Abstract
The Aquaporin-4 (AQP4) water channel contributes to brain water homeostasis in perivascular astrocyte endfeet where it is concentrated. We postulated that AQP4 is tethered at this site by binding of the AQP4 C terminus to the PSD95-Discs large-ZO1 (PDZ) domain of syntrophin, a component of the dystrophin protein complex. Chemical cross-linking and coimmunoprecipitations from brain demonstrated AQP4 in association with the complex, including dystrophin, β-dystroglycan, and syntrophin. AQP4 expression was studied in brain and skeletal muscle of mice lacking α-syntrophin (α-Syn−/−). The total level of AQP4 expression appears normal in brains of α-Syn−/− mice, but the polarized subcellular localization is reversed. High-resolution immunogold analyses revealed that AQP4 expression is markedly reduced in astrocyte endfeet membranes adjacent to blood vessels in cerebellum and cerebral cortex of α-Syn−/− mice, but is present at higher than normal levels in membranes facing neuropil. In contrast, AQP4 is virtually absent from skeletal muscle in α-Syn−/− mice. Deletion of the PDZ-binding consensus (Ser-Ser-Val) at the AQP4 C terminus similarly reduced expression in transfected cell lines, and pulse–chase labeling demonstrated an increased degradation rate. These results demonstrate that perivascular localization of AQP4 in brain requires α-Syn, and stability of AQP4 in the membrane is increased by the C-terminal PDZ-binding motif.
Aquaporin water channel proteins are responsible for the high membrane water permeabilities of the tissues where they are expressed. The physiological significance of the ten mammalian aquaporins is emerging from studies of naturally occurring mutants and mice carrying targeted disruptions in aquaporin genes, with phenotypes ranging from congenital cataracts to nephrogenic diabetes insipidus (1). Each aquaporin has characteristic subcellular localization and tissue expression patterns that determine its physiological roles, but the mechanisms responsible for these complex patterns are largely unknown. In the mammalian central nervous system, AQP4 is concentrated in astrocyte endfeet membranes facing blood–brain and brain–cerebrospinal fluid interfaces (2, 3). AQP4 is also expressed in sarcolemma of fast-twitch fibers in skeletal muscle (2) and in basolateral membranes of collecting duct epithelium (4), sites where the dystrophin protein complex is expressed (5–8).
Identified as the gene mutated in Duchenne/Becker muscular dystrophies (9), dystrophin is part of a large membrane assembly connecting the cytoskeleton to the extracellular matrix (10). Dystrophin forms a bridge between filamentous actin and the transmembrane protein β-dystroglycan, which is associated with a laminin/agrin-binding protein, α-dystroglycan. On the cytoplasmic side of the complex, dystrophin binds to dystrobrevin, providing a scaffold for binding up to four syntrophin molecules (11, 12). Syntrophins are a family of five proteins (α, β1, β2, γ1, γ2) containing two pleckstrin homology domains, a PSD95-Discs large-ZO1 (PDZ) domain, and a C-terminal syntrophin-unique region (8). The PDZ domain serves as an adaptor for recruiting membrane channels, receptors, kinases, and other signaling proteins. Tissue-specific variations in the dystrophin complex are in part caused by different syntrophins. The α-syntrophin isoform is expressed primarily in skeletal and cardiac muscle, and at lower levels in brain (11, 13).
Localization of the dystrophin protein complex in perivascular astrocyte endfeet, skeletal muscle sarcolemma, and renal collecting duct suggests association with AQP4. AQP4 has the C-terminal sequence Ser-Ser-Val (tSSV), a sequence potentially capable of binding to PDZ domains in proteins such as syntrophins (14–16). AQP4 expression is markedly reduced in the skeletal muscle of mdx mice, which lack full-length dystrophin and associated proteins (17). Here we show that expression and subcellular localization of AQP4 depend on association with the dystrophin complex through a tSSV-PDZ-mediated interaction with α-syntrophin.
Materials and Methods
Coimmunoprecipitation.
Sprague–Dawley rat cerebellum (130 mg) was homogenized by Potter–Elvehjem at 4°C in 5 ml of 7.5 mM sodium phosphate (pH 7.0)/0.25 M sucrose/5 mM EDTA/5 mM EGTA, with Complete Protease Inhibitor Cocktail (Roche Diagnostics). After a 10-min 1,000 × g spin at 4°C, supernatants were incubated 30 min at 4°C with 150 μM dithiobis(succinimidyl propionate) (Sigma–Aldrich). Then 1.0% Triton X-100, 0.5% sodium deoxycholate, and 150 mM NaCl were added for 30 min. One-third of the sample was incubated for 4 h at 4°C with anti-AQP4 antibody (3 μg), anti-syntrophin hybridoma culture supernatant (20 μl), or rabbit IgG (3 μg). Immune complexes were adsorbed to 80 μl of protein A-Sepharose CL-4B (Sigma–Aldrich), washed four times at 4°C in 20 mM Tris (pH 8.0), 1.0% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, 5 mM EDTA, and 5 mM EGTA, and once with salt-free buffer. Immune complexes were eluted with 50 μl of SDS/PAGE sample buffer at 37°C for 30 min.
Immunoblotting.
Immunoprecipitates, cell membrane fractions, and lysates were analyzed by SDS/PAGE immunoblotting (18). Skeletal muscle AQP4 was immunoprecipitated from tibialis anterior and extensor digitorum longus. Mouse monoclonal MANEX7374A was used to detect Dp71 in anti-AQP4 immunoprecipitate; rabbit polyclonal 331 was used for Dp71 detection in anti-Syn immunoprecipitate.
Antibodies.
Rabbit polyclonal antibody to AQP4 (4, 18), pan-syntrophin mouse monoclonal SYN1351 (19), α-Syn-specific rabbit polyclonal SYN17 (11), and dystrophin polyclonal 331/2723 (19) were used. Dystrophin monoclonal MANEX7374A (clone 10A11) was from Glenn Morris (North East Wales Institute, Wrexham, U.K.). Commercial monoclonal antibodies to α-dystroglycan (VIA4–1; Upstate Biotechnology, Lake Placid, NY) and β-dystroglycan (43DAG1/8D5; NovoCastra, Newcastle, U.K.) were used.
Electron Microscopy.
Mice were perfused through the heart with 4% formaldehyde in phosphate buffer containing 0.2% picric acid at pH 6.0, then pH 10.0. Brains were cut into 0.5- to 1.0-mm slices, cryoprotected, quick-frozen in liquid propane (−170°C), and subjected to freeze substitution. Specimens were embedded in methacrylate resin (Lowicryl HM20) and polymerized by UV light below 0°C. Ultrathin sections incubated with antibodies to AQP4 (0.5 μg/ml) or α-Syn (10 μg/ml) followed by goat anti-rabbit antibody coupled to colloidal gold were examined using a Philips CM 10 electron microscope at 60 kV.
Fluorescence Microscopy.
Immunofluorescence labeling of unfixed skeletal muscle was performed on 8-μm cryosections of quadriceps femoris (11, 20). AQP4-transfected cell lines grown on glass cover slips (HEK293 and MDCK cells) or Nalge Nunc Permanox chamber slides (CHO-K1 cells) were fixed with formaldehyde, permeabilized with Triton X-100, and incubated with the AQP4 antibody (0.15 μg/ml) followed by Alexa 488-conjugated goat anti-rabbit IgG (2.0 μg/ml, Molecular Probes). Confocal microscopy used a Zeiss LSM 410.
Transfected Cell Lines.
HEK293 cells and CHO-K1 cells (American Type Culture Collection, Manassas, VA) and type II MDCK cells (8) were used. HEK293 and MDCK cells were cultured in 90% Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Rockville, MD)/10% fetal bovine serum. CHO-K1 cells were cultured in 90% F-12K (Kaighn's modified F-12; Life Technologies)/10% fetal bovine serum. AQP4 expression plasmids were constructed from the pCI-neo vector (Promega) and cDNA sequences for the wild-type M1 (amino acids 1–323) and M23 (amino acids 23–323) isoforms of rat AQP4 (GenBank U14007) or for M1 and M23 isoforms lacking the C-terminal SSV (amino acids 1–320 and amino acids 23–320). Cell lines transfected with Lipofectamine Plus (Life Technologies) were selected for 14 days in medium with 700 μg/ml Geneticin and maintained in 200 μg/ml Geneticin.
Reverse Transcription (RT)-PCR.
Total RNA was isolated by extraction with phenol and guanidine isothiocyanate (Trizol; Life Technologies). Single-stranded cDNA was synthesized with reverse transcriptase and oligo(dT)18 primer (Advantage RT-for-PCR Kit; CLONTECH). cDNA was used for PCR with oligonucleotide primers to amplify segments from AQP4, β-actin, and neomycin resistance (NeoR) genes. PCR controls used RNA without RT.
Metabolic Labeling.
Stably transfected HEK293 cells in 60-mm dishes were incubated for 30 min in methionine-free DMEM supplemented with L-glutamine and for 60 min in methionine-free DMEM supplemented with L-glutamine and 0.4 mCi/ml (1 Ci = 37 GBq) L-[35S]methionine. Labeling medium was then replaced with DMEM. At chase times, cells were harvested in 1.0 ml of solubilization buffer [20 mM Tris (pH 8.0)/200 mM NaCl/1% Triton X-100/1% sodium deoxycholate/0.1% SDS/5.0 mM EDTA/5.0 mM EGTA/Complete Protease Inhibitor Cocktail (Roche)]. Lysates were incubated with 3 μg of AQP4 antibody for 1 h at 4°C. Immune complexes were precipitated with protein A-Sepharose for SDS/PAGE autoradiography.
Results
Association of AQP4 with the Dystrophin Protein Complex.
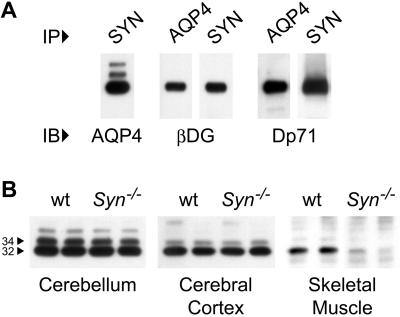
PDZ proteins bind directly to a motif of three to five residues at the C terminus of certain membrane proteins. The chemical cross-linking agent dithiobis(succinimidyl propionate) was used to stabilize this association before immunoprecipitation with anti-AQP4 or with pan-specific anti-syntrophin. After cleaving the cross-linker with DTT, SDS/PAGE immunoblotting revealed AQP4 in the anti-syntrophin immunoprecipitate (Fig. 1A). β-dystroglycan and the astrocyte isoform of dystrophin, Dp71 (6), were also present. Likewise, the anti-AQP4 immunoprecipitate contained β-dystroglycan and Dp71; however, presence of syntrophins could not be evaluated because they comigrate on SDS/PAGE with the IgG heavy chain. Coimmunoprecipitation of AQP4 with multiple dystrophin-associated proteins and the known reduction of AQP4 in the skeletal muscle of mdx mice (17) strongly suggest that these proteins reside in a large complex.
Figure 1.
Immunoblots of brain and skeletal muscle. (A) Coimmunoprecipitation (IP) of AQP4 with syntrophin and other dystrophin-associated proteins from dithiobis(succinimidyl propionate)-stabilized rat cerebellum homogenate by AQP4 or pan-syntrophin antibodies. Immunoblots (IB) showed specific reactions with antibodies to AQP4 (major band, 32 kDa), β-dystroglycan (βDG, 43 kDa), or dystrophin isoform (Dp71, 71 kDa). Nonspecific IgG immunoprecipitates were nonreactive. (B) AQP4 immunoblots of cerebellum and cerebral cortex membranes (20 μg protein per lane) and AQP4 immunoprecipated from skeletal muscle of wild-type (wt) mice or mice lacking α-Syn (Syn−/−).
AQP4 Expression in α-Syn−/− Mice.
To determine whether α-Syntrophin binds to AQP4 in vivo, we analyzed AQP4 expression in tissues of mice with targeted disruption of the gene for α-Syn (20), which is normally expressed in brain and skeletal muscle (11, 21). Dystrophin expression is normal in these mice, even though α-Syn is absent (20). Immunoblots revealed that total AQP4 expression is unchanged in brain but is markedly reduced in skeletal muscle (Fig. 1B).
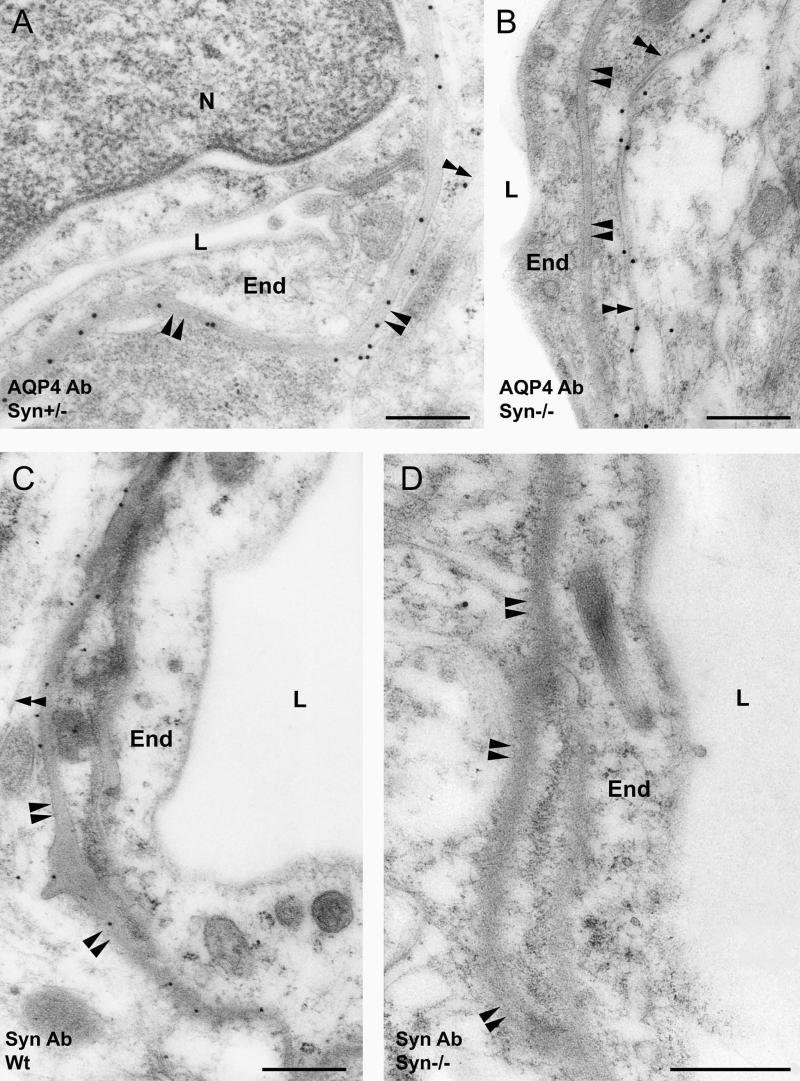
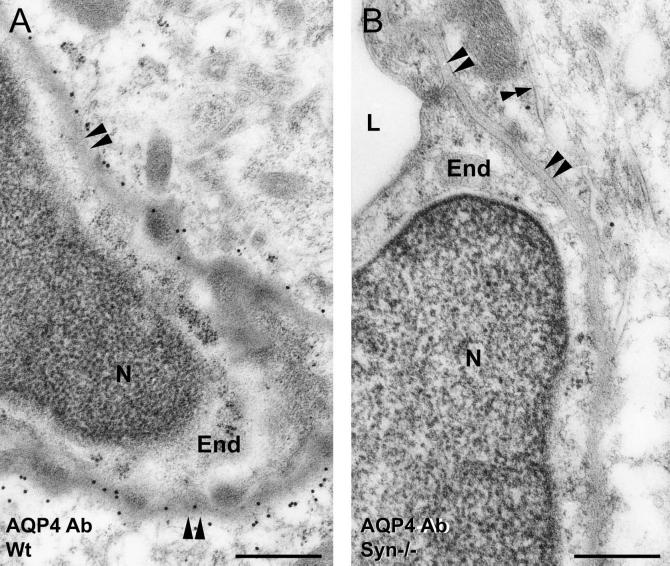
High-resolution, immunogold electron microscopy of rat brain has shown AQP4 to be concentrated in astrocyte membranes apposed to blood vessels but nearly absent in membranes away from the endothelial basal lamina (3). Cerebellum from wild-type α-Syn+/+ (data not shown) or heterozygous α-Syn+/− mice exhibits this same pattern (Fig. 2A). In contrast, the pattern is reversed in cerebellum from α-Syn−/− animals, with perivascular astrocyte processes showing stronger immunogold labeling along the membrane facing the neuropil than along the membrane facing the blood vessel (Fig. 2B). Immunogold labeling demonstrated that α-Syn in astrocytes is concentrated in membranes in direct contact with endothelial basal lamina, similar to AQP4 (Fig. 2C). α-Syn antibody labeling is entirely absent in α-Syn−/− cerebellum (Fig. 2D). Elsewhere in brain, AQP4 is concentrated in astrocyte membranes adjacent to endothelial basal lamina in cerebral cortex from wild-type mice (Fig. 3A), whereas AQP4 expression is strongly reduced in this membrane in cortex from α-Syn−/− animals (Fig. 3B).
Figure 2.
Immunogold electron micrographs of cerebellum from heterozygous (α-Syn+/−) and α-Syn-null (α-Syn−/−) mice incubated with antibodies to AQP4 (AQP4 Ab) or α-Syn (Syn Ab). (A and B) AQP4 immunolabeling of astrocyte perivascular endfeet in membranes facing endothelial basal lamina (paired arrowheads) or membranes facing neuropil (double arrowheads). (C and D) α-Syn immunolabeling is similar to AQP4 in wild-type (wt) mice but is absent in α-Syn−/− mice. End, capillary endothelial cell; N, nucleus; L, lumen. (Scale bars, 0.5 μm.)
Figure 3.
Anti-AQP4 immunogold electron micrographs of cerebral cortex from (A) wild-type (wt) and (B) α-Syn-null (α-Syn−/−) mice. Anti-AQP4 labeling of astrocyte endfeet in membranes facing endothelial basal lamina (paired arrowheads) or membranes facing neuropil (double arrowheads). End, capillary endothelial cell; N, nucleus; L, lumen. (Scale bars, 0.5 μm.)
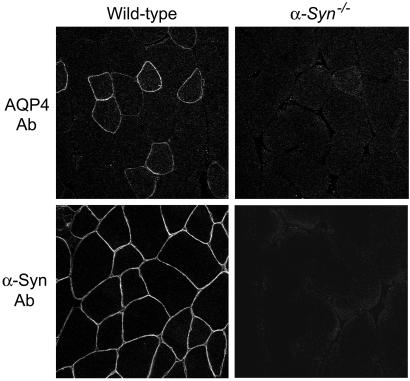
AQP4 is also expressed in sarcolemma of fast-twitch muscle fibers (2), as contained in quadriceps femoris. Immunolabeling revealed AQP4 in muscle from wild-type mice but almost none in α-Syn−/− muscle (Fig. 4), although residual AQP4 was localized to the sarcolemma. Thus, although proper distribution of AQP4 depends on α-Syn in brain, overall expression of AQP4 requires α-Syn in skeletal muscle.
Figure 4.
Confocal anti-AQP4 immunofluorescence of skeletal muscle from wild-type (wt) and α-Syn-null (α-Syn−/−) mice. Quadriceps femoris labeled with AQP4 and α-Syn antibodies reveals nearly complete loss of AQP4 from fast-twitch fibers in the absence of α-Syn.
Expression of AQP4 Lacking C Terminus.
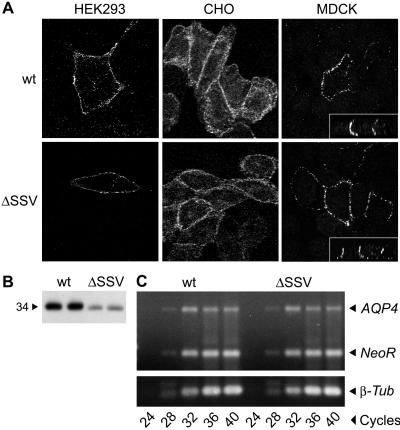
Because the C terminus of AQP4 (-SSV-COOH) conforms to the consensus for a PDZ-binding site, we reasoned that α-syntrophin may stabilize plasma membrane expression of AQP4 through a direct tSSV–PDZ interaction. To determine whether the AQP4 C terminus actually binds to syntrophins or other PDZ proteins, we compared the expression of wild-type (wt) AQP4 to that of AQP4 lacking three amino acids at the C terminus (AQP4 ΔSSV) in transfected cell lines expressing syntrophins and other dystrophin-associated proteins (8, 22). Immunofluorescence confocal microscopy demonstrated wt AQP4 and AQP4 ΔSSV in plasma membranes of the cell lines, and both were restricted to basolateral domains in polarized MDCK cells (Fig. 5A Insets). Thus, the tSSV is not essential for targeting of AQP4.
Figure 5.
AQP4 expression in cell lines transfected with wild-type (wt) AQP4 or AQP4 lacking three C-terminal residues (ΔSSV). (A) Transfected HEK293, CHO-K1, and MDCK cells labeled with anti-AQP4 and analyzed by immunofluorescence confocal microscopy show AQP4 wt and ΔSSV localized to the plasma membrane. Basolateral targeting in MDCK cells was confirmed in vertical sections (see Insets). (B) AQP4 immunoblot of membrane proteins prepared in duplicate from transfected HEK293 cells (20 μg protein per lane). (C) Expression of AQP4, neomycin resistance gene (NeoR), and β-tubulin (β-tub) assessed by RT-PCR of RNA from transfected HEK293 cells and ethidium bromide staining of agarose gels.
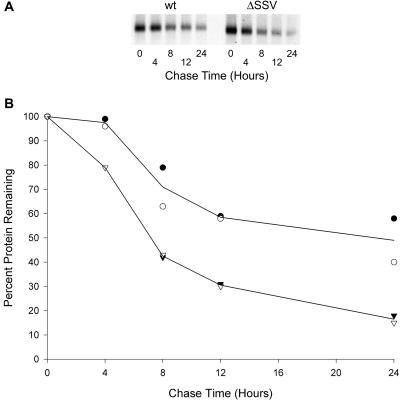
AQP4 expression in stably transfected HEK293 cells was evaluated further to determine whether the tSSV influences AQP4 stability. Immunoblots revealed a reduction in overall expression of AQP4 ΔSSV when compared with wt AQP4 (Fig. 5B). RT-PCR demonstrated that AQP4 mRNA and β-tubulin mRNA levels were equivalent in both cell lines. Similar transfection efficiencies were confirmed by levels of NeoR (Fig. 5C). Stability of wt AQP4 and AQP4 ΔSSV were also compared by [35S]methionine pulse–chase labeling of the transfected HEK293 cells. The exponential decline of signal showed a half-life of ≈8 h for AQP4 ΔSSV compared with ≈24 h for wt AQP4 (Fig. 6). Thus, the C-terminal SSV determines overall expression of AQP4 by influencing the degradation rate.
Figure 6.
Pulse–chase metabolic labeling of stably transfected HEK293 cells expressing wild-type (wt) AQP4 or AQP4 lacking three C-terminal residues (ΔSSV). (A) Autoradiograph of cell lysates analyzed by AQP4 immunoprecipitation and SDS/PAGE. (B) Graph of the amount of [35S]methionine-labeled AQP4 protein in the cells during chase. Two independent experiments (open and filled symbols) are shown. wt AQP4 (○); AQP4 ΔSSV (▿).
Discussion
Polarized expression of AQP4 at the blood–brain interface depended on the dystrophin-associated PDZ protein α-Syn. α-Syn influences the overall level of AQP4 expression in skeletal muscle, although sarcolemmal targeting of AQP4 can occur in its absence. Likewise, in transfected cell lines, the AQP4 C-terminal SSV is required for high-level AQP4 expression but not for plasma membrane targeting. Decreased expression of AQP4 ΔSSV is due to an increased degradation rate. α-Syn most likely regulates AQP4 expression by stabilizing the protein in the plasma membrane through a tSSV-PDZ-mediated interaction, similar to the mechanism stabilizing plasma membrane retention of the epithelial γ-aminobutyric acid transporter BGT1 by the PDZ protein LIN7 (23).
Targeting of AQP4 to basolateral domains may involve C-terminal di-leucine or tyrosine-based motifs (S. Le Maout, personal communication). While our work was being completed, independent studies suggested that subcellular localization of AQP4 is altered in α-Syn−/− skeletal muscle without a change in total expression estimated by immunoblot (24). Although we cannot explain this discrepancy, we have not been able to detect AQP4 in immunoblots of whole membranes from skeletal muscle without prior immunoprecipitation (18).
Consistent with a PDZ-mediated interaction of α-Syn with AQP4, ongoing studies indicate that transgenic mice overexpressing a dominant-negative mutant α-Syn (lacking the PDZ domain) also have reduced AQP4 expression in skeletal muscle (25). Nevertheless, we have not been able to detect direct binding between peptides or recombinant fusion proteins representing the AQP4 C terminus and α-Syn PDZ domain in blot overlays, ELISAs, or surface plasmon resonance assays (data not shown). We have also attempted without success to identify an intermediate linker protein by yeast two-hybrid screening and ELISAs involving the PDZ domains of various candidates (data not shown). These results are consistent with requirement for an accessory factor, a posttranslational modification, or binding by multiple sites, as was recently shown for the interaction between β2-syntrophin and the ICA512 transmembrane protein (26).
The need for chemical cross-linking before detergent solubilization to capture the interaction between AQP4 and the dystrophin complex may indicate that the association is labile. Although our AQP4-transfected cell lines express syntrophins and other dystrophin-associated proteins, we were unable to demonstrate cross-linking between AQP4 and the dystrophin-like complex in these cells (data not shown), possibly because of the differing compositions of the complex in brain and cell lines. The predominant complex in astrocytes contains Dp71 (6), whereas cell lines express primarily utrophin complexes (22, 27). A weaker association exists between utrophin and β-dystroglycan than between dystrophin and β-dystroglycan (22, 28). This observation may be explained by studies showing that adhesion-dependent phosphorylation of β-dystroglycan abolishes utrophin binding (29).
Our immunogold analyses of brain from α-Syn−/− mice indicate that α-Syn directly or indirectly anchors AQP4 in astrocyte membranes facing blood vessels. Absence of α-Syn diminishes perivascular AQP4 expression and reverses the normal concentration difference between perivascular membranes and other regions. Astrocytes may express other anchoring proteins that stabilize AQP4 in membranes facing neuropil in the absence of perivascular α-Syn. The presence of multiple AQP4 anchoring proteins in different distributions may explain why expression of AQP4 in stomach and kidney persists without α-Syn (data not shown).
Polarized expression of AQP4 in astrocytes may be critical for brain water homeostasis. We have suggested (30, 31) that AQP4 works in concert with inwardly rectifying K+ channels to allow for K+ siphoning during high neuronal activity. Absence of α-Syn may therefore interfere with the brain's capacity to handle excess K+ and the resulting osmotic gradients, even though total AQP4 expression is unchanged. In general, syntrophins and the dystrophin complex may provide the mechanism for the functional and anatomical polarization of astrocytes in the central nervous system.
Acknowledgments
We thank Richard L. Huganir and Ted Dawson for critical readings of the manuscript. This work was supported by the National Institutes of Health, the Muscular Dystrophy Association, the Norwegian Research Council, and the Jahre Foundation.
Abbreviations
- AQP4
Aquaporin-4
- Dp71
dystrophin
- tSSV
C-terminal sequence Ser-Ser-Val
- wt
wild type
- Syn
syntrophin
- PDZ
PSD95, Discs large, ZO1
- RT
reverse transcription
References
- 1.King L S, Yasui M, Agre P. Mol Med Today. 2000;6:60–65. doi: 10.1016/s1357-4310(99)01636-6. [DOI] [PubMed] [Google Scholar]
- 2.Frigeri A, Gropper M A, Umenishi F, Kawashima M, Brown D, Verkman A S. J Cell Sci. 1995;108:2993–3002. doi: 10.1242/jcs.108.9.2993. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen S, Nagelhus E A, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen O P. J Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terris J, Ecelbarger C A, Marples D, Knepper M A, Nielsen S. Am J Physiol. 1995;269:F775–F785. doi: 10.1152/ajprenal.1995.269.6.F775. [DOI] [PubMed] [Google Scholar]
- 5.Tian M, Jacobson C, Gee S H, Campbell K P, Carbonetto S, Jucker M. Eur J Neurosci. 1996;8:2739–2747. doi: 10.1111/j.1460-9568.1996.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 6.Blake D J, Hawkes R, Benson M A, Beesley P W. J Cell Biol. 1999;147:645–658. doi: 10.1083/jcb.147.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbeej M, Campbell K P. J Biol Chem. 1999;274:26609–26616. doi: 10.1074/jbc.274.37.26609. [DOI] [PubMed] [Google Scholar]
- 8.Kachinsky A M, Froehner S C, Milgram S L. J Cell Biol. 1999;145:391–402. doi: 10.1083/jcb.145.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monaco A P, Neve R L, Colletti-Feener C, Bertelson C J, Kurnit D M, Kunkel L M. Nature (London) 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 10.Worton R. Science. 1995;270:755–756. doi: 10.1126/science.270.5237.755. [DOI] [PubMed] [Google Scholar]
- 11.Peters M F, Adams M E, Froehner S C. J Cell Biol. 1997;138:81–93. doi: 10.1083/jcb.138.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newey S E, Benson M A, Ponting C P, Davies K E, Blake D J. Curr Biol. 2000;10:1295–1298. doi: 10.1016/s0960-9822(00)00760-0. [DOI] [PubMed] [Google Scholar]
- 13.Peters M F, O'Brien K F, Sadoulet-Puccio H M, Kunkel L M, Adams M E, Froehner S C. J Biol Chem. 1997;272:31561–31569. doi: 10.1074/jbc.272.50.31561. [DOI] [PubMed] [Google Scholar]
- 14.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 15.Gee S H, Madhavan R, Levinson S R, Caldwell J H, Sealock R, Froehner S C. J Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fanning A S, Anderson J M. J Clin Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frigeri A, Nicchia G P, Verbavatz J M, Valenti G, Svelto M. J Clin Invest. 1998;102:695–703. doi: 10.1172/JCI2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neely J D, Christensen B M, Nielsen S, Agre P. Biochemistry. 1999;38:11156–11163. doi: 10.1021/bi990941s. [DOI] [PubMed] [Google Scholar]
- 19.Froehner S C, Murnane A A, Tobler M, Peng H B, Sealock R. J Cell Biol. 1987;104:1633–1646. doi: 10.1083/jcb.104.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams M E, Kramarcy N, Krall S P, Rossi S G, Rotundo R L, Sealock R, Froehner S C. J Cell Biol. 2000;150:1385–1398. doi: 10.1083/jcb.150.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters M F, Kramarcy N R, Sealock R, Froehner S C. NeuroReport. 1994;5:1577–1580. [PubMed] [Google Scholar]
- 22.James M, Nguyen T M, Wise C J, Jones G E, Morris G E. Cell Motil Cytoskeleton. 1996;33:163–174. doi: 10.1002/(SICI)1097-0169(1996)33:3<163::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 23.Perego C, Vanoni C, Villa A, Longhi R, Kaech S M, Frohli E, Hajnal A, Kim S K, Pietrini G. EMBO J. 1999;18:2384–2393. doi: 10.1093/emboj/18.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokota T, Miyagoe Y, Hosaka Y, Tsukita K, Kameya S, Shibuya S, Matsuda R, Wakayama Y, Takeda S. Proc Jpn Acad. 2000;76:22–27. [Google Scholar]
- 25.Adams M E, Mueller H A, Froehner S C. J Cell Biol. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ort T, Maksimova E, Dirkx R, Kachinsky A M, Berghs S, Froehner S C, Solimena M. Eur J Cell Biol. 2000;79:621–630. doi: 10.1078/0171-9335-00095. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen T M, Le T T, Blake D J, Davies K E, Morris G E. FEBS Lett. 1992;313:19–22. doi: 10.1016/0014-5793(92)81174-k. [DOI] [PubMed] [Google Scholar]
- 28.Koga R, Ishiura S, Takemitsu M, Kamakura K, Matsuzaki T, Arahata K, Nonaka I, Sugita H. Biochim Biophys Acta. 1993;1180:257–261. doi: 10.1016/0925-4439(93)90047-5. [DOI] [PubMed] [Google Scholar]
- 29.James M, Nuttall A, Ilsley J L, Ottersbach K, Tinsley J M, Sudol M, Winder S J. J Cell Sci. 2000;113:1717–1726. doi: 10.1242/jcs.113.10.1717. [DOI] [PubMed] [Google Scholar]
- 30.Nagelhus E A, Horio Y, Inanobe A, Fujita A, Haug F M, Nielsen S, Kurachi Y, Ottersen O P. Glia. 1999;26:47–54. doi: 10.1002/(sici)1098-1136(199903)26:1<47::aid-glia5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 31.Niermann H, Amiry-Moghaddam M, Holthoff K, Witte O W, Ottersen O P. J Neurosci. 2001;21:3045–3051. doi: 10.1523/JNEUROSCI.21-09-03045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]