Abstract
Tolerance to self-antigens prevents the elimination of cancer by the immune system1,2. We used synthetic chimeric antigen receptors (CARs) to overcome immunological tolerance and mediate tumor rejection in patients with chronic lymphocytic leukemia (CLL). Remission was induced in a subset of subjects, but most did not respond. Comprehensive assessment of patient-derived CAR T cells to identify mechanisms of therapeutic success and failure has not been explored. We performed genomic, phenotypic and functional evaluations to identify determinants of response. Transcriptomic profiling revealed that CAR T cells from complete-responding patients with CLL were enriched in memory-related genes, including IL-6/STAT3 signatures, whereas T cells from nonresponders upregulated programs involved in effector differentiation, glycolysis, exhaustion and apoptosis. Sustained remission was associated with an elevated frequency of CD27+CD45RO- CD8+ T cells before CAR T cell generation, and these lymphocytes possessed memory-like characteristics. Highly functional CAR T cells from patients produced STAT3-related cytokines, and serum IL-6 correlated with CAR T cell expansion. IL-6/STAT3 blockade diminished CAR T cell proliferation. Furthermore, a mechanistically relevant population of CD27+PD-1CD8+ CAR T cells expressing high levels of the IL-6 receptor predicts therapeutic response and is responsible for tumor control. These findings uncover new features of CAR T cell biology and underscore the potential of using pretreatment biomarkers of response to advance immunotherapies.
Patients with relapsed or refractory CLL have dismal prognoses. With the possible exception of allogeneic stem cell transplantation, CLL is incurable with available therapies. Targeted inhibitors of B cell-signaling pathways, such as ibrutinib and idelalisib, have demonstrated remarkable activity in CLL but are not curative3; prolonged treatment has substantial medical, social and economic costs, and patients who become resistant have very poor outcomes4. Clinical trials of CD19-targeted T cell (CTL019) therapy have shown durable antitumor responses in CLL5, but in only 26% of patients6. This finding contrasts with relapsed or refractory acute lymphoblastic leukemia, in which anti-CD19 CAR T cells induce complete remission (CR) in over 90% of cases7. This disparity in therapeutic efficacy may be attributed to innate, humoral and cellular immune deficiencies, including inherent T cell defects that are characteristic of CLL and worsen with disease progression. To date, it has not been possible to identify patient- or disease-specific factors that predict why only certain patients with CLL have such dramatic responses to CTL019 treatment. Therefore, a detailed analysis is necessary to determine the T cell intrinsic mechanisms by which patients with CLL who have complete responses to CTL019 are able to maintain sustained antitumor effects.
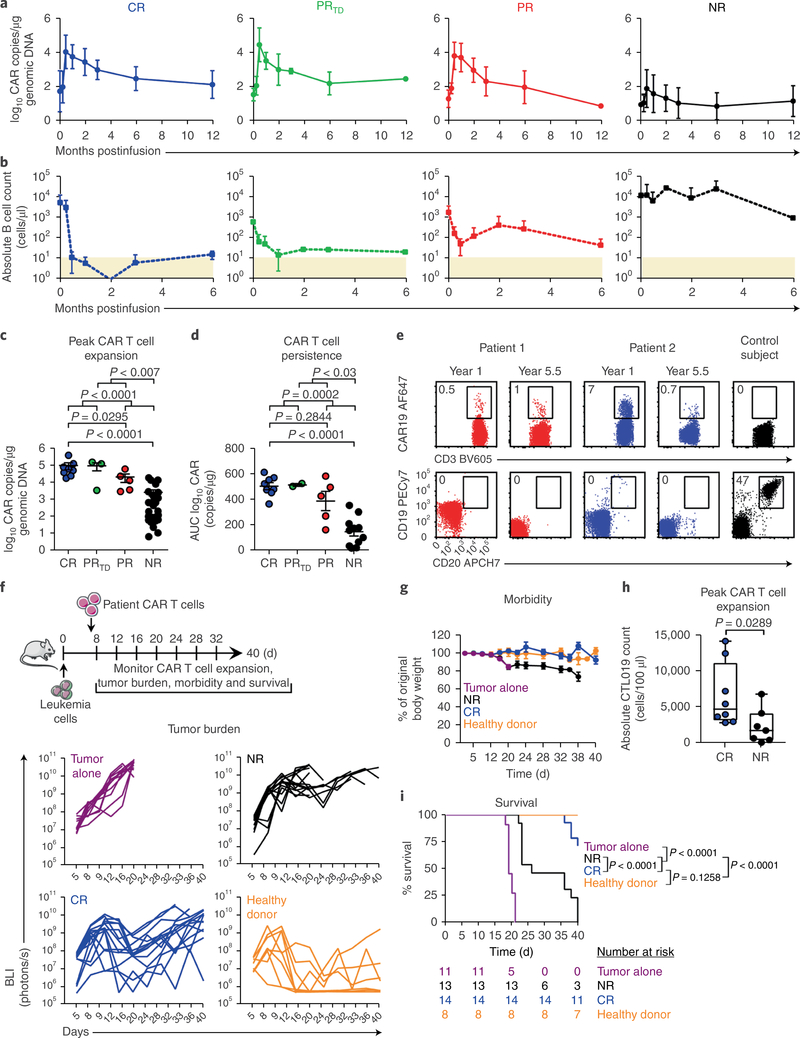
We studied 41 patients with advanced, heavily pretreated and high-risk CLL who received at least one dose of CD19-directed CAR T cells (patient characteristics in Supplementary Table 1). Some of these patients had been included in our original clinical trial6. In agreement with our previously reported findings, we were not able to identify patient- or disease-specific factors predicting which subjects responded best to CTL019 therapy6. Efficacy was not related to patient age, prior therapy, peripheral tumor burden, p53 status or other typical factors (Supplementary Table 1). Patients who responded to CTL019 exhibited dramatic in vivo expansion of CAR T cells (Fig. 1a) coincident with B cell aplasia (Fig. 1b) in the first two weeks after infusion, which was followed by a lognormal decay in peripheral blood; in contrast, nonresponding (NR) patients displayed limited or, in most cases, no in vivo T cell proliferation (Fig. 1a). NR patients, compared with responding subjects, also exhibited a limited degree of B cell aplasia in the peripheral blood during the first 6 months of therapy (Fig. 1b). The best overall response, clinical outcomes and toxicities for responding patients are summarized in Supplementary Table 2. The median peak expansion, expressed in CAR copy number per microgram genomic DNA in the peripheral blood, was 58,570 (range, 18,003–409,645 copies/μg) in patients who achieved a CR and 13,257 (range, 2,951–63,168 copies/μg) in partially responding (PR) subjects. A small subset of patients (PRTD) had highly active T cell products (Fig. 1a) but later relapsed with CLL that had transformed into aggressive B cell lymphoma. All three of these patients had a clinical cytokine-release syndrome requiring intervention and rapid clearance of bulky, extensive disease that was unrelated to lymphodepleting chemotherapy. PRTD patients exhibited a peak T cell expansion (median, 130,258 copies/μg; range, 3,480–160,977) equally robust to that in CR patients (Fig. 1c). In contrast, the peak expansion of CTL019 cells was minimal in NR patients (median, 205 copies/μg; range, 7–19,169) and was significantly lower than that in individuals in the CR, PR and PRTD categories (Fig. 1c). When expressed as area under the curve (AUC), the CAR copy number per microgram DNA from the time of peak expansion to 6 months postinfusion in evaluable patients with complete and durable remission was significantly higher than that in PR and NR subjects, thus suggesting that the infused CAR T cells persisted over the long term in CR patients (Fig. 1d). In extended follow-up analyses, we observed CR enduring beyond 5 years, and these CR patients still possessed detectable CTL019 cells in the peripheral blood (Fig. 1e). Furthermore, ongoing B cell aplasia was seen (Fig. 1e), thus demonstrating that the persisting CAR T cells remained functional. Therefore, CTL019 cells in responding patients expanded dramatically in vivo and persisted with ongoing functional activity to induce long-lasting remission. It appears that all effective CD19 CAR T cells—regardless of the costimulatory domain, the specific T cell subset enriched or the disease type treated—require in vivo cell expansion to be effective6–11.
Fig. 1|. The intrinsic potency of CAR T cells from patients with CLL drives response to therapy.
a, Expansion kinetics of peripheral blood CTL019 cells from patients with CLL. b, Peripheral blood B cell counts from matched patients. In a and b, graphs show mean± s.d.; shaded areas, detection limit (CR, n = 8; PRTD, n = 3; PR, n = 5; NR, n = 25). c, Peak expansion of CTL019 cells (log10 copies per microgram genomic DNA). d, Persistence of CTL019 cells in evaluable patients, shown by AUC calculations (log10 copies per microgram genomic DNA from day 0 to maximum expansion time by day 180). In c and d, P values were calculated with two-tailed Mann-Whitney tests. Symbols, individual patients; bars, s.e.m. (CR, n = 8; PRTD, n = 2–3; PR, n = 5; NR, n = 25). e, Flow cytometric plots of peripheral blood CAR T cells (top) and B cells (bottom) in two CR patients at 1 and 5.5 years postinfusion, compared with a control ubject (representative of 2 independent experiments). f, Top, schema of CAR T cell transfer into leukemic mice. Bottom, longitudinal tumor burden (BLI) of CTL019-treated mice (each line represents one mouse). g, Longitudinal weight loss in mice. Data are shown as mean± s.e.m. (tumor alone, n = 11; NR, n = 13; CR, n = 14; healthy donor, n = 8 for f and g). h, Peak CAR T cell counts in mice treated with CR (n = 8) or NR (n = 7) patient T cells (each symbol indicates one animal; boxes extend from the twenty-fifth to seventy-fifth percentiles; middle line, median; whiskers, minimum and maximum). i, Kaplan-Meier analysis of survival. P values were determined with two-tailed log-rank Mantel-Cox tests. The numbers of animals at risk in each group at specific time intervals are listed below the survival curve.
To evaluate whether the antitumor activity of CTL019 cells in patients with CLL is dictated by the intrinsic potency of the T cell infusion product independently of disease type and burden, we used clinically manufactured CAR T cells to treat NOD/SCID/γc−/− (NSG) mice engrafted with NALM-6 leukemia cells. CD19-directed CAR T cells generated from CR patients significantly decreased tumor progression (Fig. 1f), thereby resulting in diminished morbidity (Fig. 1g). Similarly to our clinical findings, CAR T cells from CR patients exhibited robust in vivo expansion (Fig. 1h) accompanied by significant prolongation of survival (Fig. 1i), as compared with CTL019 cells produced from NR subjects (Fig. 1h-i). These observations support the notion that the response of CLL to CAR T cell therapy is influenced by the composition of the cellular product and/or intrinsic T cell fitness.
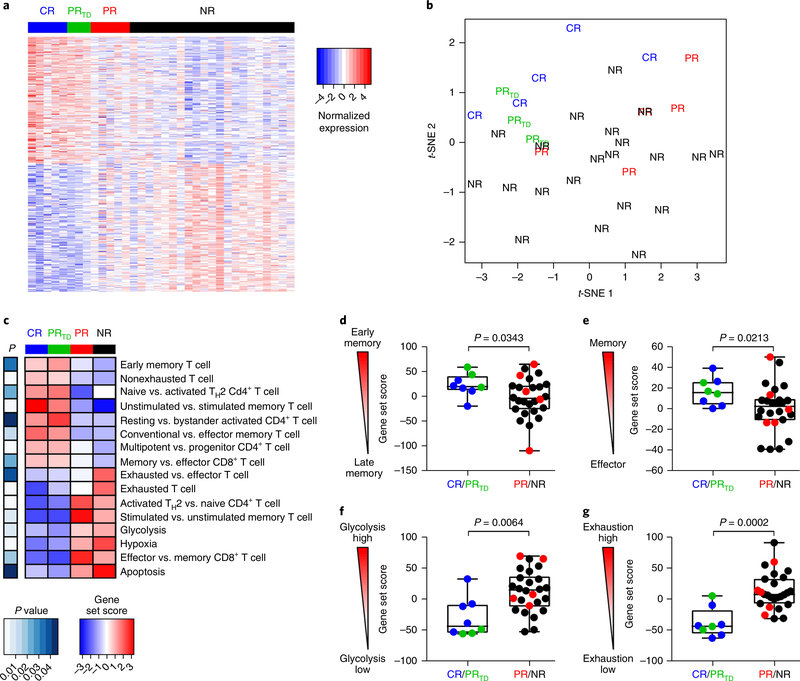
Because typical patient and disease characteristics did not explain the differences in response rates among patients with CLL, we explored other factors that might influence CAR T cell activity. The robust in vivo expansion of CTL019 cells in CR patients that led to sustained CR suggested that intrinsic T cell factors such as the differentiation state of the final CTL019 cells obtained after ex vivo expansion may be an important determinant of therapeutic efficacy. Accordingly, we compared the transcriptomes of T cells expanded from patients with complete, partial or no response to CTL019 therapy. The gene expression profiles of CAR T cells generated from CR and PRTD patients were markedly different from those generated from PR and NR patients (Fig. 2a and Supplementary Table 3). Both f-distributed stochastic neighbor embedding (Fig. 2b) and principal component analysis (Supplementary Fig. 1) confirmed that CTL019 cells derived from CR and PR patients who relapsed with transformed disease had similar expression profiles that were distinct from those of PR patients with lower levels of CAR T cell persistence as well as NR subjects. Gene set enrichment analysis revealed that CAR T cells from CR and PRTD patients were enriched in gene expression profiles involved in early memory differentiation, such as the expression of TCF7 and LEF1 (Fig. 2c-e and Supplementary Table 4). In contrast, CTL019 cells from PR and NR patients exhibited elevated expression of key regulators of late memory as well as effector T cell differentiation, apoptosis and aerobic glycolysis, which effector T cells rely on to produce energy, hypoxia and exhaustion (Fig. 2c,f,g).
Fig. 2|. Transcriptional profiles of CAR T cellular products reveal T cell-intrinsic quality attributes associated with clinical response.
a, Genes differentially expressed in CTL019 cellular infusion products from patients with CLL. Each column depicts an individual sample, and each row represents an individual gene, shaded to indicate normalized expression. The top 200 genes in either direction are shown. b, RNA-sequencing results from CAR T cell infusion products, shown as a f-distributed stochastic neighbor embedding (f-SNE) visualization. Each point depicts data from an individual patient sample labeled according to clinical response. c, Heat map of selected pathways enriched in genes significantly upregulated or downregulated in CTL019 cells from CR (blue bar), PRTD (green bar), PR (red bar) and NR (black bar) patients. For each pathway, a single sample enrichment score was calculated, and the mean was taken per response group. A color gradient ranging from dark blue to dark red indicates the mean normalized enrichment score (ranging from −3 to+3) of pathways enriched in induced (red) or repressed (blue) genes. d, Enrichment of an early memory T cell differentiation signature in preinfusion CTL019 cells from patients with CLL, for different response categories. e-g, Gene signature sets for T cell memory compared with effector (e), glycolysis (f) and exhaustion (g), as assessed by single-sample enrichment analysis. In d-g, each point represents the relative enrichment of these signatures in individual patient T cellular product samples. Boxes indicate the twenty-fifth to seventy-fifth percentiles; middle line, median; whiskers, minimum and maximum. The normalized enrichment score for each gene set is plotted on the y axis. P values were calculated with two-tailed Welch’s f tests (c-g). For a-g, CR, n = 5; PRTD, n = 3; PR, n = 5; NR, n = 21.
In accordance with our previous findings demonstrating that stimulation through a CAR can metabolically regulate T cell-fate differentiation12, we sought to functionally validate the glycolytic-gene signature that was enriched in PR and NR patient CAR T cells. CAR-specific stimulation of retrospective cellular infusion-product samples increased glycolysis (Supplementary Fig. 2a) and the uptake of a glucose analog (Supplementary Fig. 2b), thus providing evidence that T cells in patients with CLL can undergo metabolic modulation through ligation of a synthetic CAR. CAR-stimulated T cells from PR/NR relative to CR/PRTD patients demonstrated a higher uptake of a glucose analog (Supplementary Fig. 2c), a finding consistent with the transcriptional profiles of these cells (Fig. 2c,f). Furthermore, inhibition of glycolysis with 2-deoxy-D-glucose decreased effector differentiation and resulted in increased frequencies of CAR T cells with a central memory phenotype (Supplementary Fig. 2d,e). Pharmacologic inhibition of glycolysis concomitantly promoted the formation of memory CTL019 lymphocytes with enhanced proliferative capacity after re-stimulation with CD19-expressing tumor cells (Supplementary Fig. 2f). These results indicate that T cells generated from CR and PRTD patients show a gene expression profile that may confer properties of persistence as well as robust antitumor potency, and that decreasing glycolytic metabolism may be an actionable cellular manufacturing improvement for enhancing CAR T cell efficacy12.
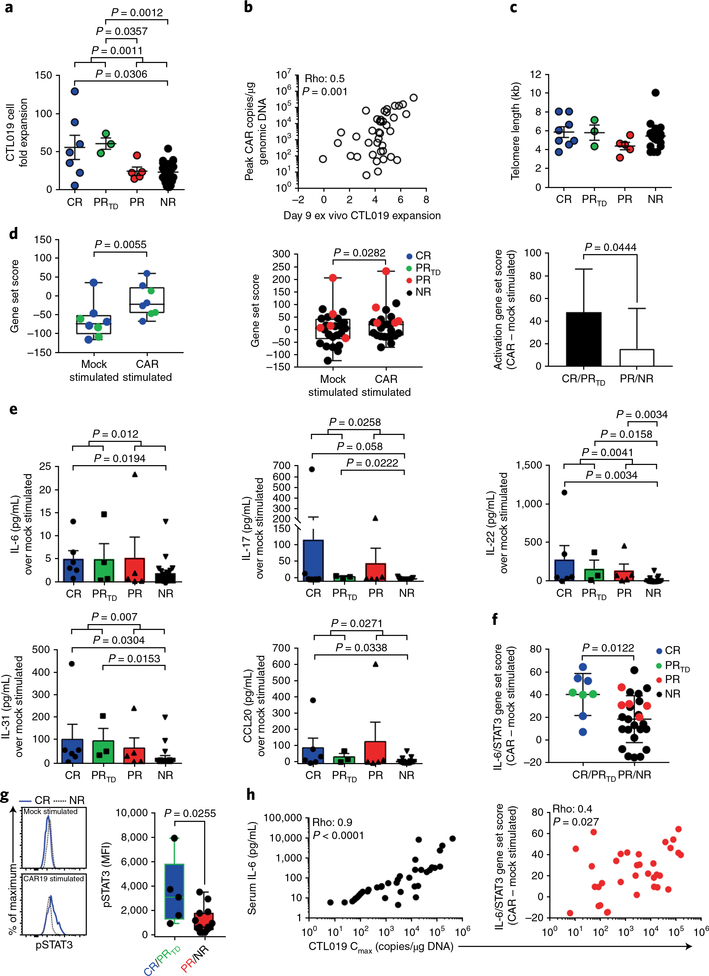
The proliferative and survival capacities of T lymphocytes that are largely a consequence of their differentiation status strongly correlate with the antitumor activity of adoptively transferred T cells13–15. We observed that CAR T cells from individuals who achieved CR and PRTD, compared with CTL019 cells obtained from PR and NR patients, exhibited superior expansion during clinical manufacturing, (Fig. 3a). Overall, the ex vivo expansion potential of patient-derived CAR T cells directly correlated with their degree of in vivo proliferation (Fig. 3b). To explore whether responses to CTL019 therapy were linked to the CAR T cell replicative history, we retrospectively analyzed the telomere lengths of preinfused CTL019 cells generated from patients with CLL. We did not observe significant differences in telomere lengths among CR, PRTD, PR and NR patients with CLL (Fig. 3c and Supplementary Fig. 3a). Telomere- length assessment by Southern blotting was concordant with qPCR- based measurements (Supplementary Fig. 3b). Similarly, telomerase activity in patient CTL019 cells after stimulation through the CAR or endogenous T cell receptor was uninformative in terms of predicting CAR T cell replicative capacity in vivo (Supplementary Fig. 3c,d). These results were consistent with a similar evaluation of expanded autologous tumor-infiltrating lymphocytes16 but were in contrast to results from a different trial in which telomere length in tumor-infiltrating lymphocytes was found to be a predictor of clinically meaningful tumor regressions17. These inconsistencies are likely to be associated with the heterogeneity of adoptively transferred cells across studies and the challenges associated with telomere-length assessment in bulk populations of lymphocytes. Thus, telomere length may not be a consistently reliable biomarker for predicting the antitumor efficacy of cellular products generated for T cell therapy.
Fig. 3|. CAR T cell replicative capacity, activation potential and IL-6/STAT3-pathway enrichment define therapeutic response and failure.
a, Ex vivo proliferation of CTL019 cells during clinical manufacturing, stratified according to clinical response (CR, n = 7; PRTD, n = 3; PR, n = 5; NR, n = 25). P values were calculated with two-tailed Mann-Whitney tests. b, Spearman’s rho correlation (two-tailed) between the ex vivo expansion potential and maximum in vivo proliferative capacity (measured by the cellular population doubling number) of adoptively transferred CTL019 cells in matched patients with CLL (n = 40). c, Comparison of telomere lengths between bulk, preinfused CTL019 cells in responding and nonresponding patients (CR, n = 8; PRTD, n = 3; PR, n = 5; NR, n = 20). Graphs in a and c show mean ± s.e.m. d, Single-sample enrichment analysis of T cell-activation gene signatures in the CTL019 gene expression profiles from CR/PRTD patients (n = 8; left) and PR and NR subjects (n = 26; middle) who received mock or CAR-specific stimulation. The rightmost panel depicts the magnitude of change in a T cell activation-associated gene-set score from mock- to CAR-stimulated CTL019 preinfusion cells from patients belonging to the respective response categories. P values were calculated with two-tailed paired t tests. e, Levels of soluble cytokines produced from CAR-stimulated CTL019 cells subtracted from baseline levels; matched, unstimulated controls in evaluable patient samples were used (CR, n = 6; PRTD, n = 3; PR, n = 5; NR, n = 21). P values were calculated with two-tailed Mann-Whitney tests. Graphs show mean with s.e.m. overlaid; symbols represent individual patient samples. f, Single-sample gene set enrichment analysis of the IL-6/STAT3 pathway in CAR-stimulated CTL019 cells from patients in each response group (P values were determined with two-tailed Welch’s t tests). Bars represent the mean ± s.e.m. g, Representative flow cytometry showing levels of pSTAT3 in preinfusion CTL019 cells from a CR and NR patient after overnight stimulation with isotype-control antibody-coated beads (mock stimulated) or beads coated with an anti-idiotypic antibody against CAR19 (CAR19 stimulated) (left). Pooled data from patients with highly functional (CR, n = 3; PRTD, n = 2) versus poorly functional (PR, n = 3; NR, n = 11) CAR T cells are shown in box plots (right). Boxes indicate the twenty-fifth to seventy-fifth percentiles; middle line, median; whiskers, minimum and maximum. Each symbol denotes an individual patient sample. The change in fluorescence intensity (ΔΜΡΙ) was calculated by subtracting the MFI of pSTAT3 in stimulated cells from that in unstimulated cells. The P value was calculated with a two-tailed Mann-Whitney test. h, Spearman’s rho correlation (two-tailed) shown between the maximum in vivo proliferative capacity of adoptively transferred CAR T cells (Cmax) and peak levels of serum IL-6 (n = 41; left) or IL-6/STAT3 gene enrichment in patient-matched CAR-stimulated (as above) CTL019 cells (n = 34; right).
To further determine whether the persistence of high frequencies of CAR T cells in CR patients was attributed to the functional quality of the cellular product, we stimulated preinfusion CTL019 cells with a novel bead-bound anti-CAR19 idiotypic antibody, which serves as a surrogate for cognate CD19 antigen12, and performed RNA-sequencing-mediated transcriptome analysis (Supplementary Table 5a,b). After anti-CD19-specific CAR stimulation, CAR T cells from CR and PRTD patients exhibited a significantly more robust increase in T cell activation genes (mean 49-fold induction) than did CAR T cells from PR and NR patients (mean 16-fold induction; Fig. 3d). As another parameter of how T cell intrinsic fitness may be associated with therapeutic efficacy, we evaluated the profiles of cytokines and chemokines produced from CTL019 cells derived from CR, PRTD, PR and NR patient infusion products after CAR stimulation. CD19-directed T cells generated from CR and PRTD subjects, compared with PR and NR subjects showed higher levels of STAT3 signaling mediators and targets, including IL-6, IL-17, IL-22, IL-31 and CCL20, a finding consistent with IL-6/STAT3-pathway upregulation in evaluable CR and PRTD patient CTL019 cells that were CAR stimulated (Fig. 3f). These findings suggest that activation of STAT3 in CAR T cells might be involved in the generation of potent, less-differentiated T lymphocytes15·18·19.
We then investigated whether levels of phosphorylated STAT3 (pSTAT3) could segregate highly active CR/PRTD patient CAR T cells from those of PR/NR subjects with poor functionality. CAR-specific pSTAT3 activity was significantly elevated in CTL019 cells from CR/ PRTD patients compared with CAR T cells expanded from PR/NR patients (Fig. 3g). In line with these findings, we observed a strong direct correlation between the maximum degree of in vivo CAR T cell expansion and serum IL-6 as well as IL-6/STAT3-pathway gene enrichment in preinfusion CTL019 cells after stimulation (Fig. 3h). Accordingly, pharmacologic blockade of STAT3 signaling in CTL019 cells diminished their proliferative capacity after serial re-stimulation with CD19-expressing leukemia cells (Supplementary Fig. 4a-c) without affecting viability (Supplementary Fig. 4b,c). Because STAT3 signaling plays a role in the generation and maintenance of memory T cells20, induction of this pathway (Fig. 3f,g) not only might indicate a demarcation of less differentiated cells in the CAR T cell product but also may be functionally important for their expansion and long-term survival after infusion.
Because CAR T cells from CR/PRTD patients expressed key regulators of cellular memory differentiation and maintenance, we explored whether the transcriptomes of premanufactured T cells from these patients differed from those of subjects exhibiting limited or very poor CAR T cell activity. Similarly to the results of our transcriptional analyses of CAR T cell infusion products, transcriptomic profiles of ex vivo T cells from CR/PRTD patients showed marked differences compared with those from nonresponders (Supplementary Fig. 5a and Supplementary Table 5c). The genes significantly upregulated in unmanipulated T cells from NR patients were enriched in the functions of T cell exhaustion, activation, glycolysis and apoptosis (Supplementary Fig. 5b and Supplementary Table 5c). These additional findings provide a framework that may inform future clinical trials in which patients most likely to respond to CAR T cell treatment could be selected before a somewhat costly and complex cellular manufacturing process.
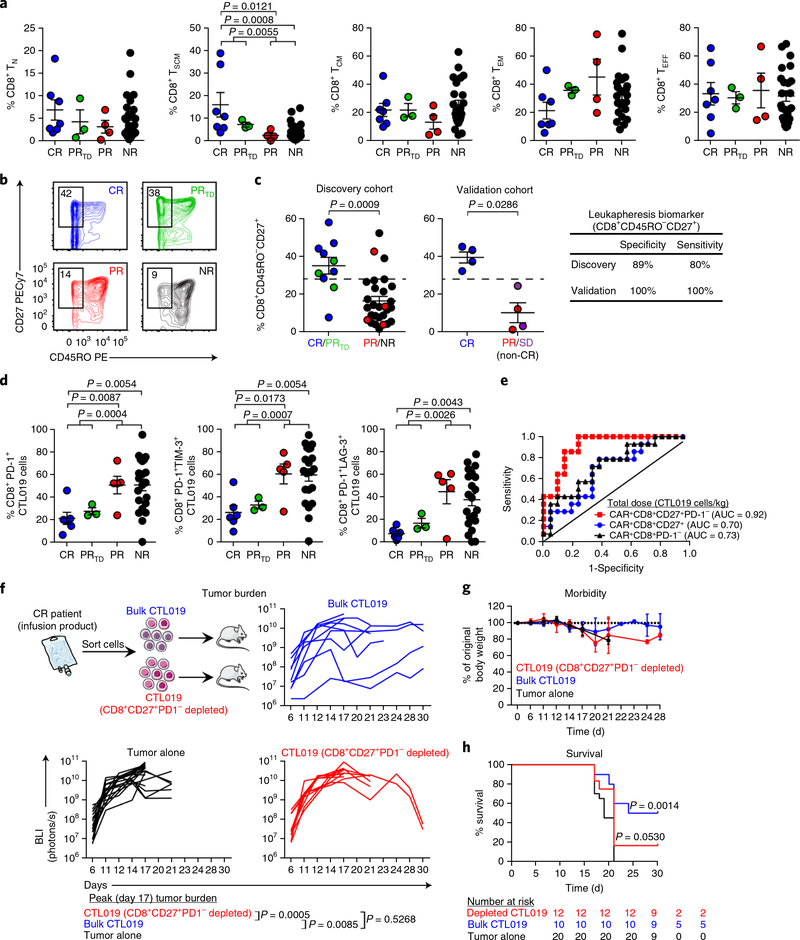
According to the results of our transcriptomic analyses, the presence of specific memory populations at the preexpansion stage of cellular therapy may be a predictive indicator of clinical response. This indicator might be used to inform efforts to enrich the most efficacious starting subset of T cells for large-scale clinical expansion. We therefore investigated whether, before ex vivo expansion, T cells collected by leukapheresis from patients with CLL who responded to CTL019 differed from nonresponders in the composition of naive, stem cell memory, central memory, effector memory or effector CD4+ and CD8+ subsets15·21. The median frequency of stem cell memory T cells was modestly higher in CR/PRTD than PR/NR patient groups (Fig. 4a). Notably, supervised analysis showed that the frequency of CD45ROCD27+CD8+ T cells from CR/PRTD patients at the time of leukapheresis was significantly higher than that in PR/NR patients (Fig. 4b,c). To confirm this finding, we performed unbiased computational analysis of flow cytometric data. Our initial approach was based on an unsupervised clustering algorithm (cluster identification, characterization and regression (CITRUS)) that is agnostic to population gating thresholds and instead uses the expression levels of all phenotypic markers to identify clinically prognostic cell subsets22. Two clusters, 48223 and 48236, were identified during the CITRUS analysis as relevant biologic segregators of CR/PRTD from PR/NR patient T cells (Supplementary Fig. 6a). Cells in both clusters expressed high levels of CD27 and low levels of CD45RO (Supplementary Fig. 6a). We found a population with a very similar predictive phenotype when comparing extreme- response CR/NR patient groups (Supplementary Fig. 6b).
Fig. 4|. The presence of mechanistically relevant T cell populations in patients can predict response to CTL019 therapy.
a, Frequencies of naive (TN), stem cell memory (TSCM), central memory (TCM), effector memory (TEM) and effector (TEFF) CD8+ T cells from patients in each response group (CR, n = 7; PRtd, n = 3; PR, n = 4; NR, n = 24) at the time of leukapheresis. b, Representative contour plots examining the proportions of CD45RO-CD27+CD8+ T cells from CR, PRtd, PR and NR CTL019 patients (3 independent experiments). c, Summary of leukapheresis CD27+CD45RO-CD8+ T cell frequencies in patients belonging to each response category in an original discovery cohort (left). A cutoff value of 28.6% (represented by the dashed lines) optimally segregated patients with highly functional (CR/PRtd; n = 10) or poorly functional (PR/NR; n = 28) CAR T cells (positive predictive value, 72.73%; negative predictive value, 92.59%). A CD45RO-CD27+CD8+ T cell frequency >28.6% segregated CR (n = 4) from non-CR (SD (stable disease) + PR; n = 4) patients with CLL in a validation cohort (middle). Clinical-response assessments for the validation cohort were conducted 3 months after CTL019 cell infusion and are based on tumor-burden status in the lymph nodes and spleen. Specificity and sensitivity measurements for each patient group are displayed in tabular form (right). d, Percentages of CD8+ T cells expressing PD-1 and coexpressing PD-1 and LAG-3 or TIM-3 in preinfusion CTL019 cells (CR, n = 6; PRtd, n = 3; PR, n = 5; NR, n = 21). For a, c and d, two-tailed Mann-Whitney tests were used for comparisons between groups. Graphs show mean and s.e.m. e, Receiver operating characteristic (ROC) curve for multiple biomarkers that define different T cell populations in the CAR T cellular product. ROC curves are based on the total dose (cells/kg) of CTL019 cells infused into CR (n = 6) versus NR patients (n = 21). AUC values, shown in parentheses, represent areas under the respective ROC curves and provide an overall measure of predictive power. f, Top, experimental setup, showing transfer of retrospective CR CAR T cell infusion products with (bulk) or without CD8+CD27+PD-1- lymphocytes (depleted) from patients with CLL into fully leukemic NSG mice (tumor alone, n = 20; bulk CTL019, n = 10; depleted CTL019, n = 12). Bottom, longitudinal tumor burden (BLI) of NALM-6-bearing mice treated with CAR T cells (each line indicates one mouse). g, Weight loss in mice, shown over time. h, Kaplan-Meier curves showing survival of mice. P values were calculated with twotailed log-rank Mantel-Cox tests. Numbers of mice at risk in each group at specific time points are listed below the survival curve.
To further substantiate the validity of this biomarker profile, we applied a second computational approach by performing logical transformation of flow cytometric data and subsequent gate determination for each marker to effectively separate positive and negative cell populations. All possible biomarker profiles were then extracted with the flowType algorithm23 to identify T cell phenotypes associated with a significant difference between patient response categories. Using this conceptually different approach, we identified several CD8+ T cell subsets with phenotypes concordant with those discovered during the CITRUS analysis, including many consistently defined by parental CD45RO- and CD27+ profiles that segregated CR from NR patients (Supplementary Table 6a). In contrast, we did not identify a differentiation phenotype that stratified CR from NR patients in leukapheresed CD4+ T cells by using this computational method (Supplementary Table 6b). A second independent cohort of eight patients with advanced CLL treated with CTL019 cells was analyzed for CD45ROCD27+CD8+ T cell frequency at the time of leukapheresis, and this biomarker identified CR patients with a high degree of sensitivity and specificity (Fig. 4c).
The CD45ROCD27+ subset of CD8+ T cells is composed of non-naive, antigen-experienced lymphocytes that persist in a resting state and possess properties of long-lived memory cells24. Furthermore, CD45RO-cD27+CD8+ T cells represent a small proportion of both total and antigen-specific CD8+ T cells that rapidly expand and acquire effector functions after antigen reexposure24–27. The levels of CCR7 were significantly lower on these cells than on both naive and central memory T cells, but were significantly higher than those on effector memory T cells (Supplementary Fig. 7a). We also examined the proliferative capacity and differentiation phenotype of purified CD45RO-CD27+CD8+ T cells over time, after stimulation with anti-CD3/CD28 agonistic antibodies. Similarly to conventional memory T cells, CD45RO-CD27+CD8+ T lymphocytes divided extensively (Supplementary Fig. 7b) and maintained a less differentiated phenotype in extended culture, even after this potent stimulus (Supplementary Fig. 7c). These cells also expressed low levels of IL-2 after acute stimulation (Supplementary Fig. 7d), a result consistent with previous reports on the ex vivo functionality of early memory T cells15,24. We then selected CD45ROCD27+CD8+ T lymphocytes for the generation of CAR-engineered T cells and demonstrated that they potently lysed CD19-expressing tumor targets. This cytotoxic effector capacity was similar to that mediated by gene-modified central memory T cells (ref. 28 and Supplementary Fig. 7e). In further support of this cell type persisting as a stable quiescent pool of memory T cells with phenotypic characteristics of both memory and effector cells24, we observed elevated levels of granzyme B (Supplementary Fig. 7f) and a lower frequency of Ki-67-expressing cells (an indicator of cycling status; Supplementary Fig. 7g) in ex vivo CD45RO-CD27+CD8+ patient T cell populations compared with their CD45RO+CD27+ counterparts. Thus, enriched CD45ROCD27+CD8+ T cells at the time of leukapheresis are associated with complete and durable responses to CTL019 therapy for CLL, a finding consistent with less differentiated T cells mediating robust antitumor activity as a result of enhanced proliferation and persistence after adoptive transfer13–15.
Additional makers such as the CD4+/CD8+ ratio10,16 of the infused cells and the differential expression of inhibitory molecules16 can affect both the survival of CAR T cells and their effector function in vivo. The typical compositional characteristics of CLL patient leukapheresis samples and CTL019 cellular infusion products as well as the total CTL019 dose infused did not segregate subjects according to clinical outcome (Supplementary Table 7). NR patients did exhibit a significantly higher CD4+/CD8+ T cell ratio after large- scale expansion of their CTL019 cells, relative to the leukapheresis time point (Supplementary Fig. 8). However, the CD4+/CD8+ T cell ratio of the CAR T cell infusion product did not differ among CR, PRTD, PR and NR subjects, thus suggesting that the CD4+/CD8+ T cell ratio within CTL019 products may not influence therapeutic efficacy in the setting of CLL (Supplementary Fig. 8). Additional clinical studies of CAR T cells in CLL with larger numbers of patients will be required to independently validate these findings.
We also examined other phenotypes that might be associated with CAR T cell activation differentiation and exhaustion, including PD-1, TIM-3 and LAG-3, all of which can function as inhibitory receptors on CD8+ T cells16·29–35. The frequencies of preinfusion CD8+ CTL019 cells expressing PD-1, TIM-3 and LAG-3 varied widely across the entire population of treated patients with CLL (Fig. 4d). CR patients, compared with PR and NR patients, had significantly lower percentages of CAR T cells with a CD8+PD-1+ phenotype (Fig. 4d). Analysis of PD-1 coexpression with LAG-3 and TIM-3 revealed that both CD8+LAG-3+ and CD8+TIM-3+ CTL019 cells coexpressing PD-1 were associated with poor responses, whereas individuals who had complete and durable remissions were infused with products containing significantly lower frequencies of these cells (Fig. 4d). Interestingly, the percentages of CD8+ T cells expressing these inhibitory receptors at the time of leuka-pheresis did not correlate with clinical outcome (Supplementary Fig. 9), thus suggesting that changes in these inhibitory phenotypes across response groups may be an activation-driven phenomenon. Because most T cells expressed CD45RO after our expansion process (data not shown), we conducted unbiased computational analysis of flow cytometry data from CTL019 infusion products to identify cell populations predictive of clinical response at the end of the cellular manufacturing process. Evaluation of 1,696 phenotypes identified the PD1CD27+ parental population of CD8+ T cells as a phenotype that strongly segregated CR from NR patients (Supplementary Table 8). In accordance with previous data17 and our collective findings, we noted a highly significant association between the likelihood of responding to CTL019 therapy and the infusion of high doses of CD27+PD-1CD8+ CAR T cells (Fig. 4e and Supplementary Fig. 10).
To determine whether the CD27+PD-1- subset was responsible for mediating tumor control, we treated fully leukemic NSG mice with retrospective CR patient CAR T cells that contained enriched or depleted CD27+PD-1CD8+ T cell populations. Removal of this population resulted in a loss of tumor control in mice, as compared with treatment with CR patient infusion products containing high frequencies and absolute numbers of CD27+PD-1CD8+ CAR T cells (Fig. 4f). The magnitude of these antitumor responses was directly associated with the degree of morbidity in these animals (Fig. 4g) as well as the overall length of their survival (Fig. 4h). Mechanistically, we found that the CD27+PD-1CD8+ subset comprised the population that upregulated pSTAT3 in response to IL-6 stimulation (Supplementary Fig. 11a), as a result of their markedly high levels of the IL-6 receptor-β chain (Supplementary Fig. 11b). Therefore, the effectiveness of CAR T cell therapy for CLL may be increased by treating patients with cellular products enriched in CD27+PD-1CD8+ cells.
We also investigated blood-based biomarkers in the setting of predicting clinical response. The observed rise in IL-15 that peaked within the first 28 d after CTL019 infusion segregated NR patients from CR (P < 0.05) and PRTD (P = 0.01) patients (Supplementary Table 9), thus suggesting that adoptively transferred genetically redirected T cells that exhibit robust expansion in vivo are responsive to homeostatic cytokines. In contrast, IL-2 levels were not significantly different across response groups (Supplementary Table 9). The higher levels of serum IL-6 seen in CR and PRTD patients compared with NR patients (P < 0.05 and P = 0.010, respectively; Supplementary Table 9) may be indicative of the superior potency and persistence of CTL019 cells in subjects who have more durable responses to this therapy, a possibility supported by the infusion of CD27+PD-1CD8+ cAr T cells that express genes and activate pathways such as STAT3 and consequently confer long-term cell persistence and durable clinical remission.
In summary, we explored the integrated genomic and phenotypic characteristics as well as the functional determinants and mechanisms associated with clinical response and resistance to CAR T cell therapy for CLL, providing evidence that intrinsic T cell fitness mediates long-term CR. Tumor burden and disease- or patient- specific characteristics alone are not sufficient to distinguish clinical outcomes and predict response. The specific baseline T cell qualities reported here, however, make it possible to delineate classes of predicted treatment outcome. A pretreatment biomarker is valuable not only in helping to prognosticate responses but also in predicting immunological failure and tailoring alternative therapies. It will be important to test whether the biomarker-driven approaches to CAR T cell therapy proposed here can be extended to other cancer types, particularly solid tumors. Our findings also highlight the utility of selecting cultures with high absolute numbers of a mechanistically relevant subpopulation responsible for mediating tumor control and consequently increasing the efficacy of genetically redirected T lymphocytes. Generation of CAR T cells with optimal differentiation potential as well as effector activity might also be achieved by minimizing culture time, using alternative cytokines or performing metabolic engineering or other approaches, such as IL-6/STAT3-pathway modulation. Finally, the ability to use biomarkers to identify the individuals most likely to respond should dramatically change patient selection and provide critical information that may allow immune modification to improve the efficacy of this approach.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41591-018-0010-1.
Methods
Patient samples.
Samples were acquired from patients with CLL who enrolled in clinical trials of single-agent CTL019 therapy, which were approved by the Institutional Review Board (IRB) of the University of Pennsylvania. All subjects provided written informed consent according to the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice. All ethical regulations were followed. These studies have been registered at ClinicalTrials.gov (identifiers NCT01029366, NCT01747486 and NCT02640209).
Vector production, T cell isolation and generation of CTL019 cells.
A lentiviral vector containing a transgene encoding the CD19-specific CAR with 4–1BB/CD3ζ domains (GeMCRIS 0607–793) was constructed and produced as previously described6,36. Autologous T cells were collected by leukapheresis and activated with anti-CD3 and anti-CD28 monoclonal-antibody-coated polystyrene beads, then transduced with the above lentiviral vector5,37. CAR T cell expansion was carried out for 9–11 d (ref. 6). Absolute cell counts during large-scale CTL019 cell culture were obtained with a Coulter counter (Beckman Coulter). The following equation was used to calculate the number of cellular population doublings during culture: At=A02n, where n is the number of doublings, A0 is the number of cells seeded, and At is the total cell number.
Correlative assays.
Pre- and postinfusion sample processing, flow cytometry, serum cytokine quantification and qPCR analyses were carried out as previously described7. These correlative studies were performed at protocol-defined time points in conjunction with disease response assessments as part of completed and ongoing clinical trials (NCT01029366, NCT01747486 and NCT02640209)6.
Cell lines.
NALM-6, K562 and IMR-90 cell lines were acquired from the American Type Culture Collection (ATCC). Low-passage working banks of cells were tested for mycoplasma with a MycoAlert kit (Lonza), according to the manufacturer’s instructions. Cell-line authentication was performed by the University of Arizona Genetics Core, on the basis of criteria established by the International Cell Line Authentication Committee. Short-tandem-repeat profiling revealed that these cell lines were above the 80% match threshold. Mycoplasma testing and authentication were routinely performed before and after molecular engineering.
Mouse systemic tumor model.
Mouse experiments were carried out with equal numbers of 8- to 12-week-old male and female NOD/SCID/IL-2Ry-null (NSG) mice (Jackson Laboratory), under an Institutional Animal Care and Use Committee protocol approved by the University of Pennsylvania. Mice were engrafted with 1 X 106 click beetle green luciferase-green fluorescent protein (CBG-GFP)-expressing NALM-6 leukemia cells through intravenous injection, then with 1 X106 CAR T cells 7 d later. Tumor burdens are very even with this cell line, and therefore, no mice were excluded before treatment. Imaging of animals and survival measurements were carried out by an operator who was blinded to treatment groups. For some experiments, mice were treated with bulk, unmanipulated retrospective CTL019 infusion products from patients with CLL. Depletion experiments were carried out by sorting bulk infusion products and matched samples with CD27+PD-1- cells removed from the CD8 compartment with a FACSAria II cell sorter (BD). Longitudinal bioluminescence imaging was performed with a Xenogen IVIS Imaging System, and absolute cell counts as well as morbidity assessments were carried out as previously described38,39.
RNA-sequencing and bioinformatics analysis.
Total RNA was purified from CTL019 cells or bulk CD3+ T cells (leukapheresis samples) with an RNeasy Mini Kit according to the manufacturers instructions (Qiagen), and integrity was verified with an Agilent TapeStation (RIN). Preparation for sequencing was then performed with a TruSeq RNA v2 prep kit (Illumina). To identify, accurately quantify and determine differential gene and transcript expression, we performed high-throughput sequencing on an Illumina HiSeq 2500 platform to a target depth of 50 million paired-end reads per sample. Reads were aligned to the reference human genome (build hg19) in STAR40. Next, mapped reads were assembled into transcripts, and transcript abundance was estimated and normalized in Cufflinks41. Finally, HTSeq was used to count the number of reads mapping to each gene42. Data normalization for differential expression analysis was carried out with the edgeR R package43. Differential gene expression between CR/PRTD and PR/NR patients was performed with the edgeR R package for fitting generalized linear models with a design matrix. Dispersions were estimated with the Cox-Reid profile-adjusted likelihood method, and differential expression was determined with a generalized-linear-model likelihood-ratio test. The gene signatures used were as follows: T cell differentiation15,44, T cell exhaustion45,46, T cell activation47–49, hypoxia50 and IL-6/STAT3 (ref. 51). For metabolic gene profiles, ‘CP:REACTOME: Reactome gene sets’ and ‘GO biological process sets’ were also selected: http://www.broadinstitute.org/gsea/msigdb/.
Flow cytometry.
Routine longitudinal measurements of the expansion and persistence of CTL019 cells, as well as peripheral B-CLL burden, were conducted as previously described with a six-parameter Accuri C6 flow cytometer (BD)5,6. For T cell deep immunophenotyping, PBMC or CTL019 infusion products (bulk T cells or specific subsets purified by flow cytometry) were preincubated with Aqua Blue dead cell exclusion dye (Invitrogen) and subsequently surface stained with commercially available flow cytometry antibodies. CAR19 protein expression was detected with an Alexa Fluor 647-conjugated anti-idiotype monoclonal antibody52. All antibodies were titrated before use, and fluorescence-minus-one controls were created for each antibody panel to set gates for positive events. Information pertaining to all flow cytometry antibodies used in this study can be found in Supplementary Table 10. Cells were acquired on a custom 17-color, 19-parameter special-order LSRFortessa instrument (BD). Data were analyzed in FlowJo software (TreeStar).
Measurement of lactate production and glucose uptake by CAR T cells from patients with CLL.
Retrospective CTL019 cells from patients with CLL were thawed and rested in 24-well tissue culture plates (BD) at 1 X106 cells/mL, then stimulated overnight with anti-idiotypic antibody-coated beads to trigger the anti-CD19BBζ CAR38. Similarly conjugated isotype antibody-coated beads were used as the mock control. Beads were added in a ratio of three beads per cell according to the transduction efficiency of each patient infusion product. Lactate levels were measured with an Enspire Multiparameter Bioanalytical System (Model 2300, PerkinElmer). Enzymatic diagnostic kits (Sigma) were used according to the manufacturer’s instructions. The signals generated from a range of concentrations of this metabolite were used to construct a standard curve. Cellular supernatants from mock- and CAR-stimulated T cells were collected and applied to the instrument.
After overnight stimulation, cells were washed and refed with fresh RPMI, and 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose (2-NBDG; Sigma Aldrich) was added at a final concentration of 500 μM. After a 30-min incubation with 2-NBDG, cells were placed on ice and stained for flow cytometry to detect CAR+ T cells that had taken up the fluorescently labeled glucose analog.
Metabolic modulation of CAR T cells.
Anti-CD19BBζ CAR T cells were produced as previously described39 in the presence or absence of the glycolysis inhibitor 2-deoxyglucose (2-DG; Sigma-Aldrich). After 9 d of culture, the differentiation phenotype of these cells was determined by flow cytometry. CAR+ T cells were sorted on a FACSAria II cell sorter (BD) and combined 1:1 with irradiated K562 cells expressing CD19 (K562-CD19)6. CTL019 cells were restimulated three times with K562-CD19 targets. During the proliferation assay, absolute cell counts were obtained with a Luna automated cell counter (Logos Biosystems).
Cytokine analyses.
CTL019 products were cultured overnight with anti-idiotypic antibody-coated beads or isotype-control antibody-coated beads as described above, and supernatants were collected. Serum was isolated through centrifugation of the whole blood of patients at baseline and at defined time points after CTL019 treatment. Serum samples were then divided into aliquots and stored at −80 °C. Measurement of cytokines in the above culture supernatants and serum samples was performed with a Luminex bead array platform (Life Technologies) according to the manufacturer’s instructions. All samples were analyzed in triplicate and compared against multiple internal standards with a nine-point standard curve. Data were acquired on a FlexMAP-3D system (Luminex), and analysis was performed in XPonent 4.0 software (Luminex) as well as through five-parameter logistic regression7.
Measurement of pSTAT3 and STAT3 blockade in CAR T cells.
CTL019 cells from infusion products of patients with CLL were thawed and stimulated with anti-idiotypic antibody-coated beads or mock beads as described above. For some experiments, cells were stimulated for 10 min with recombinant human IL-6 (Miltenyi) at a final concentration of 10 ng/mL. In cell-subset-evaluation experiments, populations of interest were stained for surface markers and sorted on a FACSAria II (BD) cell sorter before acute stimulation with IL-6. After stimulation, cells were fixed with Phosflow Lyse/Fix Buffer (BD) for 12 min at 37 °C, then permeabilized with Phosflow Perm Buffer III (BD). Intracellular staining was carried out for 60 min at room temperature with an anti-STAT3 (pY705) antibody (BD). Samples were immediately acquired on an LSRFortessa instrument (BD).
Blockade of the STAT3 pathway was carried out through generation of anti- CD19BBζ CAR T cells39 in the presence or absence of the STAT3-specific inhibitor Stattic (Selleckchem) at a final concentration of 5 pM. An equivalent amount of dimethyl sulfoxide (DMSO) served as a vehicle-alone negative control. The ability of Stattic to inhibit STAT3 signaling was assessed through Phosflow staining as described above. Serial restimulation assays using sorted CAR T cells and irradiated K562-CD19 cells were also carried as described above. Absolute cell numbers and viability were simultaneously measured during the course of CAR T cell expansion with a Luna automated cell counter (Logos Biosystems).
Computational analyses of flow cytometric data.
We used two conceptually different data-driven approaches for biomarker identification. The aim was to identify cellular phenotypes that correlated with clinical response to CTL019 therapy. Pregating was first performed to limit analyses to viable singlet populations of lymphocytes. Pregated data were subsequently used for data mining and biomarker identification with the automated biostatistical algorithms CITRUS22 (Cytobank) and flowType23 (based in R+; The R Foundation). CITRUS is agnostic to preestablished gating thresholds and instead uses the expression levels of all phenotypic makers, beginning with unsupervised identification of subsets of phenotypically similar cells. These clusters are then hierarchically ordered by clustering cell events that express similar makers. This algorithm is based on the assumption that clinically predictive cell populations of a given phenotype will be identified as events that robustly recur in the aggregate data22. The CITRUS algorithm conservatively dictates by default that T cell populations of interest must comprise at least 5% of measured events22.
FlowType uses a clustering algorithm to partition every channel/marker density to separate a positive and a negative cell population; this procedure is based on the assumption that the there are two distinct populations23. These partitions are then combined to produce a set of multidimensional phenotypes. This algorithm generates a total number of 3 × n possible phenotypes, where n represents the number of phenotypic markers. In this study, we limited the length of any given phenotype to be defined by a maximum of four markers to allow for the identification of biologically meaningful phenotypes. We chose this number because too many markers would extract populations consisting of very low cell counts, thus rendering their predictive relevance difficult to interpret in our patient cohorts. These processes allowed us to examine many possible phenotypes collectively in leukapheresis (premanufactured) and CAR T cell infusion product (postmanufactured) samples. The most statistically significant phenotypes were selected for manual confirmation that was carried out using FlowJo software (TreeStar).
Telomere-length and telomerase-activity assessments.
CTL019 cells or IMR- 90 cells were thawed and subjected to genomic DNA isolation with an iPrep Purification Instrument (Invitrogen). For Southern blot analysis, 2 μg genomic DNA was digested overnight at 37 °C with the restriction enzymes HinfI and RsaI. Digested DNA samples were separated on a 0.8% agarose gel under 50 V at 4 °C for 17 h. After electrophoresis, Southern blot procedures were performed with a TeloTAGGG Telomere Length Assay kit (Roche). After depurination in 0.4% hydrochloric acid for 15 min at room temperature, the DNA gel was rinsed for 30 min in denaturation buffer containing 0.5 M sodium hydroxide and 1.5 M sodium chloride. Separated DNA fragments were subsequently blotted onto a nylon membrane through an overnight capillary-transfer procedure with 20X saline sodium citrate buffer at room temperature. After blotting, the nylon membrane was fixed, washed and probed according to the manufacturer’s instructions (Roche). Finally, telomeres were visualized by autoradiography. The sizes of the telomeres were determined by densitometry analysis.
Measurement of relative telomere length by qPCR was carried out as previously described53. Briefly, two sets of PCR reactions were performed in triplicate, and each reaction contained 10 ng of the genomic-DNA template. The first set amplified telomere-repeat DNA (T reaction), and the second amplified a singlecopy gene (36B4; S reaction). The PCRs were performed with a ViiA 7 Real-Time PCR System, and the accumulation of amplified DNA was detected through SYBR green fluorescence (Applied Biosystems). Each plate contained dilutions of a reference sample of genomic DNA to generate a standard curve for determining the relationship between threshold cycle (Ct) values and input genomic-DNA mass for T and S reactions. Test samples were normalized to the standard curve to generate relative T and S values and T/S ratios.
Telomerase activity was measured in CTL019 cellular infusion products unstimulated or stimulated with CD3/CD28-coated beads (Dynabeads; ThermoFisher Scientific) compared with a bead-bound anti-idiotypic antibody that recapitulates triggering of the anti-CD19BBζ CAR by CD19 (ref. 38). This assay was based on a SYBR green quantitative telomeric-repeat amplification protocol (Q-TRAP)54,55 in which telomeric-repeat sequences are added to the 3’ end of an oligonucleotide. Briefly, T cells were collected and lysed as previously described54,55, and total protein concentrations were measured with a BCA protein kit (Pierce) according to the manufacturer’s instructions. Telomerase-positive 293 human embryonic kidney cells were used as a standard. Telomerase-mediated extension of an oligonucleotide primer by the cell extracts and amplification of the telomerase- extended primer products was carried out as described above. Ct values for each sample were determined from amplification plots (i.e., log increase in fluorescence versus cycle number) and compared against standard curves generated from the serially diluted 293 cell lysates.
Functional evaluations of the CD27+CD45ROCD8+ T cell subset.
Primary bulk human T cells were sorted into subsets on the basis of expression of CD27 and CD45RO, with a FACSAria II cell sorter (BD). Cells were stimulated with anti- CD3/CD28 antibody- or isotype-control antibody-coated beads for functional assessments of proliferation through carboxyfluorescein succinimidyl ester (CFSE) dilution and cytokine production, as previously described39. CTL019 cells were generated from cell subsets, and cytolytic capacity was determined with a luciferase-based assay incorporating NALM-6 (CBG-GFP) tumor targets39
Statistical analyses.
Descriptive statistics were computed for study variables. Normality was assessed for all data with the DAgostino-Pearson omnibus test. When the sample size was too small to adequately examine normality, nonparametric statistics were used. Specific statistical tests used for different study variables are described below and in the figure legends. With the sample sizes available in each response group (n = 11 for CR/PRTD and n = 30 for PR/NR), we had 80% power to detect a minimum effect size of 1.0 (in s.d. units) using a two-sided unpaired t test, and 74% power to detect the same effect size if the nonparametric Mann-Whitney test was used. Although we recognize that this effect size is large, and we do not have prior data available to determine whether such an effect size is biologically meaningful, the results of the current study may be useful for planning future confirmatory investigations.
The current study is a secondary correlative investigation using samples collected from existing clinical trials. Thus, the sample sizes in this report were determined by the original clinical trial designs and sample availability; no additional inclusion/exclusion criteria were applied. The clinical trials were singletreatment studies; the comparison groups in the current study were defined by the observed clinical responses. Investigators were blinded to clinical responses, because correlative assays were conducted on deidentified subject samples. Estimates of variation within each group of data are presented with error bars in figures indicating either the s.d. or s.e.m. Descriptive statistics on the measures of central tendency and variability of the data distribution, including the minimum, twenty-fifth percentile, median, seventy-fifth percentile, maximum, mean, s.d. and s.e.m., are listed for the appropriate figures in Supplementary Table 11. In cases in which the distribution of data was normal, parametric tests (for example, Welch’s t test) were used to accommodate the possibility of unequal variance.
The significance of associations among the four response groups (CR, PRtd, PR and NR) and characteristics listed in Supplementary Tables 1, 7 and 9 were examined using Fisher’s exact test for categorical variables or the nonparametric Kruskal-Wallis exact test for continuous variables. The MannWhitney exact test was used for comparisons between two individual or combined response groups. Assessments of differential gene expression/enrichment and T cell functional markers were compared with a t test for paired or unpaired samples. CTL019 expansion levels were measured through PCR-based detection of transferred cells above the lower detection limit (25 copies^g DNA). Peak (that is, maximum) levels of expansion, in vitro CTL019 expansion, telomere length, cytokine/chemokine production by stimulated CAR T cells, T cell differentiation and inhibitory phenotypes as well as serum cytokine levels in patients were compared with the Mann-Whitney exact test between two individual or combined (for example, CR/PRTD versus PR/NR) response groups. To measure the predictive power of each phenotype identified through computational approaches, we used a t test for unpaired samples to examine differences in cell frequencies (i.e., the number of cells of a particular phenotype divided by the total number of cells in the parent population) between clinical response groups. ROC curves based on different T cell populations from patients with CLL, alone or in combination, were generated; the predictive values were then evaluated through examination of the AUC. In the case of the CD45RO-CD27+CD8+ T cell leukapheresis biomarker, the optimal cutoff frequency satisfied the criteria maximizing the Youden index as well as the shortest distance to top left corner (0, 1) of the ROC curve (i.e., perfect classification). A Spearman’s rho (ρ) test was used to perform the nonparametric correlations presented in main and supplementary figures. A Wilcoxon sign-rank test was used for evaluating significant differences in the CD4/CD8 ratio between leukapheresis samples and patient-matched CTL019 products. Descriptions of statistical tests for all other data can be found in the figure legends.
Analyses were performed with SAS (SAS Institute Inc.), Stata 14.0 (StataCorp) or GraphPad Prism 6 (GraphPad Software). All tests were two sided. A P value <0.05 was considered statistically significant. The present study was exploratory, given its post hoc nature. When appropriate, exact unadjusted and adjusted P values are reported in figures and/or in Supplementary Table 11. The Holm-Sídák method was used to compute adjusted P values when controlling for multiple comparisons.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary.
Data availability.
Patient-related data not included in the paper were generated as part of completed or ongoing clinical trials and may be subject to patient confidentiality. Therefore, these data may be restricted. With regard to RNA- sequencing data, counts of sequencing reads are presented in the supplementary information. All associated raw data files have been submitted to the database of Genotypes and Phenotypes (dbGaP; accession numbers pending). Any other data that support the findings of the study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank the patients for their participation in the clinical trials from which research samples were obtained. We also acknowledge A. Fesnak, A. Lamontagne, A. Malykhin, C. Corl, Y. Ohayon and other members of the Clinical Cell and Vaccine Production Facility for cell manufacturing and testing. In addition, we are grateful to V. Gonzalez, J. Finklestein, F. Nazimuddin, J.-M. Navenot, M. Bogush, Y. Tanner, N. Kengle, K. Marcucci, A. Chew, C. Pletcher, P. Hallberg and R. Schretzenmair for contributions to correlative studies and/or other research support. D. Campana, C. Imai and others at St. Jude Children’s Research Hospital designed, developed and provided, under materialtransfer agreements, the CAR used in this study. B. Jena and L. Cooper (MD Anderson Cancer Center) are acknowledged for providing the CAR anti-idiotype detection reagent. The functional anti-idiotypic antibody that was used for in vitro CAR stimulation experiments was a kind gift from Novartis Pharmaceutical Corporation. This work was supported by funding from NCI T32CA009140 (J.A.F.) R01CA165206 (D.L.P. and C.H.J.), P01CA214278 (C.H.J), a Stand Up to Cancer Phillip A. Sharp Innovation in Collaboration Award (C.H.J) and Novartis.
Footnotes
Competing interests
J.A.F., S.F.L., F.B., R.M.Y., B.L.L., J.L.B., D.L.P., C.H.J. and J.J.M. hold patents related to CTL019 cell therapy. These authors declare no additional interests. The remaining authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41591–018-0010–1.
References
- 1.Makkouk A & Weiner GJ Cancer immunotherapy and breaking immune tolerance: new approaches to an old challenge. Cancer Res. 75, 5–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vesely MD, Kershaw MH, Schreiber RD & Smyth MJ Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol 29, 235–271 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Brown JR et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110delta, for relapsed/refractory chronic lymphocytic leukemia. Blood 123, 3390–3397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain P et al. Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood 125, 2062–2067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalos M et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med 3, 95ra73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter DL et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med 7, 303ra139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maude SL et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turtle CJ et al. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor- modified T cells. Sci. Transl. Med 8, 355ra116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee DW et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose- escalation trial. Lancet 385, 517–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turtle CJ et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest 126, 2123–2138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kochenderfer JN et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J.Clin. Oncol 33, 540–549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawalekar OU et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 712 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Gattinoni L et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest 115, 1616–1626 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinrichs CS et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc. Natl. Acad. Sci. USA 106, 17469–17474 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gattinoni L et al. A human memory T cell subset with stem cell-like properties. Nat. Med 17, 1290–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radvanyi LG et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin. Cancer Res 18, 6758–6770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg SA et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirth TC et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity 33, 128–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betz UA & Müller W Regulated expression of gp130 and IL-6 receptor alpha chain in T cell maturation and activation. Int. Immunol. 10, 1175–1184 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Siegel AM et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35, 806–818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lugli E et al. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat. Protoc 8, 33–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ & Nolan GP Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci (7SA 111, E2770–E2777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aghaeepour N et al. Early immunologic correlates of HIV protection can be identified from computational analysis of complex multivariate T-cell flow cytometry assays. Bioinformatics 28, 1009–1016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lécuroux C et al. Identification of a particular HIV-specific CD8+ T-cell subset with a CD27+ CD45RO-/RA+ phenotype and memory characteristics after initiation of HAART during acute primary HIV infection. Blood 113, 3209–3217 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Dunne PJ et al. Epstein-Barr virus-specific CD8+ T cells that re-express CD45RA are apoptosis-resistant memory cells that retain replicative potential. Blood 100, 933–940 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Rufer N et al. Ex vivo characterization of human CD8+ T subsets with distinct replicative history and partial effector functions. Blood 102, 1779–1787 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Precopio ML et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med 204, 1405–1416 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kueberuwa G et al. CCR7+ selected gene-modified T cells maintain a central memory phenotype and display enhanced persistence in peripheral blood in vivo. J. Immunother. Cancer 5, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones RB et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J. Exp. Med 205, 2763–2779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber DL et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Blackburn SD et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmadzadeh M et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114, 1537–1544 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fourcade J et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med 207, 2175–2186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grosso JF et al. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J. Clin. Invest 117, 3383–3392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grosso JF et al. Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J. Immunol 182, 6659–6669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milone MC et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther 17, 1453–1464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laport GG et al. Adoptive transfer of costimulated T cells induces lymphocytosis in patients with relapsed/refractory non-Hodgkin lymphoma following CD34+-selected hematopoietic cell transplantation. Blood 102, 2004–2013 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Ruella M et al. The addition of the BTK inhibitor Ibrutinib to anti-CD19 chimeric antigen receptor T cells (CART19) improves responses against mantle cell lymphoma. Clin. Cancer Res. 22, 2684–2696 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Fraietta JA et al. Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood 127, 1117–1127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Pyl PT & Huber W HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luckey CJ et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 103, 3304–3309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinney EE, Lee JC, Jayne DR, Lyons PA & Smith KG T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry EJ et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Abbas AR et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 6, 319–331 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Bangs SC et al. Human CD4+ memory T cells are preferential targets for bystander activation and apoptosis. J. Immunol 182, 1962–1971 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Ramirez K et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity 36, 921–932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter SC et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 67, 3 441–3449 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Campia I et al. An autocrine cytokine/JAK/STAT-signaling induces kynurenine synthesis in multidrug resistant human cancer cells. PLoS One 10, e0126159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jena B et al. Chimeric antigen receptor (CAR)-specific monoclonal antibody to detect CD19-specific T cells in clinical trials. PLoS One 8, e57838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cawthon RM Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim NW et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Herbert BS, Hochreiter AE, Wright WE & Shay JW Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat. Protoc 1, 1583–1590 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Patient-related data not included in the paper were generated as part of completed or ongoing clinical trials and may be subject to patient confidentiality. Therefore, these data may be restricted. With regard to RNA- sequencing data, counts of sequencing reads are presented in the supplementary information. All associated raw data files have been submitted to the database of Genotypes and Phenotypes (dbGaP; accession numbers pending). Any other data that support the findings of the study are available from the corresponding author upon reasonable request.