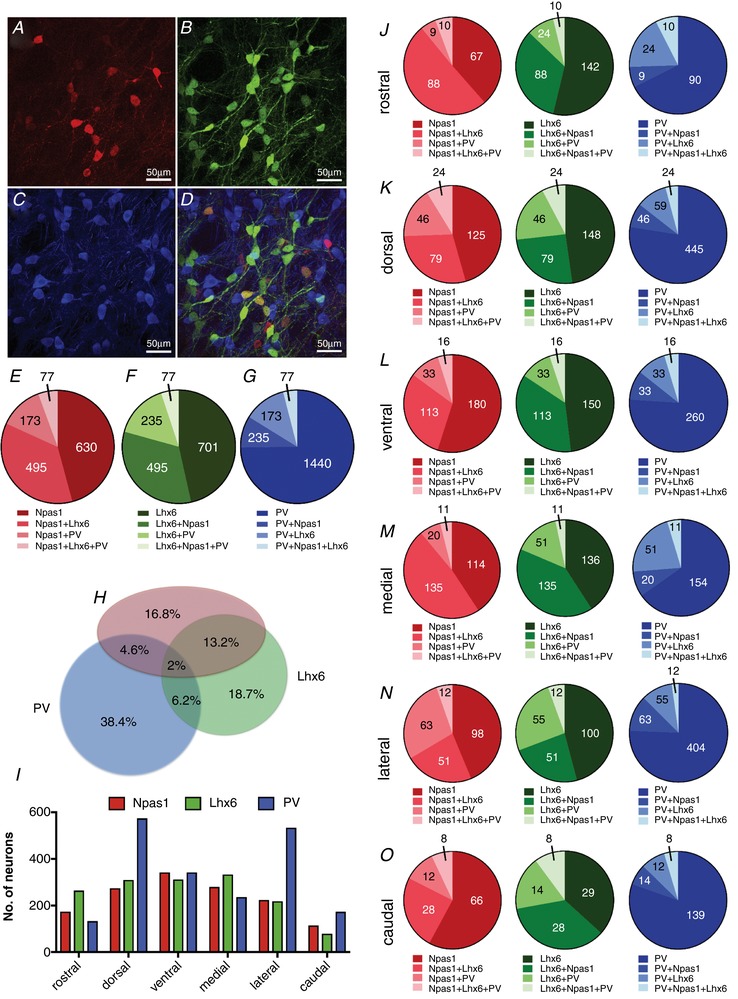
A, representative image of Tdtm (Npas1) immunofluorescence. B, representative image of EGFP (Lhx6) immunofluorescence. C, representative image of PV immunofluorescence. These images have the background fluorescence subtracted. D, merged image showing the overlap among neurons with different biomarkers. E, number of Npas1 neurons expressing different molecular markers (percentages are given in the text). F, number of Lhx6 neurons expressing different molecular markers (percentages are given in the text). G, number of PV neurons expressing different molecular markers (percentages are given in the text). H, proportional Euler diagram showing the percentages of different expression profiles of all GPe GABAergic neurons expressing Npas1, Lhx6 and PV biomarkers. I, distribution of Npas1‐, Lhx6‐ and PV‐positive neurons in subregions of the GPe. Number of images measured by subregion: rostral, 9; dorsal, 22; ventral, 21; medial, 18; lateral, 18; caudal, 8. J–O, GPe subregional distribution of Npas1, Lhx6 and PV neurons
We next used double hemizygous transgenic Lhx6–EGFP and NPas1–Tdtm mice to perform a triple immunostaining experiment for GFP, Tdtm and FoxP2 (in blue). We were able to count 1232 Npas1‐positive neurons (∼616 neurons per mouse), 1367 Lhx6‐positive neurons (∼683 neurons per mouse) and 1112 FoxP2‐positive neurons (∼556 neurons per mouse) (Fig. 3 A–D). Out of the NPas1‐positive neurons, 18.9 ± 4.7% expressed Npas1 alone, 26 ± 0.28% co‐expressed Lhx6, 51.4 ± 7.1% co‐expressed FoxP2 and 3.7 ± 2.1% co‐expressed Lhx6+FoxP2 (Fig. 3 E). Out of the Lhx6‐positive neurons: 68.5 ± 9.6% expressed Lhx6 alone, 23.6 ± 7.6% co‐expressed Npas1, 4.7 ± 2.5% co‐expressed FoxP2 and 3.4 ± 0.5% co‐expressed Npas1+FoxP2 (Fig. 3 F). Out of the FoxP2‐positive neurons: 33.2 ± 12.8% expressed FoxP2 alone, 5.6 ± 0.2% co‐expressed Lhx6, 57 ± 8.9% co‐expressed Npas1 and 4.1 ± 4.0% co‐expressed Npas1+Lhx6 (Fig. 3 G). Figure 3 H shows a proportional Euler diagram for the percentages of different expression profiles of all GPe GABAergic neurons observed in this experiment. The proportional distributions of NPas1‐, Lhx6‐ and FoxP2‐expressing neurons were relatively uniform across different subregions of the GPe (Fig. 2 I). The relative subregional distribution of neurons and their overlap is shown in Fig. 3 J–O.
Figure 3. Molecular signature and overlap among Npas1, Lhx6 and FoxP2 biomarkers in neurons of the GPe.
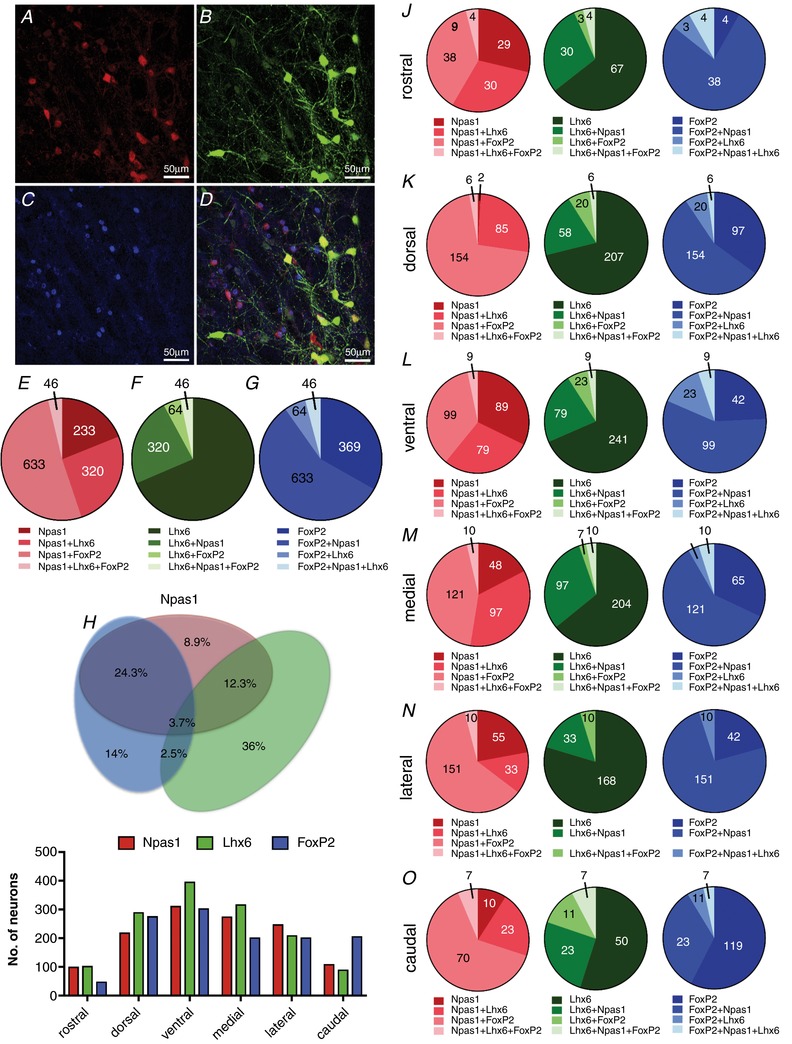
A, representative image of Tdtm (Npas1) immunofluorescence. B, representative image of EGFP (Lhx6) immunofluorescence. C, representative image of FoxP2 immunofluorescence. These images have the background fluorescence subtracted. D, merged image showing the overlap among the biomarkers. E, number of Npas1 neurons expressing different molecular markers (percentages are given in the text). F, number of Lhx6 neurons expressing different molecular markers (percentages are given in the text). G, number of FoxP2 neurons expressing different molecular markers (percentages are given in the text). H, proportional Euler diagram for the percentages of different expression profiles of all GPe GABAergic neurons expressing Npas1, Lhx6 and FoxP2 biomarkers. I, distributions of Npas1‐, Lhx6‐ and FoxP2‐positive neurons in subregions of the GPe. Number of images measured by subregion: rostral, 3; dorsal, 16; ventral, 15; medial, 14; lateral, 14; caudal, 8. J–O, GPe subregional distribution of Npas1, Lhx6 and FoxP2 neurons
Electrophysiological characteristics of previously identified GPe neurons
The majority of recent studies have defined the identity of GPe neurons by molecular phenotype and neuronal connectivity. A direct comparison between these parameters and the electrophysiological characteristics has remained unexplored. Most studies have compared the electrophysiological signature between two subtypes of molecularly identified GPe neurons: Lhx6‐ and PV‐expressing neurons (Mastro et al. 2014) or between two subtypes recorded in vivo and defined by two electrophysiological parameters, firing rate and ISI CoVar, such as the arkypallidal or prototypical neurons (Abdi et al. 2015; Dodson et al. 2015). Hernandez et al. (2015) have presented a better description of the electrophysiological signature of Npas1, Lhx6, PV and choline acetyltransferase (ChAT)‐positive neurons, but this work lacked the FoxP2‐positive neuron electrophysiological data and the recordings were performed at non‐physiological temperatures. Here we present a comprehensive description and analysis of four molecularly identified GABAergic neurons of the GPe: FoxP2, Npas1, Lhx6 and PV neurons.
In Fig. 4, example traces showing the firing rate, the action potential shape and the responses to current steps of molecularly identified GPe neurons are presented. While FoxP2, Npas1 and Lhx6 neurons have similar firing rates (Fig. 4 A–C) and action potential shapes (Fig. 4 E–G), PV neurons have higher firing rates (Fig. 4 D), lower input resistance, and faster depolarization slope as can be noted in the inset phase plot graph in Fig. 4 H compared to the graphs inset in Fig. 4 E–G. In addition, FoxP2 neurons show a larger sag than the other neurons (Fig. 4 L).
Figure 4. Representative electrophysiological traces of identified GPe neurons.

Each column represents one type of GPe neuron coded by different colours. A–D, example traces of the firing during 1 s. E–H, average traces and standard deviation of the action potential shape in the 5th minute of recording after forming the whole‐cell configuration. Phase plot analyses of action potentials are inserted (arrow represents the beginning of the action potential). I–L, example voltage traces for –200, –150 and –100pA current injections for 1 s.
To summarize and compare the electrophysiological signatures of FoxP2, Npas1, Lhx6 and PV neurons, we ran a Kruskal–Wallis non‐parametric test followed by Dunn's multiple comparisons tests (Fig. 5). PV neurons are the most distinct group of neurons when compared to the other subtypes, presenting higher firing rate, sag and capacitance, but lower resistance, sag ratio and ISI interval. The FoxP2 neurons show the most extreme differences relative to PV neuron electrophysiological characteristics. FoxP2 neurons have lower firing rate, sag and capacitance, but higher sag ratio.
Figure 5. Molecularly identified GPe neurons differ in their electrophysiological signatures.

Box plots show the median and interquartile ranges, and the whiskers show 10th to 90th percentiles. A, autonomous firing rate. PV neurons were significantly different from all other cell classes (P < 0.01). Npas1 neurons presented higher firing rate than FoxP2 and Lhx6 neurons (P < 0.05). B, firing rate coefficient of variation (CoVar) was not strongly different among the subtypes of neurons; only Npas1 neurons showed lower firing rate CoVar than FoxP2 neurons (P < 0.05). C, sag or I h shows significant variability among the recorded neurons. Lhx6 neurons show smaller sag than FoxP2 neurons (P < 0.05) and PV neurons have the smallest sag among all the other identified neurons (P < 0.05). D, membrane resistance was smaller in PV neurons (P < 0.05). E, sag ratio was smaller in PV neurons when compared to FoxP2 and Npas1 (P < 0.05), but not to Lhx6. In addition, Lhx6 neurons have smaller sag ratio than FoxP2 neurons (P < 0.05). F, interspike interval (ISI) differences followed the firing rate differences, as expected. G, ISI CoVar was lower in Npas1 when compared to FoxP2 (P < 0.05), but no other significant difference was observed. H, action potential (AP) threshold was significantly higher in Npas1 and Lhx6 neurons when compared to FoxP2 neurons only (P < 0.05). I, AP width was not different among FoxP2, Npas1 and Lhx6, but was lower in PV neurons (P < 0.05). J, capacitance was higher in PV neurons when compared to all other subtypes (P < 0.05). Lhx6 neurons also had higher capacitance than FoxP2 and Npas1 neurons (P < 0.05). Comparisons were made by Kruskal–Wallis ANOVA test followed by Dunn's multiple comparison test. Horizontal bars represent statistically significant differences between 2 clusters (P < 0.05).
Cluster analysis for electrophysiological properties of unidentified GPe neurons
The previous description of the electrophysiological properties of labelled GPe neurons indicates that it is possible to characterize the neuronal subtypes using their electrophysiological signatures. There are experimental circumstances where it may not be possible to use transgenic mice to identify neurons, and it is even more difficult to have all the neuronal subtypes labelled in the same experimental mouse. Thus, a comprehensive classification of the GPe neurons based on electrophysiological properties will help scientists to perform physiological, anatomical and pharmacological studies in specific neurons of the GPe.
Among the electrophysiological variables we examined, no variable showed a normal distribution (Table 1), and thus all variables were considered for the correlation analysis. Since the electrophysiological properties vary greatly in mean and variance, all measurements were z‐scored to standardize the electrophysiological data to a mean of 0 and variance of 1, before the cluster analysis.
Table 1.
Shapiro–Wilk normality test and Spearman rank order correlations among variables numbered (No.) from 1 to 10
| Normality (W) | Correlation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| Firing rate | 0.84 * | 1 | |||||||||
| Firing rate CV | 0.85 * | 2 | −0.48 * | ||||||||
| Sag | 0.87 * | 3 | 0.39 * | −0.13 | |||||||
| Resistance | 0.92 * | 4 | −0.51 * | 0.17 * | −0.68 * | ||||||
| ISI mean | 0.97 * | 5 | −0.53 * | 0.54 * | −0.16 * | 0.24 * | |||||
| ISI CV | 0.63 * | 6 | −0.41 * | 0.87 * | −0.13 | 0.11 | 0.54 * | ||||
| AP threshold | 0.78 * | 7 | −0.16 * | −0.18 * | −0.08 | 0.04 | 0.00 | −0.18 * | |||
| AP width | 0.98 * | 8 | −0.12 | −0.07 | −0.13 | 0.33 * | 0.27 * | −0.09 | 0.09 | ||
| Sag ratio | 0.89 * | 9 | −0.33 * | 0.11 | −0.97 * | 0.55 * | 0.12 | 0.11 | 0.15 * | 0.10 | |
| Capacitance | 0.92 * | 10 | 0.19 * | −0.12 | 0.48 * | −0.62 * | −0.07 | −0.02 | 0.10 | −0.37 * | −0.42 * |
* P < 0.05. Numbers in bold font represent correlation > |−0.40|.
First, we selected variables that showed weak correlations (r |< ±0.40|, Table 1). Firing rate was not strongly correlated with sag, AP threshold, AP width, sag ratio or capacitance. However, as expected, sag and resistance are strongly correlated with sag ratio, so we decided to use sag ratio as this would better represent the data. In addition, sag ratio is strongly correlated with capacitance, so we did not use capacitance. Thus, firing rate, sag ratio, AP width and AP threshold were considered for the first unsupervised hierarchical cluster analysis (data not shown). AP threshold did not contribute to separating four different clusters.
We decided to use only firing rate, sag ratio and AP width for the next cluster analysis (Fig. 6 A–E). The Thorndike procedure indicated that four clusters could explain most of the variability observed using these variables. We then analysed other variables not included in the cluster analyses as an internal evaluation of the clusters (Fig. 6 F–L). Clusters predicted significant differences in variables not included in the cluster analysis, but those differences did not capture much of the variability among the pre‐labelled neuron data. For example, AP width is not different between low frequency FoxP2, Npas1 and Lhx6 neurons (Fig. 5 I). However, the cluster analysis showed that AP width is different between one cluster (which contains most of the FoxP2 neurons: neurons with low firing rate and big sag) and another one (fits any other low frequency‐firing neuron). Thus, the use of AP width added complexity into the cluster analysis and did not create clusters that can describe the differences among the different subtypes of GPe neurons.
Figure 6. Cluster analysis based on firing rate, sag ratio and AP width.
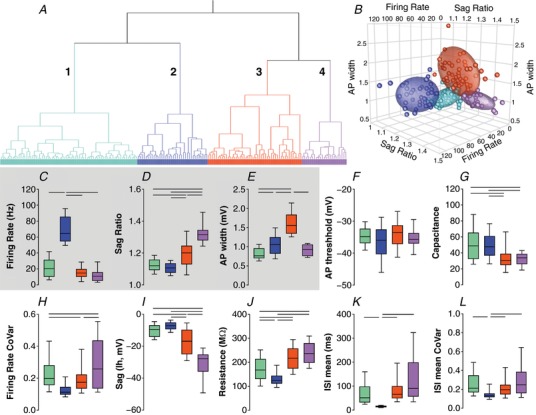
Colours outline 4 clusters: cluster 1 in light green, cluster 2 in blue, cluster 3 in red and cluster 4 in purple. A, Ward's method of hierarchical unsupervised clustering applied to 177 neurons recorded in the GPe of C57BL/6J mice. The x‐axis of the dendogram shows individual neurons and the y‐axis represents the linkage distance between the clusters. B, 3D scatter plot graph showing the distribution of the firing rate, sag ratio and AP width of individual neurons and the 60% elliptical shadow coverage of each cluster identified by the analysis. C–E, box plots showing the median and interquartile range with whiskers showing 10th–90th percentiles of the variables used for the cluster analysis. F–L, variables not included in the cluster analyses used to allow internal evaluation of the quality of this cluster. Horizontal bars represent statistically significant differences between 2 clusters (Kruskal–Wallis test followed by Dunn's multiple comparisons test).
We tried to use other combinations of electrophysiological variables based on information in the literature about different kinds of neurons in the GPe. In vivo recordings point to the importance of the firing rate and ISI CoVar as electrophysiological characteristics for arkypallidal and prototypical neurons (Abdi et al. 2015; Dodson et al. 2015). Analysis using these variables detected three clusters (Fig. 7 A–E), but these clusters could not explain important differences observed in labelled neurons, such as the capacitance, sag, resistance and sag ratio. In addition, we observed that the capacitance is significantly different for FoxP2, NPas1, Lhx6 and PV neurons (Fig. 5 J). Thus, we ran a cluster analysis using the firing rate, sag ratio and capacitance, even though sag ratio and capacitance were more highly correlated. This analysis detected four clusters (data not shown). Although the clusters showed differences in other variables and predicted the classification of labelled neurons, it did not add extra information that was not already observed in the following cluster analysis.
Figure 7. Cluster analysis based on firing rate and ISI mean CoVar.
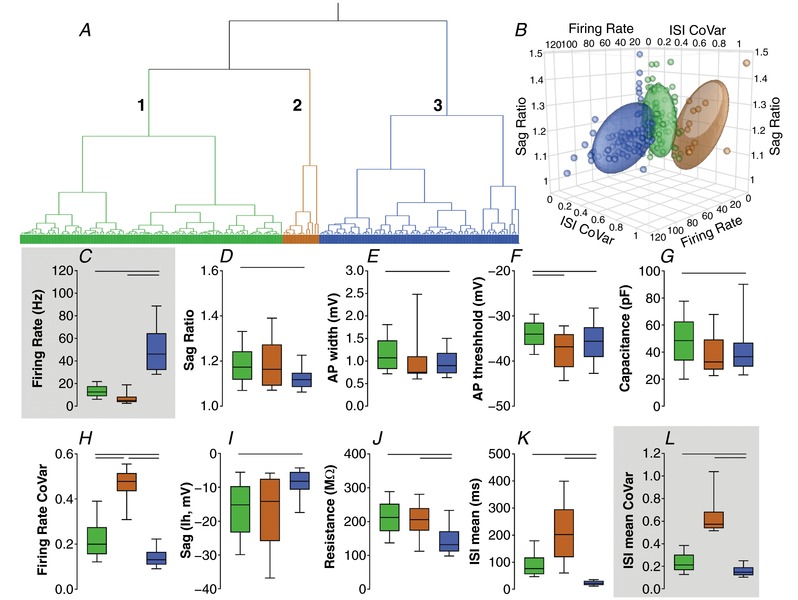
Colours outline 3 clusters: cluster 1 in light green, cluster 2 in brown and cluster 3 in blue. A, Ward's method of hierarchical unsupervised clustering applied to 177 neurons recorded in the GPe of C57BL/6J mice. The x‐axis of the dendogram shows individual neurons and the y‐axis represents the linkage distance between the clusters. B, 3D scatter plot graph showing the distribution of the firing rate, ISI mean CoVar and sag ratio of individual neurons and the 60% elliptical shadow coverage of each cluster identified by the analysis. C–E, box plots showing the median and interquartile range with whiskers showing 10th–90th percentiles of the variables used for the cluster analysis. F–L, variables not included in the cluster analyses used to allow internal evaluation of the quality of this cluster. Horizontal bars represent statistically significant differences between 2 clusters (Kruskal–Wallis test followed by Dunn's multiple comparisons test).
We then ran an unsupervised hierarchical cluster analysis using firing rate and sag ratio data (Fig. 8). The Thorndike procedure indicated that three clusters could explain most of the variability observed with these measures (Fig. 8 A and B). Cluster 3 has higher firing rate than clusters 1 and 2 (Fig. 8 C). Cluster 2 has higher sag ratio than clusters 1 and 3 (Fig. 8 D). We analysed other variables not included in the cluster analyses as an internal evaluation of the clusters (Fig. 8 F–L). Kruskal–Wallis tests followed by Dunn's multiple comparisons detected that cluster 3 had lower action potential width than clusters 1 and 2 (P < 0.05, Fig. 8 E). No significant action potential threshold difference was observed (Fig. 8 F). Cluster 2 showed lower capacitance and sag than clusters 1 and 3 (P < 0.05; Fig. 8 G–I). Cluster 3 had lower firing rate CoVar, resistance, ISI mean and ISI CoVar when compared to clusters 1 and 2 (P < 0.05, Fig. 8 H–L).
Figure 8. Cluster analysis based on firing rate and sag ratio.

Colours outline 3 clusters: cluster 1 in yellow, cluster 2 in purple, cluster 3 in blue. A, Ward's method of hierarchical unsupervised clustering applied to 177 neurons recorded in the GPe of C57BL/6J mice. The x‐axis of the dendrogram shows individual neurons and the y‐axis represents the linkage distance between the clusters. B, 3D scatter plot graph showing the distribution of the firing rate, sag ratio and AP width of individual neurons and the 60% elliptical shadow coverage of each cluster identified by the analysis. C–D, box plots showing the median and interquartile ranges with whiskers showing 10th–90th percentiles of the variables used for the cluster analysis. E–L, variables not included in the cluster analyses used to allow testing of the quality of this cluster. Horizontal bars represent statistically significant differences between 2 clusters (Kruskal–Wallis test followed by Dunn's multiple comparisons test).
To compare the results of the cluster analysis to an independent analysis of different neuronal subtypes, i.e. labelled neurons, we plotted the firing rate and sag ratio data from pre‐labelled neurons (FoxP2, NPas1, Lhx6 and PV) on top of the 75% elliptical shadow coverage of each cluster (Fig. 9). As mentioned before, FoxP2 and PV neurons are considered the biomarkers that best segregate at least two types of GPe neurons. Clusters created by this analysis predicted electrophysiological characteristic differences observed in pre‐labelled neurons. Cluster 2 characteristics align with the FoxP2 neuron electrophysiological signature (Fig. 9 A), while cluster 3 recapitulates the characteristics of PV neurons (Fig. 9 D). In addition, it can be seen in Fig. 6 that the overlap between clusters 2 and 3 is very small, which corroborates the data indicating that FoxP2 and PV are separate neuronal subtypes. In addition, our immunohistochemistry data show that Npas1 is also highly co‐localized with FoxP2 (51% of Npas1‐positive neurons also express the FoxP2 biomarker). In accordance with these data, most of the Npas1 neurons align with cluster 2 (Fig. 9 B); the same cluster has electrophysiological signatures similar to FoxP2 neurons. Although some previous work has shown substantial overlap between Lhx6 and PV neurons, our immunofluorescence data indicate overlap of only 12–15% (Fig. 2 F–G). This is corroborated by the segregation of the electrophysiological data from Lhx6 and PV neurons in at least two different clusters (1, yellow, and 3, blue, as observed in Fig. 7 C and D, respectively). Some of the Lhx6 data overlaps with cluster 2 (the one which contains the FoxP2 data), which recapitulates the immunofluorescence considering that FoxP2 and Lhx6 neurons overlap by 4–6% (Fig. 3). In future experiments, neurons with characteristics that fall within the 10% and 90% percentile values for firing rate and sag ratio presented in Table 2 can be safely classified as fitting within each cluster.
Figure 9. 2D scatter plot graph showing the distribution of the firing rate and sag ratio of labelled neurons, FoxP2, Npas1, Lhx6 and PV (in each column); and the 75% elliptical shadow coverage of each cluster identified by the analysis shown in Figure 6 .
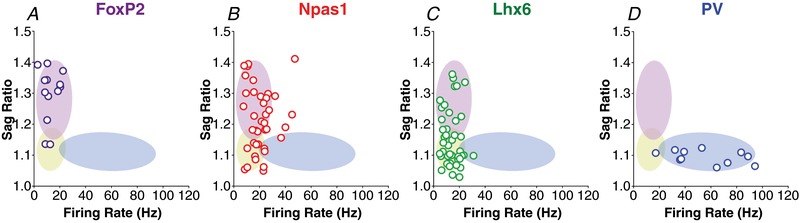
Colours outline 3 clusters: cluster 1 in yellow, cluster 2 in purple, cluster 3 in blue. Firing rate and sag ratio explain the variability among different labelled neurons of the GPe, but AP width does not add further information.
Table 2.
Descriptive statistics of clusters 1, 2 and 3 based on cluster analysis of firing rate (Hz) and sag ratio of GPe neurons from wild‐type mice shown in Fig. 6
| Cluster | ||||||
|---|---|---|---|---|---|---|
| 1 (yellow) | 2 (purple) | 3 (blue) | ||||
| Mean | Percentile 10: 90 | Mean | Percentile 10: 90 | Mean | Percentile 10: 90 | |
| Firing rate (Hz) | 13.08 | 4.2: 22.65 | 15.47 | 5.01: 28.43 | 57.29 | 31.60: 92.93 |
| Sag ratio | 1.11 | 1.05: 1.17 | 1.28 | 1.19: 1.41 | 1.11 | 1.06: 1.16 |
Discussion
We have developed a classification model to identify GPe neurons of wild‐type mice using an unbiased statistics‐based approach derived from electrophysiological characteristics. The cluster method is based on GPe neurons recorded from C57BL/6J mouse brain slices with post hoc validation using analysis of variables not included in the cluster analysis and data from neurons identified by a specific biomarker. We observed that three reliable clusters were built using the firing rate and sag ratio as variables in the unsupervised hierarchical cluster analysis. The good agreement between these clusters with respect to significant differences in other electrophysiological variables and grouping the data for identified PV, FoxP2, Npas1 and Lhx6 neurons provide strong validation that the groups classified by cluster analysis are meaningful. Thus, we propose that the GPe GABAergic neurons are best classified into three subgroups. (1) The arkypallidal neurons, grouped as cluster no. 2, marked mostly by FoxP2 and Npas1 markers, and partially by the Lhx6 marker, presumably projecting mostly to the striatum (Glajch et al. 2016), and presenting low firing rates and large sag. (2) The low‐frequency prototypical neurons, grouped as cluster no. 1, marked mostly by Lhx6 and partially by the Npas1 marker. Previous studies indicate that these neurons project mostly to the STN with a minor projection to striatal interneurons (Mastro et al. 2014; Corbit et al. 2016). These neurons have low firing rates and small sag. And (3) the high‐frequency prototypical neurons with high firing rates and small sag, grouped as cluster no. 3, marked by PV, and projecting predominantly to the STN as indicated in previous studies (Mastro et al. 2014).
Different research groups have demonstrated that FoxP2 and PV are the most segregated biomarkers expressed in neurons of the GPe (Dodson et al. 2015; Hernandez et al. 2015). We did not perform the immunostaining for these two biomarkers in the same set of brain slices, but our electrophysiological data and cluster segregation support the fact that these two subgroups represent two different subtypes of neurons. FoxP2 is probably the best biomarker for arkypallidal neurons and PV is a good biomarker for prototypical neurons, especially those with high firing rates. We also agree with the literature about the strong co‐expression of FoxP2 and Npas1. Although we observed about 57% of FoxP2‐positive neurons co‐expressing Npas1, levels of 80% were observed in rostral, lateral and caudal parts of the GPe, which is in agreement with the levels observed in previous work (Dodson et al. 2015; Hernandez et al. 2015). Although incomplete in its overlap with FoxP2 expression, Npas1 is also a good marker for arkypallidal neurons. Indeed, both FoxP2 and Npas1 neurons have strong projections to the spiny projection neurons of the striatum, characteristics of the arkypallidal neuron (Abdi et al. 2015; Glajch et al. 2016).
Our immunohistochemistry data agree with others regarding the strong segregation between FoxP2‐ and Lhx6‐expressing neurons (about 95% of segregation) (Dodson et al. 2015; Hernandez et al. 2015). There is also a consensus that Lhx6 and NPas1 overlap in about 30% of the GPe neurons (Hernandez et al. 2015). This means that Lhx6 is unlikely to mark arkypallidal neurons, making it a useful marker for prototypical neurons. Lhx6 neurons send very strong projections down to the STN (Mastro et al. 2014), consistent with a prototypical classification, but they also innervate interneurons in the striatum (Corbit et al. 2016).
We observed that about 15% of Lhx6 neurons co‐express PV. As mentioned before, the reported overlap between neurons expressing these biomarkers shows some variability across different laboratories and studies. Mastro et al. (2014) found only 2% overlap between Lhx6 and PV, while Hernández et al. (2015) observed that about 25% of Lhx6‐expressing neurons also express PV, and Dodson et al. (2015) reported that about 55% of Lhx6 neurons also expressed PV. This disparity has spurred debate in the field, particularly with respect to the nature of Lhx6‐expressing neurons. Differences in immunohistochemistry protocols used to label these neurons could account for some of the discrepancies in these findings. The age of the mice included in the experiments is also likely to be an important difference. We used brains of 30‐ to 45‐day‐old mice, an age range similar to that examined by Mastro et al. (2014). Hernández et al. (2015) used 55‐ to 80‐day‐old mice and Dodson et al. (2015) used older mice, of 90–120 days. The influence of these different factors on classification of neurons based on some of the molecular markers should be explored in more detail in future studies. In the present study, however, the focus was on classification of neurons based on electrophysiological properties with the markers being used mainly for the verification of cluster analysis.
We observed differences in the GPe subregional distribution of the Lhx6 and PV neurons, as the PV neurons are more concentrated than Lhx6 neurons in the dorsolateral part of the GPe. Similar results were reported by others (Mastro et al. 2014; Hernandez et al. 2015). The Lhx6 and PV neurons also differ in their sensitivity to ethanol‐induced decreases in firing rate (Abrahao et al. 2017).
As we suggest, the electrophysiological signatures are one of the strongest pieces of evidence that Lhx6 and PV represent different subclasses of prototypical neurons. Indeed, studies from different groups reported significant differences in the electrophysiological signatures of Lhx6 and PV neurons (Mastro et al. 2014; Hernandez et al. 2015). Thus, although Lhx6 and PV are both biomarkers for prototypical neurons, they may represent subtypes of prototypical neurons with Lhx6 neurons having low firing rate and PV neurons, higher firing rate. These neurons also have contrasting functions within the basal ganglia circuitry, as a recent study showed that optical inhibition of pallidal Lhx6 neurons or excitation of pallidal PV neurons in dopamine‐depleted mice can both restore locomotion, indicating that these neurons have opposite functions in older mice (Mastro et al. 2017).
The electrophysiological signatures appear to add key features that can be used to distinguish between neuronal subtypes. Unsupervised cluster analysis classification methods appear ideal to identify subtypes of neurons present in GPe and other brain regions using quantitative criteria obtained by unbiased algorithms. It is also important that the results of cluster analysis are thoroughly validated for statistical significance and evidence of natural groups. Previous work used cluster analysis based on spike width, spike threshold, the spike frequency at depolarized pulse and the maximum spike frequency to classify the GPe neurons in three clusters (Mizutani et al. 2017). However, the authors did not use labelled neurons to verify their clusters. Our classification of the GPe neurons into three clusters recapitulates the expected difference among these clusters in other electrophysiological variables not included in the cluster analyses. The proposed clusters also agree with the levels of segregation or overlap among the specific biomarkers included in this work (FoxP2, Npas1, Lhx6 and PV). For example, FoxP2 and Npas1 are highly co‐expressed in GPe neurons, which was recapitulated by the cluster showing extensive overlap between the electrophysiological data of neurons expressing these two biomarkers. Thus, we believe that electrophysiological characteristics can be used to confidently classify GPe neuron subtypes when other markers are not available. This approach will be especially useful in studies such as those in which in vivo manipulations are performed in wild‐type mice, or when fluorescent cell markers cannot be used due to expression of fluorophores for other purposes (e.g. live cell imaging, photometry or cell marking for chemo‐ or optogenetics). This approach will also be useful in cases where there is a lack of one mouse expressing all desired markers, or when strains or species are used where transgenics are not available. On the other hand, it is important to bear in mind that the clustering approach has its own limitations. For example, there are always a few outlier neurons showing characteristics that do not fit neatly into a given cluster. Thus, it is important to have relatively strict criteria and large sample sizes when using this approach.
Scientists must understand the unique characteristics of different classes of neurons to better understand the brain. Different approaches have been used to classify neurons in many parts of the brain by their morphological, physiological and molecular properties (Armananzas & Ascoli, 2015). Although we focused on specific biomarkers and electrophysiological characteristics of the GPe neurons, future analysis may also consider other measures to classify GPe neurons, including single‐cell anatomy/morphology and transcriptomic analysis. These features can offer a high throughput alternative and reveal even richer cellular diversity.
Additional information
Competing interests
The authors declare no conflict of interest.
Author contributions
K.P.A. and D.M.L. conceived and designed the work. K.P.A. collected and analysed the data. Both authors refined the manuscript and its intellectual content. Both authors have read and approved the final version of this manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by the Division of Intramural Clinical and Biological Research of the National Institute on Alcohol Abuse and Alcoholism project number ZIA AA000407. K.P.A.’s fellowship was also supported by the 2014 Ciências sem Fronteiras program (Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior—CAPES 2496/13‐5, Brazil). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We acknowledge Dr C. Savio Chan for providing the Npas1‐Cre‐2A‐tdTomato BAC mice and Dr Aryn Gittis for providing the Tg(Lhx6‐EGFP)BP221Gsat mice. We also thank Dr Margaret Davis for expert assistance with immunostaining.
Biography
Karina Possa Abrahao got her PhD at the Universidade Federal de São Paulo, in Brazil. She moved to the USA in 2013 to get her postdoctoral training in the National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism, under the supervision of D.M.L. She is currently starting her own lab in the Universidade Federal de São Paulo, Brazil. Her current scientific interests are: (1) study of neuronal subtypes within the basal ganglia, (2) how ethanol can affect specific synapses and neuronal subtypes in the basal ganglia, and (3) understanding the neuronal substrates of habitual/compulsive ethanol drinking.

Edited by: Jaideep Bains & Katalin Toth
References
- Abdi A, Mallet N, Mohamed FY, Sharott A, Dodson PD, Nakamura KC, Suri S, Avery SV, Larvin JT, Garas FN, Garas SN, Vinciati F, Morin S, Bezard E, Baufreton J & Magill PJ (2015). Prototypic and arkypallidal neurons in the dopamine‐intact external globus pallidus. J Neurosci 35, 6667–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahao KP, Chancey JH, Chan CS & Lovinger DM (2017). Ethanol‐sensitive pacemaker neurons in the mouse external globus pallidus. Neuropsychopharmacology 42, 1070–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin RL, Young AB & Penney JB (1989). The functional anatomy of basal ganglia disorders. Trends Neurosci 12, 366–375. [DOI] [PubMed] [Google Scholar]
- Armananzas R & Ascoli GA (2015). Towards the automatic classification of neurons. Trends Neurosci 38, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P & Sabatini DM (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Tanaka KF, Hen R & Rayport S (2011). Functional connectome of the striatal medium spiny neuron. J Neurosci 31, 1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJ & Stanford IM (2000). Electrophysiological and morphological characteristics of three subtypes of rat globus pallidus neurone in vitro . J Physiol 527, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit VL, Whalen TC, Zitelli KT, Crilly SY, Rubin JE & Gittis AH (2016). Pallidostriatal projections promote beta oscillations in a dopamine‐depleted biophysical network model. J Neurosci 36, 5556–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR ( 1990). Primate models of movement disorders of basal ganglia origin. Trends Neurosci 13, 281–285. [DOI] [PubMed] [Google Scholar]
- Dodson PD, Larvin JT, Duffell JM, Garas FN, Doig NM, Kessaris N, Duguid IC, Bogacz R, Butt SJ & Magill PJ (2015). Distinct developmental origins manifest in the specialized encoding of movement by adult neurons of the external globus pallidus. Neuron 86, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G & Heintz N (2013). The neuron identity problem: form meets function. Neuron 80, 602–612. [DOI] [PubMed] [Google Scholar]
- Flandin P, Kimura S & Rubenstein JL (2010). The progenitor zone of the ventral medial ganglionic eminence requires Nkx2‐1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci 30, 2812–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glajch KE, Kelver DA, Hegeman DJ, Cui Q, Xenias HS, Augustine EC, Hernandez VM, Verma N, Huang TY, Luo M, Justice NJ & Chan CS (2016). Npas1+ pallidal neurons target striatal projection neurons. J Neurosci 36, 5472–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman DJ, Hong ES, Hernandez VM & Chan CS (2016). The external globus pallidus: progress and perspectives. Eur J Neurosci 43, 1239–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez VM, Hegeman DJ, Cui Q, Kelver DA, Fiske MP, Glajch KE, Pitt JE, Huang TY, Justice NJ & Chan CS (2015). Parvalbumin+ neurons and Npas1+ neurons are distinct neuron classes in the mouse external globus pallidus. J Neurosci 35, 11830–11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover BR & Marshall JF (1999). Population characteristics of preproenkephalin mRNA‐containing neurons in the globus pallidus of the rat. Neurosci Lett 265, 199–202. [DOI] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW & Carpenter AE (2011). Improved structure, function and compatibility for CellProfiler: modular high‐throughput image analysis software. Bioinformatics 27, 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita H & Kitai ST (1991). Intracellular study of rat globus pallidus neurons: membrane properties and responses to neostriatal, subthalamic and nigral stimulation. Brain Res 564, 296–305. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt DA, Cody PA, Lovinger DM & Davis MI (2015). Brain BLAQ: Post‐hoc thick‐section histochemistry for localizing optogenetic constructs in neurons and their distal terminals. Front Neuroanat 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet N, Micklem BR, Henny P, Brown MT, Williams C, Bolam JP, Nakamura KC & Magill PJ (2012). Dichotomous organization of the external globus pallidus. Neuron 74, 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro KJ, Bouchard RS, Holt HA & Gittis AH (2014). Transgenic mouse lines subdivide external segment of the globus pallidus (GPe) neurons and reveal distinct GPe output pathways. J Neurosci 34, 2087–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro KJ, Zitelli KT, Willard AM, Leblanc KH, Kravitz AV & Gittis AH (2017). Cell‐specific pallidal intervention induces long‐lasting motor recovery in dopamine‐depleted mice. Nat Neurosci 20, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Takahashi S, Okamoto S, Karube F & Fujiyama F (2017). Substance P effects exclusively on prototypic neurons in mouse globus pallidus. Brain Struct Funct 222, 4089–4110. [DOI] [PubMed] [Google Scholar]
- Nobrega‐Pereira S, Gelman D, Bartolini G, Pla R, Pierani A & Marin O (2010). Origin and molecular specification of globus pallidus neurons. J Neurosci 30, 2824–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC & Eliceiri KW (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Y, Bevan MD, Shink E & Bolam JP (1998). Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience 86, 353–387. [DOI] [PubMed] [Google Scholar]
- Thorndike RL ( 1953). Who belongs in a family? Psychometrika 18, 267–276. [Google Scholar]
- Ward JH ( 1963). Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58, 236–244. [Google Scholar]


