Abstract
Background
Long non-coding (lnc) RNAs plays an important role in chronic myeloid leukemia (CML). In this study, we aimed to uncover the mechanism of the lncRNA maternally expressed 3 (MEG3) and its target microRNA-147 (miR-147) in CML.
Methods
Sixty CML patients and 10 healthy donors were included in the study. The methylation of MEG3 and miR-147 promoter was determined by methylation-specific PCR. The relationship of MEG3 and miR-147 was explored by luciferase assay. The interactions of proteins were studied by RNA pull-down assay, RNA immunoprecipitation and co-immunoprecipitation.
Findings
Patients in accelerated phase CML (CML-AP) and blast phase CML (CML-BP) showed lower expressions of MEG3 and miR-147 and higher expressions of DNMT1, DNMT3B, MBD2, MECP2 and HDAC1 compared to the controls. These patients also showed a higher degree of methylation of MEG3 and miR-147 while there was a reduction after chidamide treatment. Furthermore, the overexpression of MEG3 and miR-147 inhibited cell proliferation both in vivo and in vitro, promoted apoptosis and decreased the expressions of DNMT1, DNMT3A, DNMT3B, MBD2, HDAC1 and MECP2. We also found MEG3 interacted with DNMT1, JAK2, STAT3, HDAC1, and TYK2, and JAK2 was bound to STAT3, STAT5 and MYC. More interestingly, JAK2 was bound to TYK2 by the bridge of MEG3.
Interpretation
LncRNA MEG3 and its target miR-147 may serve as a novel therapeutic target for CML blast crisis, and chidamide might have a potential clinical application in treating CML blast crisis.
Keywords: lncRNA MEG3, Chronic myeloid leukemia blast crisis, Chidamide, miR147 and epigenetic
Research in Context
Evidence before this Study
Chronic myeloid leukemia (CML) is a bone marrow clone malignancy. In clinic, tyrosine kinase inhibitors (TKIs) were primarily chosen for CML treatment, while some patients showed resistance to or cannot tolerate TKI administration. Therefore, it is urgent to explore novel therapeutic targets or potential medication. Development of CML is a complex process, many molecules play important roles, including long non-coding RNA (lncRNA) and microRNA (miRNA). It has been shown that lncRNA maternally expressed 3 (MEG3) can inhibit cancer cell proliferation and is associated with poor prognosis of cancer patients. MEG3 can inhibit cell proliferation in CML. MEG3 may be an important molecule in the progression of diseases including CML, while the regulation mechanism was not clear. MiR147, which is downregulated in colon cancer, suggesting that miR147 can act as tumor suppressor. The role of miR-147 in CML was unknown. In the study, we aimed to explore the role and regulation mechanism of MEG3 and miR1-47 in CML development to explore the novel therapeutic targets.
Added Value of this Study
MEG3 may be an important molecule in the progression of CML. In the study, we explored the regulation mechanism of MEG3 on CML progression. Chidamide is a novel histone deacetylase inhibitor to treat cutaneous T-cell lymphoma. In this study, we detected the epigenetic regulation of MEG3 and regulation mechanism to further uncover the pathology of CML blast crisis, and the potential treatment effect of chidamide on CML blast crisis.
Implications of all the Available Evidence
We found in our study that lncRNA MEG3 and its target miR-147 may serve as a novel therapeutic target for CML blast crisis, and chidamide might also have a potential clinical application in treating CML blast crisis.
Alt-text: Unlabelled Box
1. Introduction
Chronic myeloid leukemia (CML) is a bone marrow clone malignancy characterized by the formation of the breakpoint cluster region (BCR) and Abelson murine leukemia (ABL) fusion gene that encodes BCR-ABLp210 [1, 2]. The pathology of CML comprises three phases, namely chronic phase (CML-CP), accelerated phase (CML-AP), and blast phase (CML-BP). The CML-AP and CML-BP are the hallmarks of advanced CML [3]. In clinic, tyrosine kinase inhibitors (TKIs) were primarily chosen for CML treatment, which improves the overall survival and the five-year progression-free survival of CML patients [4]. However, some patients showed resistance to or cannot tolerate TKI administration [5]. Therefore, it is urgent to explore novel therapeutic targets or potential medication.
Development of CML is a complex process, many molecules play important roles, including long non-coding RNA (lncRNA) [6]. For example, a study showed that lncRNA maternally expressed 3 (MEG3) can inhibit cell proliferation of CML [7]. Specifically, MEG3 can inhibit cancer cell proliferation by binding to the PRC2 complex, and low levels of MEG3 were associated with a poor prognosis of cancer patients [8]. Moreover, the function of MEG3 was also reported in many cancer cells lines [9, 10]. Therefore, MEG3 may be an important molecule in the progression of diseases including CML. Therefore, in the study, we explored the regulation mechanism of MEG3 on CML progression. Besides lncRNAs, microRNAs (miRNAs) can also regulate the progression of diseases [[11], [12], [13], [14]].
In addition to molecules, many genetic and molecular biological events were involved in the development of diseases including epigenetic regulation [15]. Epigenetics are the study of heritable changes in gene expression, without any alteration of the DNA sequence [16]. The abnormal methylation of promoter regions plays an important role in tumorigenesis, including human choriocarcinoma and squamous cell lung cancer [17, 18]. Furthermore, the study of lncRNA-associated methylation has gained increased attention [19, 20].
In the cells where methylation regulation occurs, the expressions of methylation related genes were significantly changed, such as DNMT1, DNMT3A, DNMT3B, MBD2, HDAC1, and MECP2. Histone deacetylases (HDACs) also involved epigenetic regulation. HDAC expression was increased in the majority of tumor cells and induces tumorigenesis. HDAC inhibitors (HDACis) are a novel family of drugs that achieved positive results in the treatment of hematological malignancies [[21], [22], [23]] and certain solid tumors [24]. HDACis interfere with the function of HDACs and reverse the effect of their overexpression [25, 26]. Chidamide is a novel HDACi that is approved to treat cutaneous T-cell lymphoma [26]. However, whether chidamide influences the epigenetic regulation of MEG3 and miR-147 in KCL22 and K562 cells remains largely unknown.
Therefore, we aimed to detect the epigenetic regulation of MEG3 and regulation mechanism to further uncover the pathology of CML blast crisis, and the potential treatment effect of chidamide on CML blast crisis in the study.
2. Materials and Methods
2.1. Collection of Bone Marrow Samples
The bone marrow samples of 60 CML patients and 10 healthy donors were collected at Department of Hematology of the Second Hospital of Hebei Medical University between May 2016 and June 2017 (Table 1). The bone marrow samples from 10 healthy donors were used as controls. In addition, we collected 3 CML-AP patients and 3 CML-BP patients to study the role of chidamide for treatment of CML. Peripheral blood mononuclear cells were isolated by lymphocyte separation. The inclusion criteria of patients were as follows: (i) diagnosis of CML via bone marrow morphology, immunology, molecular biology, and cytogenetic analyses; (ii) clear pathological staging; and (iii) availability of intact clinical data. The exclusion criteria were as follows: (i) significant organ dysfunction; (ii) pregnancy; and (iii) failure to provide informed consent. No chemotherapy was administered before the collection of the specimens. The study was approved by the Ethics Committee of the Department of Hematology of the Second Hospital of Hebei Medical University, and each patient signed an informed consent.
Table 1.
Characteristics of the patients included in the study.
| Item | CML-CP (n = 30) | CML-AP (n = 15) | CML-BP (n = 15) |
|---|---|---|---|
| Age (years), median (range) | 41.4(9–65) | 49.1(13–69) | 51.9(20–69) |
| Male/female, (n/n) | 20/10 | 9/6 | 10/5 |
| WBCs × 109/median (range) | 221.4(30.2–517) | 263.5(47.4–396) | 69.5(27.4–224) |
| Hemoglobin level (g/l) | 94(76–120) | 75(61–105) | 62.4(52–79) |
| Platelet count,109/median (range) | 518(99–809) | 305(52–725) | 35.5(19–71) |
AP, accelerated phase; BP, blast phase; CML, chronic myeloid leukemia; CP, chronic phase; WBC, white blood cells.
2.2. Cell Culture and Drug Treatment
The KCL22 and K562 cells were maintained in our laboratory. The KCL22 cells were cultured in Iscove's modified Dulbecco's medium (IMDM; Gibco, Beijing, China) containing 10% fetal bovine serum (FBS, Clark Bio, Claymont, DE, USA), 100 units/ml penicillin, and 100 μg/ml streptomycin. The K562 cells were maintained in RPMI 1640 medium (Gibco, Beijing, China) supplemented with 10% FBS and penicillin/streptomycin. Chidamide was purchased from MedChemExpress (Monmouth Junction, NJ, USA). The KCL22 and K562 cells or the patient-derived cells were seeded into 6-well plates (5 × 105/well) and were treated with different concentrations of chidamide based on its EC50.
2.3. Construction of the Lentiviral Vector
The full-length cDNA sequence of MEG3 was synthesized by Invitrogen. The cDNA was cloned into the pLVX-hMEG3-Puro lentiviral overexpression vector (Life Invitrogen, Carlsbad, CA, USA). MiR-147 mimics and miR-147 control were also synthesized by Invitrogen (Carlsbad, CA, USA).
2.4. Cell Transfection
The KCL22 and K562 cells were seeded into 6-well plates (5 × 105/well) and were transfected with LV-MEG3 or LV-control or miR-147 controls or miR-147 mimics using lipo3000 (Life Invitrogen, Carlsbad, CA). Purinamycin was used to screen the positive cells. MEG3 targeting locked nucleic acid-antisense oligonucleotide (LNA-ASO) and negative control LNA-ASO were designed and synthesized by Saibaisheng Bioengineering Co, Ltd. (Beijing, China). The KCL22 cells were transfected with the LNA-ASO at a concentration of 80 nM, using the Oligofectamine transfection reagent (Life Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cells were harvested for analysis at 36 h after the transfection. The ASO sequences are listed in Table 2.
Table 2.
Primer sequences of MEG3 ASO and NC ASO.
| MEG3 | ASO | GACCTTAGGCTAGGTGC |
| NC | ASO | GCTCCCAACTGTTCCAA |
ASO, acid-antisense oligonucleotide; NC, negative control; MEG3, maternally expressed 3.
2.5. Cell Proliferation Assay
The KCL22 and K562 cells were seeded into 96-well plates (1 × 104 cells/well) and were both either transfected with LV-MEG3 or LV-control, or transfected with the miR-147 mimics or miR-147 controls. A total of 10 μl of 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, St. Louis, MO, USA, USA) was added to each well, and the 96-well plates were incubated at 37 °C in a humidified 5% CO2 atmosphere for 4 h. The absorbance was then read at 490 nm in a microplate reader (Thermo Fisher USA). The patient-derived cells were also assayed for proliferation using MTT after treatment with chidamide, as described above.
2.6. Apoptosis Assay
The KCL22 and K562 cells were seeded into 6-well plates (5 × 105/well) and were both either transfected with LV-MEG3 or LV-control, or transfected with the miR-147 mimics or miR-147 control. Apoptosis was determined using an AnnexinV-FITC and PI kit (BD Biosciences, Franklin Lakes, NJ, USA) and was analyzed using a BD FACSCanto II system (BDBiosciences). The patient-derived cells were also assayed using the apoptosis kit after treatment with chidamide, as described above.
2.7. Real-Time Polymerase Chain Reaction (RT-PCR)
The total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and was reverse-transcribed to cDNA using RevertAid First Strand cDNA Synthesis Kit according to the manufacturer's instructions (Thermo Fisher, USA). The reverse transcription reactions were conducted under the following conditions: 42 °C for 60 min; 25 °C for 5 min; and 70 °C for 5 min. The qPCR mixtures had a final volume of 20 μl and contained 1 μl of template cDNA, 0.5 μl of each sense and antisense primer, 10 μl of SYBR Green Mix (Invitrogen), and 8 μl of diethyl pyrocarbonate (DEPC)-treated water. The concentrations of primers for DNMT1, DNMT3A, DNMT3B, MBD2, MeCP2, and HDAC1 were 0.25 μM. The RT-qPCR conditions were as follows: denaturation at 95 °C for 5 min followed by 45 cycles of 95 °C for 15 s; 60 °C for 35 s; and 72 °C for 20 s in an ABI 7500 Real-Time Rotary Analyzer (Applied Biosystems, Thermo Fisher Scientific, Shanghai, China). The 2−ΔΔCT method was used to calculate the relative gene expression. Each reaction was repeated for at least three times, independently. The primers were synthesized by Invitrogen and are shown in Table 3.
Table 3.
Primer sequences for RT-PCR.
| Gene | Primer | Product |
|---|---|---|
| MEG3 | Forward: 5′-CTCCCCTTCTAGCGCTCACG-3′ | 154 bp |
| Reverse: 5′-CTAGCCGCCGTCTATACTACCGGCT-3′ | ||
| DNMT1 | Forward: 5′-AGACACTCGCTCAGCTTCTTG-3′ | 116 bp |
| Reverse: 5′-CAATTGCTGCTGGGATTCATC-3′ | ||
| DNMT3A | Forward: 5′-CTGCTTTGTATTCCCTTTTGCA-3′ | 141 bp |
| Reverse: 5′-TTGATTTCTCCTGGCTGTCTC-3′ | ||
| HDAC1 | Forward: 5′-CCTCCCGAGTGAAGTCATCGTGG-3′ | 181 bp |
| Reverse: 5′-GGACAGGTGCTTCATCAGCTCG-3′ | ||
| DNMT3B | Forward: 5′-ACCACACGTTCCTAAGCTGG-3′ | 124 bp |
| Reverse: 5′-TCCCTGCACGCAGAGATTTT-3′ | ||
| MECP2 | Forward: 5′-AACTACAACTTCTTCCCTCGCAA-3′ | 109 bp |
| Reverse: 5′-CAAAGTTATGTCCACTGTCTCT-3′ | ||
| MBD2 | Forward: 5-CATCACCGTAGTCCCATTG-3′ | 175 bp |
| Reverse: 5-AAATTCCCCACTTCGCTTGG-3′ | ||
| JAK2 | Forward: 5′-CCACCCTTGTGCTCTTCCCTG-3′ | 100 bp |
| Reverse: 5′-TCTGCCACCCGAGTGTAACCA-3′ | ||
| STAT3 | Forward:5′-TCCCTGCACGCAGAGATTTT-3′ | 131 bp |
| Reverse: 5′-TTGATTTCTCCTGGCTGTCTC-3′ | ||
| STAT5 | Forward: 5′-AACCCTGCTGTTTGGCTTAAC-3′ | 127 bp |
| Reverse: 5′-CGTACTTGCTGTACTCGCTCTTC-3′ | ||
| ACTB | Forward: 5′-GAGCTACGAGCTGCCTGAC-3′ | 121 bp |
| Reverse: 5′-GGTAGTTTCGTGGATGCCACAG-3′ | ||
| miR-147 | 5′-CCCCTATCACGATTAGCATTAA-3′ | 82 bp |
| U6 | 5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′ | 74 bp |
2.8. Detection of miR-147 Levels
The RNA in bone marrow mononuclear cells (BMMCs), KCL22 and K562 cell lines was extracted with Trizol (Invitrogen; Thermo Fisher Scientific, Inc., USA) and afterwards purified with the RNeasy Maxi kit (Qiagen, Germany) according to the provided instructions. Reverse transcription reactions and RT-qPCR were performed using miScript Reverse Transcription kit (Qiagen, Germany) and miScript SYBR® Green PCR kit (Qiagen, Germany). The mixtures for PCR to detect the level of miR-147 were as follows: 2 × All-in-OneTM qPCR Mix 10 μl, MiR-147 Primer 2 μl, Universal Adaptor PCR Primer 2 μl, cDNA 2 μl, and DEPC-treated water 4 μl. The concentration of primer was 0.2 μM. The programs for PCR were as follows: 95 °C for 1 min, and then 40 cycles of 95 °C for 10 s, 55 °C for 30 s and 70 °C for 30 s. After normalization to U6, the relative expression level of mature miR-147 was calculated with 2−ΔΔCT method.
2.9. Methylation-Specific PCR
Genomic DNA was extracted, and the specific steps were performed according to the manufacturer's instructions (Shanghai Generay Biotech Co. Ltd., Shanghai, China). The DNA concentration was detected, and the sulfite conversion of DNA was performed according to the instructions of the EZ DNA methylation-Gold kit (Zymo USA Inc., Fleming Island, FL, USA). For all the reactions, the reaction mix contained bisulfite-modified DNA (2 μl), Zymo Taq PerMix (12.5 μl), water (8.5 μl), upstream primer (1 μl), and downstream primer (1 μl). The PCR conditions were as follows: 95 °C for 10 min followed by 35 cycles of 30 s at 95 °C; 45 s at 54 °C for annealing; 45 s at 72 °C; and a final extension step of 7 min at 72 °C. The PCR products separated by 2% agarose gel (GoldView; Saibaisheng Bioengineering Co., Ltd., Beijing, China) electrophoresis as follows: MEG3 and miR-147 methylated (M) and MEG3 and miR-147 unmethylated (U) positive and negative methylation; MEG3 and miR-147 M and MEG3 and miR-147 U partial methylation; and MEG3 and miR-147 M and MEG3 and miR-147 U positive and negative unmethylated. The primers were synthesized by Saibaisheng Bioengineering Co, Ltd. and are shown in Table 4.
Table 4.
Primer sequences of the methylated MEG3 and miR-147 gene.
| Gene | Primer | Product |
|---|---|---|
| MEG3, M-MSP | Forward: 5′-GCGAATTATTATTTATATAGCCTTC-3′ | 162 bp |
| Reverse: 5′-TCACGCGCTACGAAGGGAAACG-3′ | ||
| MEG3, U-MSP | Forward:5′-GTGAGGTGTTATTACCGTATAGTTTGG-3′ | 162 bp |
| Reverse: 5′-TTCACACATACAGGTTCCAAACAAT-3′ | ||
| miR-147, M-MSP | Forward: 5′-CGTAGCTACTGCTATTATCGTTC-3′ | 174 bp |
| Reverse: 5′-TTATGCCTTGGGCTACGTACT-3′ | ||
| miR-147, U-MSP | Forward:5′-GGTACGTATGCTTGGGCTTATG-3′ | 174 bp |
| Reverse: 5′-ATTGGCGCTTAGCTAGGCTA-3′ |
M, methylated; MSP, methylation-specific polymerase chain reaction; U, unmethylated.
2.10. Western Blotting
The cells were lysed using radio immune precipitation assay (RIPA) buffer, and the total protein was isolated. The protein concentration was determined using a Bio-Rad DC protein assay kit II (Bio-Rad, Hercules, CA, USA). The proteins were separated by 8–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the bands were electro-transferred to a polyvinylidene fluoride (PVDF) membrane (Amersham Pharmacia, Piscataway, NJ, USA). The PVDF membrane was blocked with 5% non-fat skimmed milk and was probed with antibodies against DNMT1 (Mouse monoclonal antibody [60B1220.1]) (1:1000), DNMT3A (Rabbit monoclonal antibody [EPR18455]) (1:1000), DNMT3B (Rabbit monoclonal antibody [EPR3523]) (1:1000), MECP2 (Mouse monoclonal antibody [Mec-168]) (1:1000), MBD2 (Rabbit monoclonal [EPR18361]) (1:1000), JAK2 (Rabbit monoclonal antibody [EPR108(2)]) (1:1000), STAT3 (Rabbit monoclonal [EPR787Y]) (1:1000), STAT5A (Rabbit monoclonal antibody [E289]) (1:1000), bcl-2 (Rabbit monoclonal antibody [E17]) (1:1000), TYK2 (Rabbit polyclonal antibody) (1:1000), ACTB (Rabbit polyclonal antibody) (1:5000) (from Abcam, Cambridge, MA, USA), phosphor(p)JAK2 (Rabbit polyclonal antibody) (1:1000) (Tyr221, Cell Signaling Technology), pSTAT3 (Rabbit monoclonal antibody [D3A7]) (1:1000) (Tyr705, Cell Signaling Technology), and pSTAT5A (Rabbit monoclonal antibody [D47E7]) (1:1000) (Tyr694, Cell Signaling Technology). Subsequently, the membrane was incubated with the appropriate dilutions of the corresponding secondary antibodies at 37 °C for 1 h, rinsed three times with TBS for 5 min, and analyzed by chemiluminescence. The quantification of the immunocomplexes was performed using the Imagequant 5.1 software (Amersham-Pharmacia's Biotech, Shanghai, China). β-actin was used as the loading control.
2.11. Luciferase Assay
To assay the interaction between MEG3 and miR-147, wide type 3′-UTR region of MEG3 and the mutant 3′-UTR of MEG3 were cloned into the firefly luciferase gene reporter vector pmiRGLO (Promega, Madison, WI, USA). The plasmid was synthesized by Invitrogen. The pmirGLO-MEG3 or pmirGLO-MEG3-MUT was co-transfected with miR-147 mimics or miRNA control (RiboBio, Guangzhou, China) into the 293 cells. The luciferase assays were performed using the dual-luciferase reporter assay system kit (Promega) according to the manufacturer's instructions. The luciferase expression was analyzed by Modulus single-tube multimode reader (Promega). The relative luciferase expression equaled the expression of the Renilla luciferase divided by the expression of the firefly luciferase. All the assays were repeated for at least 3 times.
2.12. RNA Pull-Down Assay
The MEG3 binding proteins were determined using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific Waltham, MA, USA) according to the manufacturer's protocol. The MEG3 sense and MEG3 antisense lncRNAs were generated and incubated with the proteins from the KCL22 cells, and 100 ml of a silver staining solution were added to the MEG3 sense and MEG3 antisense lncRNA and protein mixture. The proteins were then collected for the mass spectrometry analysis.
2.13. RNA Immunoprecipitation
RNA immunoprecipitation was performed using the Magna RIP kit (EMD Millipore Co., Billerica, MA, USA). The complete KCL22 cell lysates were prepared in accordance with the manufacturer's instructions and contained phosphatase and proteinase inhibitors. Next, we evaluated the connection and configuration of the antibody beads. The antibody beads were suspended and mixed with the sample after thawing and were then incubated at 4 °C overnight. The immunoprecipitation was completed after the suspension was placed on the magnetic frame and was washed with buffer for at least 6 times. Finally, the immune co-precipitation products were collected, and the RNA was extracted and purified to determine the abundance of the target RNA.
2.14. Co-Immunoprecipitation
The protein was obtained from the cells using RIPA cell lysis buffer (Invitrogen) followed by quantification, using the bicinchoninic acid assay. The interacting proteins were precipitated using a JAK2 antibody (Abcam, USA), and SDS-PAGE was performed. The target protein was pulled down according to the electrophoresis bands. The protein in the gel had a reductive alkylation reaction, and the protein was digested in the gel. The samples were analyzed by Liquid Chromatography Coupled with Tandem Mass Spectrometry (LC MS/MS).
2.15. Mass Spectrometry
The gel was cut into small pieces of 1 mm × 1 mm square. The small gels were digested using in-gel trypsin in EP tubes for 15 h at 37 °C after the gels were washed and bleached. After the digestion reaction, 400 μl of peptide extract (50% ACN, 0.1% TFA) was added to the EP tubes immediately for 30 min at 37 °C. Then the product was transferred to a new EP tube and performed peptide extraction. After the reaction, the samples were lyophilized in a freeze-dryer. The freeze-dried sample was dissolved in 20 μl of formic acid solution (0.1%), and centrifuged at 20,000 g for 30 min. To perform the MS analysis, 10 μl of supernatant was added to the sample bottle. The MS analysis was performed on a Q Exactive (Thermo Fisher Scientific, San Jose, CA, USA). The liquid phase was UPLC water, with a C18 column that was 3 μm and 250 mm × 75 μm (Eksigent). The settings were as follows: 1.0 h chromatographic gradient; 3.0 μl/min chromatographic flow rate; and 10 Strongest Ions in the MS Spectrum for MS/MS Analysis. The protein database was UNIPROT_ human, with a total of 20,214 protein sequences, and it was searched to identify the protein identities from the immunoprecipitation.
2.16. Xenograft Animal Experiments
Athymic male mice were purchased from the Animal Center of the Chinese Academy of Science (Shanghai, China) and were maintained in laminar flow cabinets under specific pathogen-free conditions. This study was approved by the Ethics Committee of the Department of Hematology of the Second Hospital of Hebei Medical University. The KCL22 cells were stably transfected with LV-MEG3 or empty vectors and were harvested from the cell culture plates. These cells were xenografted into BALB/c male nude mice. The tumor volumes and weights were measured every 5 days in the mice. The tumor volumes were measured as length × width 2 × 0.5. At 35 days after the injection, the mice were sacrificed, and the tumor weights were measured and used for further analysis.
2.17. Statistical Analysis
The data are expressed as mean ± standard deviation (SD) and were analyzed using the SPSS 19.0 software (IBM Corp, Armonk, NY, USA). Significant differences between the groups were analyzed using a Student's t-test or a one-way ANOVA for more than two subgroups. A chi-squared test was applied to compare the rates, and P < 0.05 was considered a statistically significant difference.
3. Results
3.1. MEG3 Expression and Promoter Methylation in Patients with Different Phases of CML
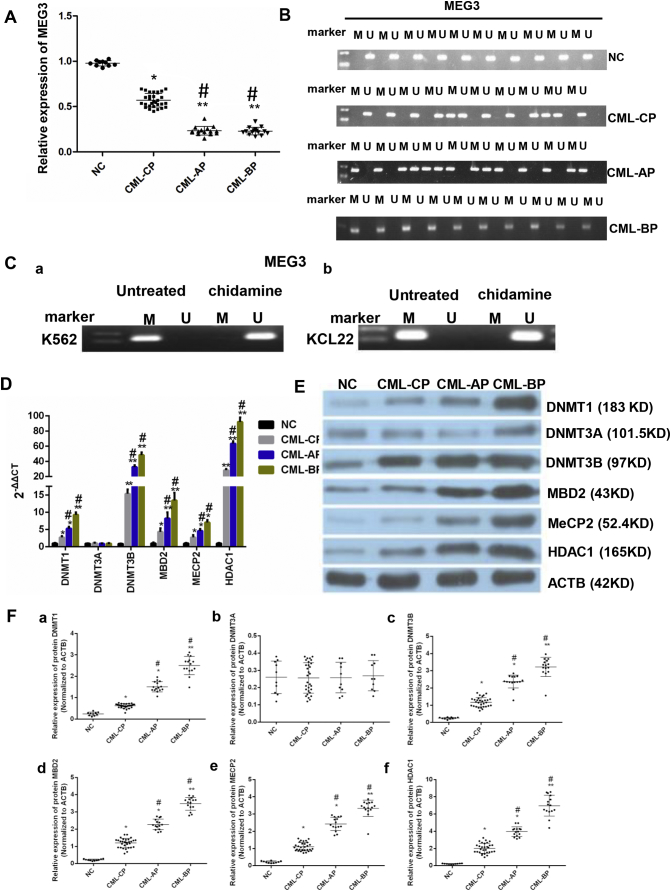
The expression levels of MEG3 were lower in the patients of different phases of CML than that in the healthy donors (P < 0.05, one-way ANOVA) (Fig. 1A). The expression levels of MEG3 in CML-AP patients and CML-BP patients were similar (P > 0.05, one-way ANOVA), but were lower than that in CML-CP patients and healthy donors. The MEG3 promoter was not methylated in the healthy donors, but showed methylation in 6 CML-CP patients (20%) (N = 30). On the other hand, all CML-AP samples showed methylation of MEG3 promoter, and methylation can be detected in 100% of the CML-BP samples (Fig. 1B).
Fig. 1.
Expression levels of MEG3 and methylation related genes in different stages of CML and in healthy donors and the methylation status of MEG3 in different stages of CML and in healthy donors. A. The MEG3 mRNA level was lower in the CML-AP and CML-BP patients than that in the CML-CP patients and the healthy donors. B. The methylation of the MEG3 promoter was not detected in the normal controls. Methylation can be detected in 20% of the CML-CP samples. All CML-AP samples showed methylation of the MEG3 DNA promoter, and methylation can be detected in 100% of the CML-BP samples. C. After treatment with chidamide, the MEG3 was unmethylated in both K562 (a) and KCL22 (b) cells. D. The mRNA expressions of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 in NC, CML-CP, CML-AP, and CML-BP groups were determined by RT-PCR. E. The protein levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 in NC (n = 10), CML-CP (n = 30), CML-AP (n = 15), and CML-BP (n = 15) groups were determined. The protein expression levels were determined by immunoblotting with ACTB as a control. F. Protein expression of DNMT1 (a), DNMT3A (b), DNMT3B (c), MBD2 (d), MECP2 (e), HDAC1 (f) in the bone marrow cells from CML patients and normal controls. The one-way ANOVA was used for data analysis. *: P < 0.05 compared to NC group. **: P < 0.01 compared to NC group. #: P < 0.05 compared to CML-CP group. M, methylated; U, unmethylated.
3.2. MEG3 Promoter Methylation after Chidamide Treatment in KCL22 and K562 Cells
Next, we assessed the MEG3 promoter methylation in KCL22 (a) and K562 (b) cells before and after chidamide treatment. The MEG3 promoter was methylated in the non-treated group, but was not in the treated group (Fig. 1C).
3.3. The Methyltransferase and HDAC1 mRNA and Protein Expression in Patients with Different Phases of CML
Then, we examined the mRNA expression of the DNA methyltransferases and HDAC1 in the CML samples by RT-qPCR. The mRNA expressions of DNA (cytosine-5)-methyltransferase (DNMT) 1, DNMT3B, MBD2, MECP2 (two methyl-CpG-binding proteins), and HDAC1 were higher in the CML-BP patients than in the healthy donors (P < 0.05, one-way ANOVA). The mRNA expression of DNMT3A, among any of the CML phases and healthy samples, was not statistically different (P > 0.05, one-way ANOVA). The mRNA expressions of DNMT1, DNMT3B, MBD2, MECP2, and HDAC1 were the highest in the CML-BP samples (Fig. 1D).
The protein expressions of DNMT1, DNMT3A, MBD2, DNMT3B, MECP2, and HDAC1 were semi-quantitatively determined in the bone marrow samples of CML-CP, CML-AP, CML-BP patients and healthy donors. Consistent with the RT-PCR results, the protein levels of DNMT1, DNMT3A, MBD2, DNMT3B, MECP2, and HDAC1 were higher in the CML-AP and CML-BP patients than that in the CML-CP patients and healthy donors (P < 0.05, one-way ANOVA). The DNMT3A (b) level was not significantly different in the samples from the CML patients or the healthy donors (P > 0.05, one-way ANOVA), and the highest expression of DNMT1 (a), DNMT3B (c), MBD2 (d), MECP2 (e), and HDAC1 (f) was found in the CML-BP patients (Fig. 1E and F).
3.4. Changes in mRNA and Protein Levels in CML Blast Cells after Chidamide Treatment
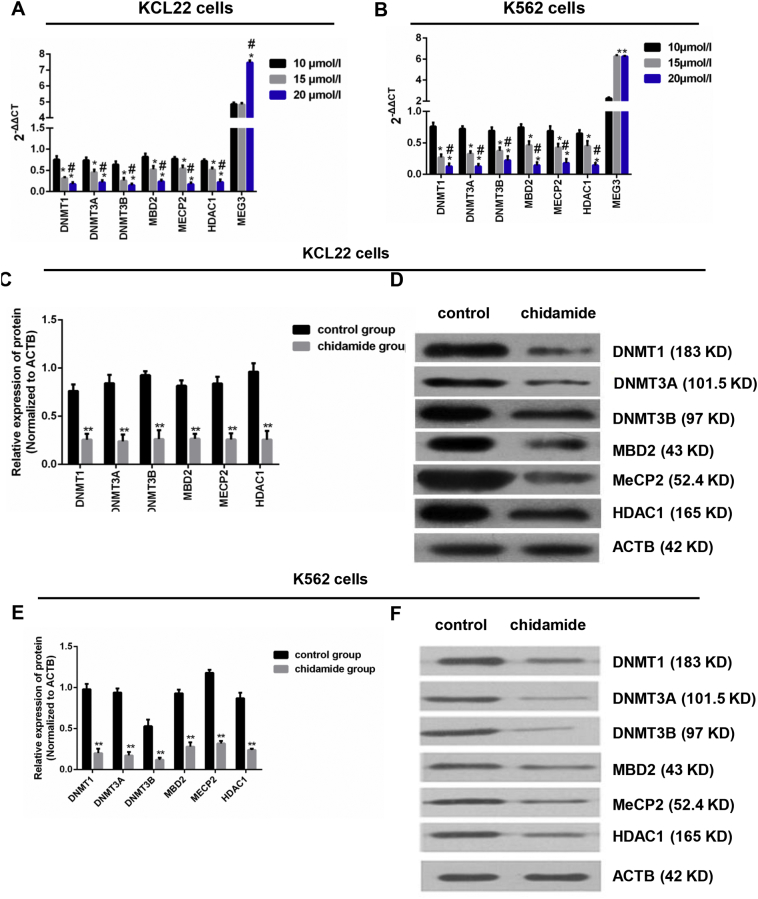
To determine the effect of HDACi on epigenetic regulation, the KCL22 and K562 cells were treated with three different concentrations of chidamide (10, 15, and 20 μmol/l). The mRNA expressions of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 were decreased with the increase of concentration. The mRNA expression of MEG3 was significantly higher in the treatment of 20 μmol/l chidamide than that in the treatment of 10 μmol/l and 15 μmol/l chidamide in KCL22 cells (P < 0.05, one-way ANOVA), and was significantly higher in the treatment of 20 μmol/l chidamide and 15 μmol/l chidamide than that in the treatment of 10 μmol/l chidamide in K652 cells (P < 0.05, one-way ANOVA) (Fig. 2A and B).
Fig. 2.
Changes in mRNA and protein levels in CML blast cells after chidamide treatment. A & B. The mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 after the KCL22 and K562 cells were treated by 10, 15, and 20 μmol/l chidamide were detected by RT-PCR. C. Chidamide treatment decreased the protein levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 in KCL22 cells. D. Western blot was used to detect the protein levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 in KCL22 cells. E. Chidamide treatment decreased the protein levels of these six proteins in K562 cells. F. Western blot was used to detect the protein levels of these six proteins in K562 cells. For Western blot, ACTB was used as a control. For Fig. 2A and B, the one-way ANOVA was used for data analysis. *: P < 0.05 compared to 10 μmol/l group. **: P < 0.01 compared to 10 μmol/l group. #: P < 0.05 compared to 15 μmol/l group. For Fig. 2C and E, the Student's t-test was used for data analysis. **: P < 0.01 compared to control group.
Next, we treated the cells with the highest concentration of chidamide (20 μmol/l) and found that, by a semi-quantitative analysis, the protein expressions of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 were lower in the treated group than that in the non-treated group in both KCL22 and K652 cells (P < 0.05, Student's t-test) (Fig. 2C–F).
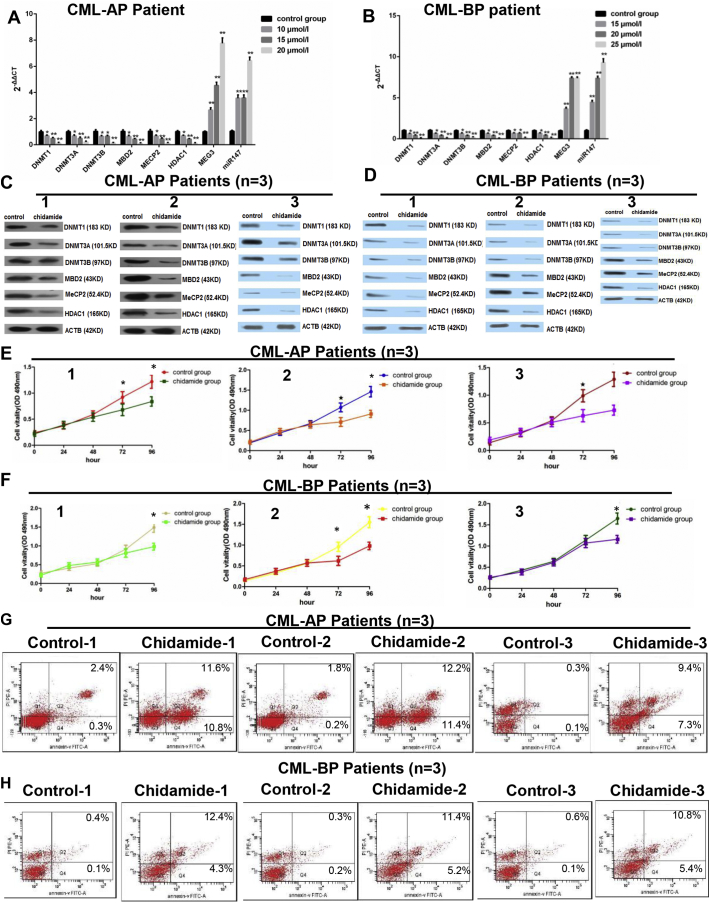
To assess the effect of chidamide on CML treatment, we treated the CD34+ cells which were isolated from the peripheral blood mononuclear cells of 3 CML-AP patients and 3 CML-BP patients. The expressions of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, HDAC1, MEG3 and miR-147, as well as cell viability and cell apoptosis were determined. We found that treatment with chidamide (all three concentrations) decreased the mRNA and protein levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 in CML-AP and CML-BP patients, while the expression levels of MEG3 and miR-147 were increased (Supplementary Fig. 1A–D). The treatment of chidamide also decreased cell viability in 3 CML-AP and 3 CML-BP patients (Supplementary Fig. 1E, F) and accelerated cell apoptosis (Supplementary Fig. 1G and H). These results indicated that chidamide might be potentially used for treating CML blast crisis.
Supplementary Fig. 1.
Chidamide treatment in CML patient-derived cells. A cohort of CML patients was recruited, the bone marrow samples were collected from 3 CML-AP patients and 3 CML-BP patients, and then the peripheral blood mononuclear cells were isolated via a lymphocyte separation. A and B. These patient-derived cells were treated with chidamide at 3 doses (10 μmol/l, 15 μmol/l or 20 μmol/l) and the mRNA expressions of DMNT1, DMNT3A, DMNT3B, MECP2, HDAC1, MEG3 and miR-147 in CML-AP and CML-BP patients were assessed by real-time PCR. C and D. The protein expression levels of these targets were also assessed after chidamide treatment in CML-AP and CML-BP patients using a western blot. E and F. The effect of chidamide treatment on the proliferation of the patient-derived cells was assessed by an MTT assay. G and H. The effect of chidamide treatment on the apoptosis of the patient-derived cells was assessed by an AnnexinV-FITC and PI kit. *: P < 0.05 compared to the control group. **: P < 0.01 compared to the control group. For Supplementary 1A and B, the one-way ANOVA was used for data analysis. For Supplementary 1E and F, Student's t-test was used for data analysis. The comparison of cell apoptosis rate between control group and chidamide group was analyzed using chi-squared test.
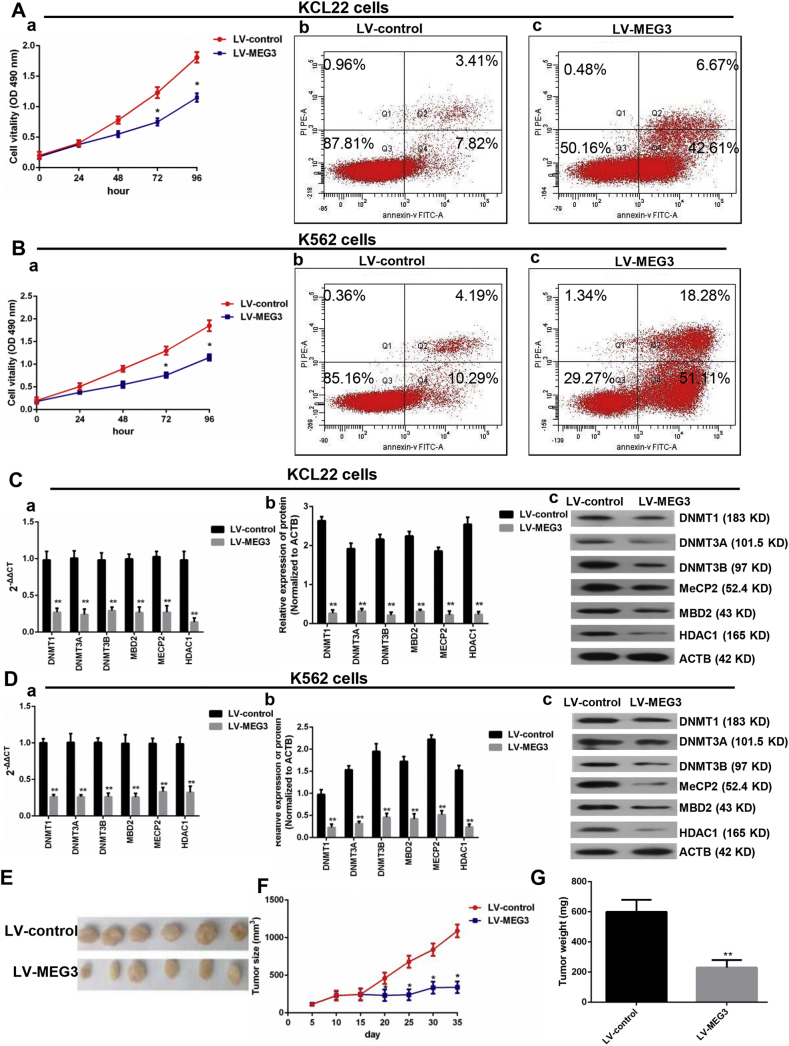
3.5. Effect of MEG3 Overexpression on CML Blast Cells In Vivo and In Vitro
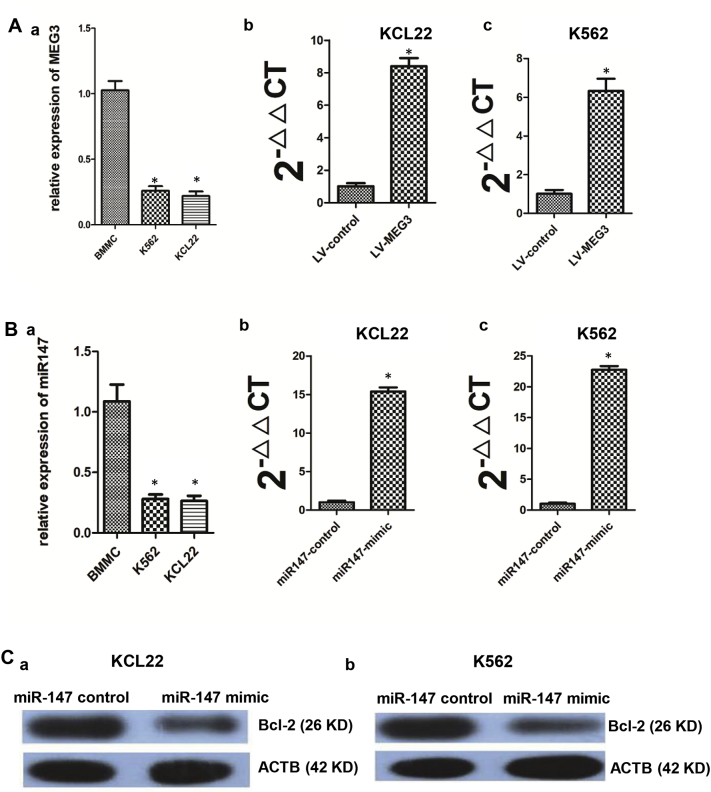
To determine the role of MEG3 in CML blast crisis, we first determined the mRNA levels of MEG3 in the BMMCs of healthy donors and in the K562 and KCL22 cells, and the results indicated that the mRNA level of MEG3 was lower in the KCL22 and K562 cells than in the BMMCs (a) (P < 0.05, one-way ANOVA) (Supplementary Fig. 2A). An overexpression vector for MEG3 was synthesized and was then transfected into the KCL22 (b) and K562 (c) cells, and MEG3 expression in the overexpression group was 7.3-fold and 6.4-fold higher than that in the empty vector group (P < 0.05, Student's t-test) (Supplementary Fig. 2A). Cell proliferation (a) and apoptosis (b and c) assay results indicated that the overexpression of MEG3 inhibited proliferation (P < 0.05, Student's t-test) and induced apoptosis (P < 0.05, chi-squared test) in the KCL22 and K562 cells (Fig. 3A and B). To further detect whether MEG3 affected the proliferation of the KCL22 cells in vivo, the KCL22 cells were stably transfected with LV-MEG3 or a control vector and were inoculated into nude mice. Thirty-five days after the injection, the tumors that formed in the control group were substantially larger than those in the LV-MEG3 group (P < 0.05, Student's t-test) (Fig. 3E, F). Moreover, the tumor weight, at the end of the experiment, was markedly lower in the control vector group compared with the LV-MEG3 group (P < 0.05, Student's t-test) (Fig. 3G). Next, we determined the effect of MEG3 overexpression on the expression of DNA-modifying genes, and found that the mRNA (a) and proteins (b and c) expressions of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 were significantly decreased (P < 0.05, Student's t-test) (Fig. 3C and D).
Supplementary Fig. 2.
The expression levels of MEG3, miR-147 and bcl-2 after treatment of LV-MEG3 or miR-147 mimic. A. The MEG3 expression levels in BMMC (a), KCL22, and K562 (c) cells and in the KCL22 (b) and K562 (c) cells after being treated by LV-MEG3. B. The miR-147 expression levels in BMMC (a), KCL22, and K562 cells and in the KCL22 (b) and K562 (c) cells after being treated by miR-147 mimic. C. The protein level of bcl-2 decreased after the overexpression of miR-147 in KCL22 (a) and K562 (b) cells. *: P < 0.05 compared to BMMC group or control group. For the panel (a) of Supplementary Fig. 2A and B, the one-way ANOVA was used for data analysis. For panel (b and c) of Supplementary Fig. 2A and B, Student's t-test was used for data analysis.
Fig. 3.
Effect of MEG3 overexpression on CML blast cells in vivo and in vitro. A. Overexpression of MEG3 inhibited KCL22 proliferation (a) and accelerated cell apoptosis (b and c). B. Overexpression of MEG3 inhibited K562 proliferation (a) and accelerated cell apoptosis (b and c). C. In the KCL22 cells, the mRNA (a) and protein (b and c) levels of the DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 were decreased in the MEG3 overexpression group. D. In the K562 cells, the mRNA (a) and protein (b) levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 decreased in the MEG3 overexpression group. E, F and G. Overexpression of MEG3 inhibited tumor growth. Student's t-test was used for data analysis. The comparison of cell apoptosis rate between LV-control group and LV-MEG3 group were analyzed using chi-squared test. *: P < 0.05 compared to LV-control group. **: P < 0.01 compared to LV-control group.
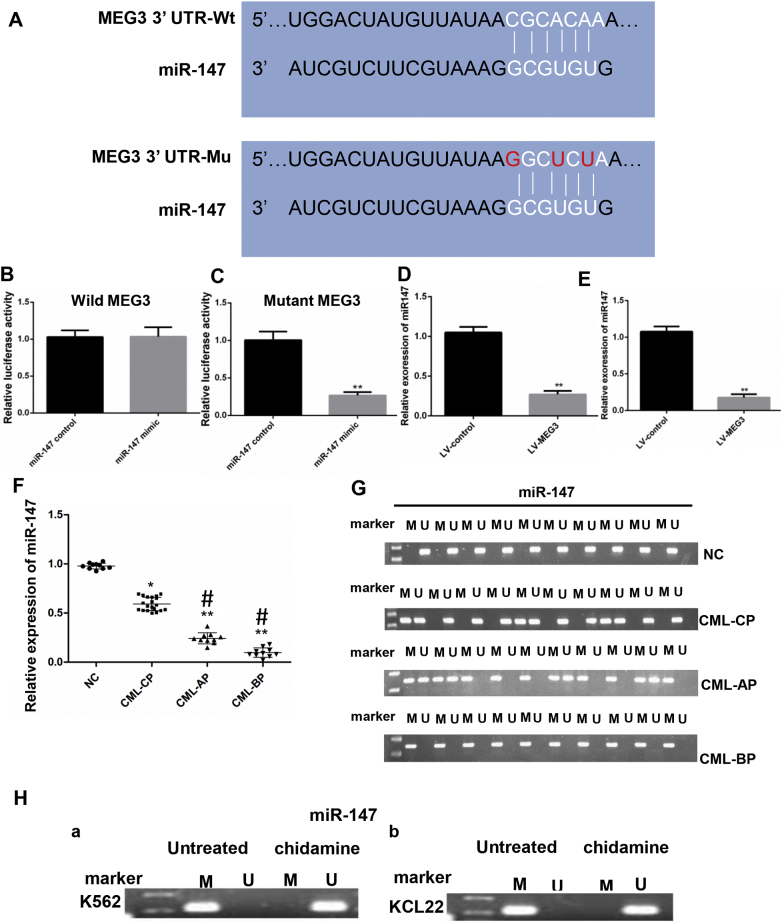
3.6. Interaction of MEG3 with miR-147
LncRNAs and miRNAs usually act as a competing endogenous RNA (ceRNA). We searched the starBase2.0 database to identify the miRNAs that interact with MEG3. Results showed that miR-147 was the potential miRNA. A dual luciferase reporter assay was performed to verify the interaction between miR-147 and MEG3, and for this purpose, we generated luciferase reporter plasmids with wild-type or mutant MEG3 (Fig. 4A). The transfection of 293 cells with miR-147 mimics had no effect on the luciferase reporter activity of the wild MEG3 (P > 0.05, Student's t-test) (Fig. 4B), but significantly decreased the luciferase reporter activity of mutant-type MEG3 (P < 0.05, Student's t-test) (Fig. 4C). Furthermore, MEG3 overexpression decreased the expression of miR-147 (P < 0.05, Student's t-test) (Fig. 4D, E). In addition, we also detected the expression and methylation of miR-147 in healthy donors, CML-CP, CML-AP, and CML-BP patients. The results showed that the level of miR-147 was significantly lower in the samples of the different phases of CML than that in healthy donors (P < 0.05, one-way ANOVA) (Fig. 4F). MiR-147 was not methylated in the healthy donors and showed methylation in 8 (37.5%) of CML-CP patients (N = 30) (Fig. 4G). Next, we assessed miR-147 promoter methylation in K562 (a) cells KCL22 (b) before and after chidamide treatment. The promoter was methylated in the non-treated group, but was not in the treated group (Fig. 4H).
Fig. 4.
The relative luciferase activity was detected by a dual luciferase assay. A. The RNAup algorithm predicted the potential binding of miR-147 to MEG3, with a considerable sequence complementary in the indicated regions. B. The relative luciferase activity was not different between the miR-147 mimics group and the miR-147 control group. C. The relative luciferase activity was lower in the miR-147 mimic group compared to the miR-147 control group. D. In the KCL22 cells, MEG3 overexpression decreased the expression level of miR-147; E. In the K562 cells, MEG3 overexpression decreased the expression level of miR-147. F. The mRNA level of miR-147 was lower in CML-AP and CML-BP patients than that in the CML-CP patients and the healthy donors. G. The methylation of the miR-147 DNA promoter was not detected in the normal controls, and methylation can be detected in 37.5% of the CML-CP samples. All CML-AP samples showed methylation of the miR-147 promoter, and methylation can be detected in 100% of the CML-BP patients. H. After treatment with chidamide, the miR-147 was unmethylated in both the K562 (a) and KCL22 (b) cells. For 4B, C, D and E, Student's t-test was used for data analysis. *: P < 0.05 compared to NC group. **: P < 0.01 compared to NC group. For 4F, the one-way ANOVA was used for data analysis. *: P < 0.05 compared to NC group. **: P < 0.01 compared to NC group. #: P < 0.05 compared to CML-CP group. M, methylated; U, unmethylated.
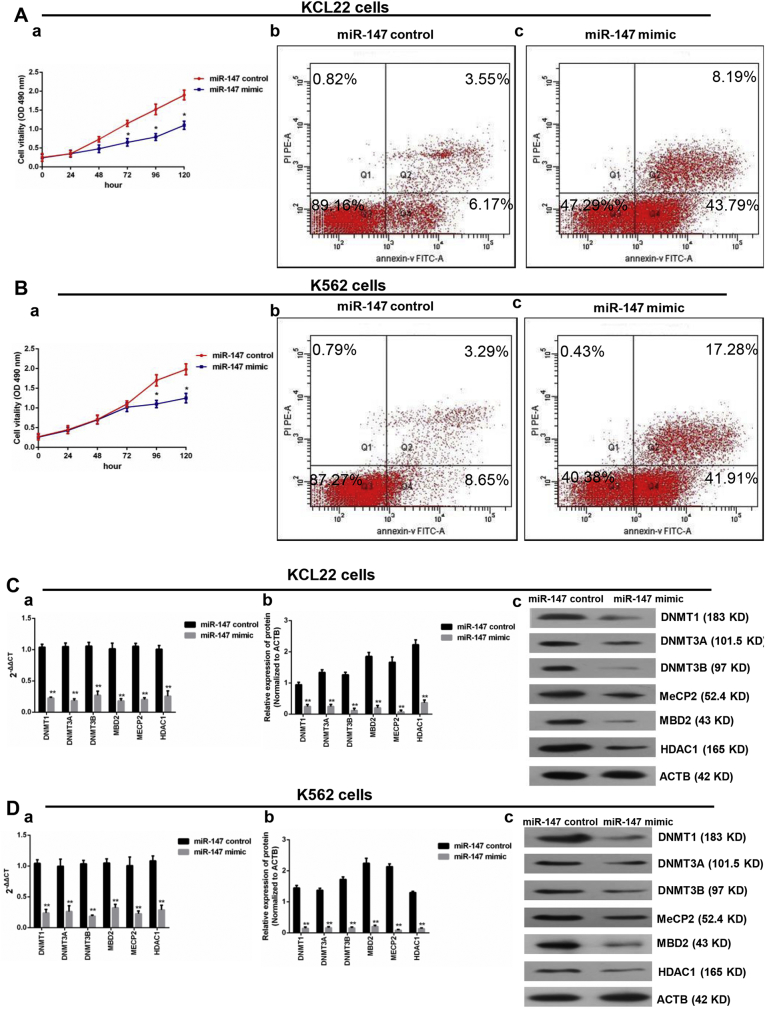
3.7. MiR-147 Overexpression Inhibited Proliferation, Promoted Apoptosis, and Affected the Expression of Epigenetically-Associated Genes
To determine the biological role of miR-147 in CML blast crisis, we first assessed the miR-147 mRNA levels in the BMMCs from healthy donors as well as in K562 and KCL22 cells, and the results indicated that the miR-147 level was lower in KCL22 and K562 cells than that in the BMMCs (a) (P < 0.05, one-way ANOVA) (Supplementary Fig. 2B). The miR-147 mimic and the miR-147 control were transfected into KCL22 and K562 cells. MiR-147 overexpression efficiently (16.3-fold in K562 cells (c) and 22.7-fold in KCL22 cells (b)) increased the expression of miR-147 compared to the negative control (P < 0.05, Student's t-test) (Supplementary Fig. 2B). The proliferation and apoptosis assay results indicated that the overexpression of miR-147 inhibited proliferation (P < 0.05, Student's t-test) and induced apoptosis (P < 0.05, chi-squared test) in KCL22 and K562 cells (Fig. 5A and B). Additionally, PCR and Western blot assays were performed to determine whether miR-147 affected the expression of epigenetically-associated genes. DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDCA1 expressions were significantly decreased in the overexpression group compared with that in the control group (P < 0.05, Student's t-test) (Fig. 5C and D). In addition, the protein level of bcl-2 was decreased in the overexpression group compared to that in the control group in KCL22 (a) and K562 cells (b) (Supplementary Fig. 2C).
Fig. 5.
MiR-147 overexpression inhibited proliferation, promoted apoptosis, and affected the genes expression. A. The KCL22 cells was transfected with miR-147 control and miR-147 mimics. Compared with the control group, transfection with miR-147 mimics inhibited proliferation (a). The apoptosis rate of the miR-147 mimics group (Q2 + Q4 = 59.19%) was higher than that of the control group (Q2 + Q4 = 11.94%) (b and c). B. The K562 cells were transfected with miR-147-control and miR-147 mimics. Compared with the control group, transfection with the miR-147 mimics inhibited proliferation (a). The apoptosis rate of the miR-147 mimics group (Q2 + Q4 = 51.98%) was higher than that of the control group (Q2 + Q4 = 10.02%) (b and c). C. In the KCL22 cells, the mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 decreased in the miR-147 mimics group (a); and the DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 protein levels decreased in the miR-147 mimics group (b and c). D. In the K562 cells, the mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 decreased in the miR-147 mimics group (a); and the DNMT1, DNMT3A, DNMT3B, MBD2, MECP2, and HDAC1 protein levels decreased in the miR-147 mimics group (b and c). Student's t-test was used for data analysis. The comparison of cell apoptosis rate between miR-147 control group and miR-147 mimic group was analyzed using chi-squared test. *: P < 0.05 compared to miR-147 control group. **: P < 0.01 compared to miR-147 control group.
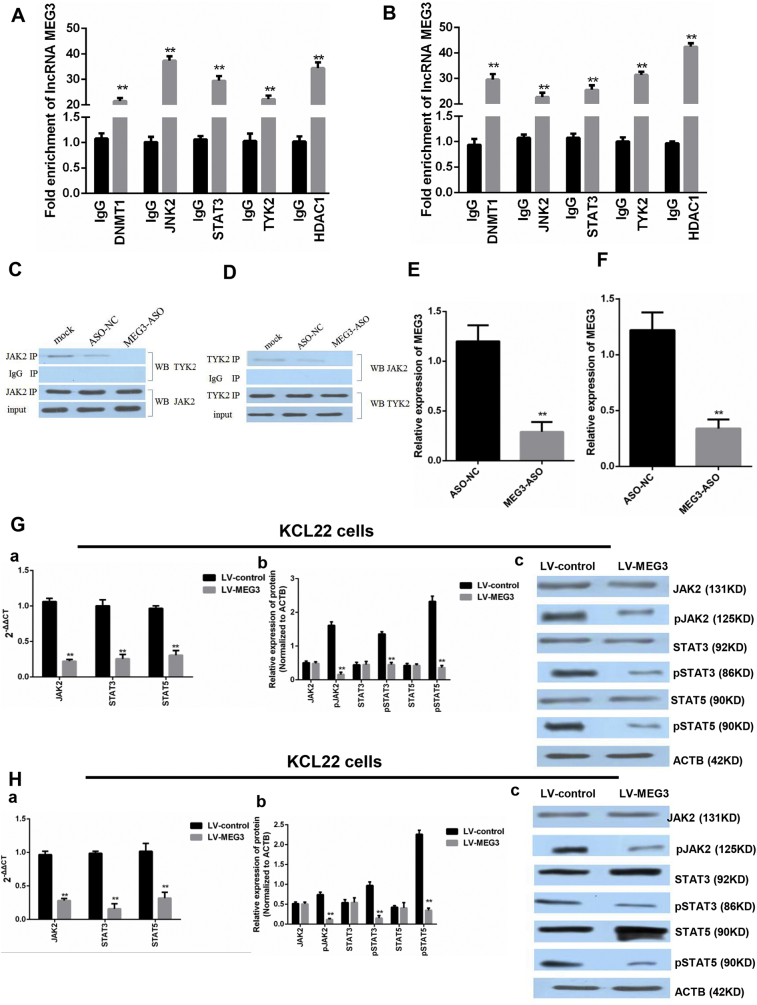
3.8. MEG3 Interacting Proteins in Chronic Myeloid Leukemia Blast Cells
RNA pulldown assays, mass spectrometry (MS) analysis, and RNA immunoprecipitation were performed to detect the MEG3 binding proteins in KCL22 cells. The MS analysis identified 2865 proteins in the sense group and 1852 proteins in the antisense group. Of the 586 proteins that were only found in the sense group, Janus kinase 2 (JAK2), signal transducer and activator of transcription 3 (STAT3), DNMT1, and tyrosine kinase 2 (TYK2) were associated with CML occurrence and development and were confirmed to interact with MEG3 by RNA immunoprecipitation (P < 0.05, Student's t-test) (Fig. 6A and B).
Fig. 6.
RIP-PCR was performed to test if DNMT1, JAK2, TYK2, STAT3 and HDAC1 were bound to MEG3. A. In the K562 cells, the fold enrichment of lncRNA MEG3 was higher in the DNMT1, JAK2, TYK2, STAT3, and HDAC1 groups than that in the IgG group (**: P < 0.01 compared to the IgG group). Student's t-test was used for data analysis. B. In the KCL22 cells, the fold enrichment of lncRNA MEG3 was higher in the DNMT1, JAK2, TYK2, STAT3, and HDAC1 groups than that in the IgG group (**: P < 0.01 compared to the IgG group). Student's t-test was used for data analysis. C and D. The pull-down assays showed that MEG3 retrieved JAK2 and TYK2 in the KCL22 cells. The retrieved proteins were detected by immunoblotting. In the KCL22 cells, JAK2 interacted with TYK2. The knockdown of MEG3, but not si-NC, abrogated this interaction. E. In the KCL22 cells, the expression of MEG3 was lower in the ASO-MEG3 group (**: P < 0.01 compared to the ASO-NC group). Student's t-test was used for data analysis.
F. In the K562 cells, the expression of MEG3 was lower in the ASO-MEG3 group (**: P < 0.01 compared to the ASO-NC group). Student's t-test was used for data analysis. G. In the KCL22 cells, the overexpression of MEG3 inhibited the mRNA (a) and phosphorylated protein (b and c) levels of JAK2, STAT3, and STAT5 (**: P < 0.01 compared to LV-control group). Student's t-test was used for data analysis. H. In the K562 cells, the overexpression MEG3 decreased mRNA (a) and phosphorylated protein (b and c) levels of JAK2, STAT3, and STAT5 (**: P < 0.01 compared to LV-control group). Student's t-test was used for data analysis.
3.9. Co-Immunoprecipitation MS and Co-Immunoprecipitation WB
Of the 543 proteins identified in the experimental group and the 306 proteins identified the in the control group, 216 proteins were only found in the experimental group and interacted with JAK2. Of these proteins, STAT3, STAT5, and MYC are known to be associated with the proliferation and apoptosis of cancer cells, and the Co-IP WB results showed that JAK2 interacted with TYK2 through MEG3 (Fig. 6C and D).
3.10. MEG3 Overexpression Substantially Reduced the Phosphorylation of JAK2 and STATs
To determine the effect of MEG3 on JAK2/STAT3 signaling, we overexpressed MEG3 and then performed qPCR and WB. In the KCL22 and K562 cells, the expression of MEG3 was both lowered in the MEG3-ASO group (P < 0.01, Student's t-test) (Fig. 6E and F). However, the overexpression of MEG3 decreased the mRNA expression (a) and proteins (b and c) phosphorylation of JAK2, STAT3, and STAT5 (P < 0.05, Student's t-test) (Fig. 6G and H).
3.11. STAT3 Regulated the Expression of MEG3
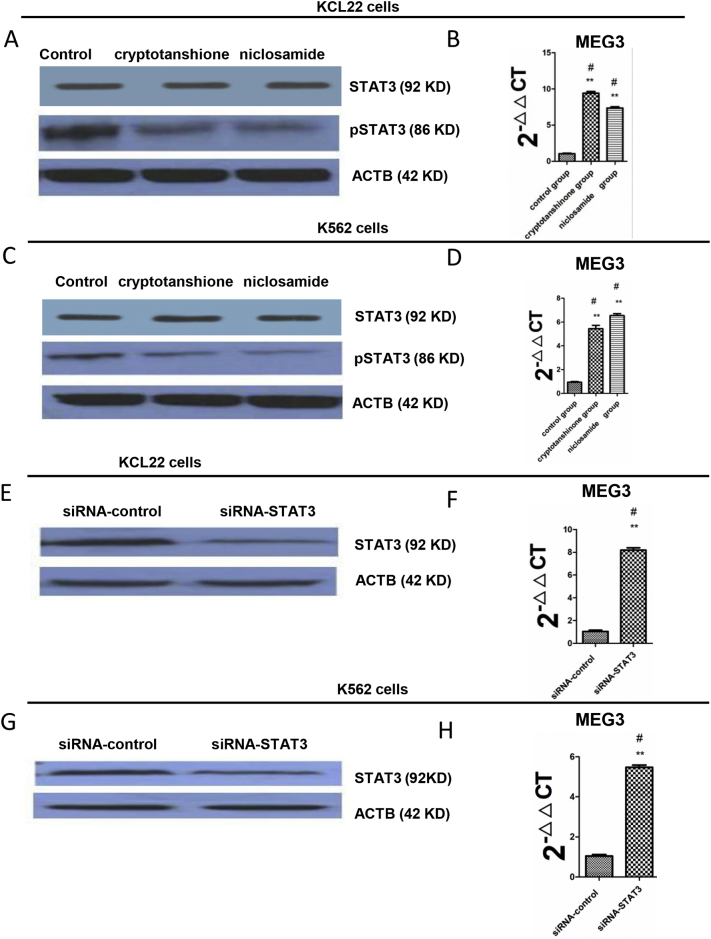
To explore the relationship between STAT3 and MEG3, we first treated K562 and KCL22 cells with cryptotanshione and niclosamide and found a decreased expression of pSTAT3 (Fig. 7A and C). Then, we detected the expression of MEG3 and found that treatment with cryptotanshione and niclosamide significantly increased MEG3 mRNA expression in both cell types (Fig. 7B and D). In addition, we used a siRNA targeting STAT3 and confirmed that STAT3 protein levels were reduced in both K562 and KCL22 cells (Fig. 7E and G). Furthermore, the knockdown of STAT3 increased the mRNA expression of MEG3 in both cell lines (Fig. 7F and H). These data revealed an inverse relationship between STAT3 and MEG3.
Fig. 7.
STAT3 regulated MEG3. A. The KCL22 cells were treated with cryptotanshione and niclosamide (8 μmol/l and 1.5 μmol/l, respectively, for 48 h). A western blot was performed to assess pSTAT3 with ACTB as the loading control. B. MEG3 mRNA levels were assessed by real-time PCR after cryptotanshione and niclosamide treatment and were compared to the control group. C. The K562 cells were treated with cryptotanshione and niclosamide (6 μmol/l and 2 μmol/l, respectively, for 48 h). A western blot was performed to assess pSTAT3 with ACTB as the loading control. D. MEG3 RNA levels were assessed by real-time PCR after cryptotanshione and niclosamide treatment and were compared to the control group. E. STAT3 was knocked down in the KCL22 cells using siRNA and the level of STAT3 was assessed by a western blot with ACTB as the loading control. F. MEG3 expression was assessed by real-time PCR after STAT3 knockdown. G. STAT3 was knocked down in the K562 cells using siRNA and the level of STAT3 was assessed by a western blot with ACTB as the loading control. H. MEG3 expression was assessed by real-time PCR after STAT3 knockdown. For 7B and C, the one-way ANOVA was used for data analysis. For 7F and G, Student's t-test was used for data analysis. **: P < 0.01 compared to the control group.
4. Discussion
CML is a hematopoietic stem cell clonal disorder characterized by a Philadelphia chromosome translocation, resulting in the generation of the Bcr-Abl oncogene encoding a constitutive kinase activity, which induces malignancy in white blood cells [3]. It accounts for approximately 15–20% of newly diagnosed leukemia in adults. Although TKIs, such as imatinib, have made a remarkable success in controlling CML, a significant proportion of CML patients still developed drug resistance to TKIs [4]. Therefore, it is crucial to comprehensively understand the underlying molecular mechanisms about the initiation and progression of CML and find novel therapeutic targets for the treatment of this disease. In this study, we investigated the important roles of a long noncoding RNA MEG3 and its target miR-147 in the regulation of the proliferation and apoptosis of cancer cells as well as the DNA methylation and the expression of histone deacetylation-related genes during the development of CML.
Accumulating evidence has suggested that lncRNAs play diverse roles in human tumorigenesis. Depending on the expression of these lncRNAs in specific tumors, they can function as suppressors or promoters. MEG3 is one of the most important lncRNAs, which has been demonstrated to be involved in multiple biological processes of normal human cells, such as gonadotrophs [27]. Recently, its low expression has been observed in many human cancers, such as bladder, bone marrow, breast, and liver cancers [28]. Consistent with these findings, we also found that the expression level of MEG3 was significantly lower in the different phases of CML patients than that of healthy donors. Notably, our results showed that the MEG3 promoter was methylated in CML-BP patients, while that was not found in healthy donors. Consistently, the methylated MEG3 promoter was also not found in KCL22 and K562 cells after the treatment of chidamide. Thus, these results indicated that there might be a close relationship between the methylation status of MEG3 promoter and its expression level in the regulation of leukemia development, which needs to be investigated in future studies.
MicroRNAs (miRNAs) are a conserved family of small noncoding RNA molecules that post-transcriptionally regulate gene expression. MiR-147 is a recently identified cancer-related miRNA. However, studies have showed that miR-147 could be either oncogene or tumor suppressive gene in different types of human cancers. For example, MiR-147 was found to be up-regulated in human gastric and liver cancers, while was found to be down-regulated in human breast cancer [29]. Our findings showed that the expression level of miR-147 was significantly lower in the different phases of CML patients than that of healthy donors. Consistent with the result of MEG3, chidamide treatment also reversed the methylation status of MiR-147 promoter in KCL22 and K562 cells. Interestingly, the results of dual luciferase reporter assay showed that lncRNA MEG3 and miR-147 could regulate each other in a negative manner. One possible explanation of this is that there might be an unknown gene that is targeted by miR-147, and the effect of MEG3 and miR-147 on cell apoptosis might be mediated by this gene [30]. For example, Han et al., found that MEG3 aggravated hypoxia injury in PC12 cells by down-regulating miR-147, and miR-147 further negatively regulated Sox2 expression [31]. It should be noted that our co-immunoprecipitation results showed that JAK2 could interact with TYK2 through MEG3, suggesting that JAK2 might be the gene contributing to the negative interaction between MEG3 and mi147.
DNA methylation and histone deacetylation are two key mechanisms that affect gene expression and gene promoters [16]. The DNMT family of proteins, which includes DNMT1, DNMT3A, and DNMT3B, are mainly involved in the mediation of CpG island methylation [32], whereas the HDAC family regulates histone deacetylation [33]. Consequently, a complex which includes MBD2, MECP1, and HDACs was formed via the DNA methylation, histone deacetylation and the methylation-dependent binding of MECP2 [34, 35]. Given that the methylation status of MEG3 and MiR-147 could be significantly regulated by HDAC inhibitor chidamide, we next detected the expression levels of the methylation-related genes including DNMT1, DNMT3A, DNMT3B, MBD2, HDAC1 and MECP2. The expressions of these proteins were higher in the CML-AP and CML-BP patients than that in the CML-CP patients and healthy individuals. Thus, we speculated that the lower expression levels of MEG3 and miR-147 in the patients with CML were possibly mediated by histone deacetylation and DNA methylation.
We next treated two CML blast phase cell lines KCL22 and K562 with different concentrations of chidamde and found that chidamide treatment led to increased MEG3 and miR-147 levels, which were negatively correlated with the mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2, MECP2 and HDAC1. The effect of chidamide was also confirmed in CD34+ cells isolated from the peripheral blood mononuclear cells of CML patients. Furthermore, MEG3 and miR-147 overexpression inhibited proliferation and promoted apoptosis in these cells. Thus, we speculated that the activity of chidamide on the proliferation and apoptosis of cancer cells might be strongly associated with its inhibitory effects on HDAC1 expression, MBD2/MECP2 complex formation and MEG3 demethylation.
The JAK/STAT signaling pathway plays an important role in regulating cellular processes, such as apoptosis, differentiation, proliferation and migration [36]. The abnormal activation of JAK/STAT led to the occurrence and development of various types of cancers, including hematological malignancies and solid tumors [[37], [38], [39]]. To further study the regulatory role of MEG3 on cell proliferation and apoptosis during the development of CML blast crisis, RNA pulldown and immunoprecipitation assay followed by the mass spectroscopy analysis were performed. Our results showed that DNMT1, JAK2, STAT3, and TYK2 could bind with MEG3. Moreover, TYK2 and STAT3 interacted with JAK2, while knockdown of MEG3 abolished this interaction. Thus, we concluded that MEG3 might function as a bridge between JAK2 and TYK2. Indeed, in vitro cell assay showed that the overexpression of MEG3 decreased the protein levels of phosphorylated JAK2, STAT3, and STAT5. Therefore, we believed that MEG3 can regulate STAT3, at least partly, by inhibiting the phosphorylation of JAK/STAT. Another interesting finding of this study was that when we treated the K562 and KCL22 cells with cryptotanshione, niclosamide and siRNA to decrease the level of STAT3, the expression of MEG3 was significantly increased. These results indicate that there might be a negative feedback loop between MEG3 and STAT3, which needs to be confirmed in animal studies.
In conclusion, our results revealed that MEG3 might bind with miR-147 and regulate the progression of leukemia. We also provided a mechanism by which MEG3 and miR-147-mediated DNA methylation, histone deacetylation and JAK/STAT pathways might contribute to the antitumor effects of chidamide on the development of CML. Therefore, targeting the MEG3-miR-147 axis might represent a novel therapeutic application in leukemia.
The following are the supplementary data related to this article.
Abbreviations
- MEG3
maternally expressed 3
- CML
chronic myeloid leukemia
- CP
chronic phase
- AP
accelerated phase
- BP
blast phase
- LV
lentiviral vector
- RIP
RNA immunoprecipitation
- CoIP
co-immunoprecipitation
- RT-qPCR
real time polymerase chain reaction
- MSP
methylation-specific PCR
- BCR
breakpoint cluster region
- ABL
Abelson murine leukemia
- TKIs
tyrosine kinase inhibitors
- Esrp2
epithelial splicing regulatory protein 2
- HDACs
histone deacetylases
- HDACi
HDAC inhibitors
- IMDM
Iscove's modified Dulbecco's medium
- FBS
fetal bovine serum
- LNA-ASO
locked nucleic acid-antisense oligonucleotide
- MTT
3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide
- DEPC
diethyl pyrocarbonate
- RIPA
radio immune precipitation assay
- SDS-PAGE
sulfate polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
- SD
standard deviation
- DNMT
DNA (cytosine-5)-methyltransferase
- bcl-2
B-cell lymphoma 2
- STAT5
signal transducer and activator of transcription 5
- STAT3
signal transducer and activator of transcription 3
- JAK2
Janus kinase 2
Funding
None.
Acknowledgments
Acknowledgements
None.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
ZL, LY, XL and JL made substantial contributions to design this study. LY contributed to patient collection. XW and YP contributed to solve the experimental technique problems. ZL did all the experiments and wrote the manuscript. LY, XL and JL analyzed the data.
References
- 1.Geary C.G. The story of chronic myeloid leukaemia. Br J Haematol. 2000;110:2–11. doi: 10.1046/j.1365-2141.2000.02137.x. [DOI] [PubMed] [Google Scholar]
- 2.Melo J.V., Barnes D.J. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 3.Deininger M.W.N., Goldman J.M., Melo J.V. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 4.Deininger M., Buchdunger E., Druker B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 5.Soverini S., Mancini M., Bavaro L., Cavo M., Martinelli G. Chronic myeloid leukemia: the paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol Cancer. 2018;17:49. doi: 10.1186/s12943-018-0780-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tano K., Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Yang L., Liu X., Nie Z., Luo J. Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomed Pharmacother. 2018;104:181–192. doi: 10.1016/j.biopha.2018.05.047. [DOI] [PubMed] [Google Scholar]
- 8.Terashima M., Tange S., Ishimura A., Suzuki T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292:82–99. doi: 10.1074/jbc.M116.750950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y.Y., Feng H.M. MEG3 suppresses human pancreatic neuroendocrine tumor cells growth and metastasis by down-regulation of MiR-183. Cell Physiol Biochem. 2017;44:345–356. doi: 10.1159/000484906. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Q., Lin Z., Xu J., Lu Y., Meng Q., Wang C. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting beta-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018;9:253. doi: 10.1038/s41419-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasgupta P., Kulkarni P., Majid S., Varahram S., Hashimoto Y., Bhat N.S. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018 doi: 10.1158/1535-7163.MCT-17-0925. [molcanther.0925.2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang W., Huang T., Zhou Y., Zhang J., Lung R., Tong J. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018;9:92. doi: 10.1038/s41419-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M., Zhang Y., Zhang J., Cai H., Zhang C., Yang Z. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis. 2018;9:189. doi: 10.1038/s41419-017-0218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan T.T., Ren L.L., Shen C.Q., Wang Z.H., Yu Y.N., Liang Q. miR-508 defines the stem-like/mesenchymal subtype in colorectal cancer. Cancer Res. 2018;78:1751–1765. doi: 10.1158/0008-5472.CAN-17-2101. [DOI] [PubMed] [Google Scholar]
- 15.Roman-Gomez J., Jimenez-Velasco A., Agirre X., Castillejo J.A., Navarro G., San Jose-Eneriz E. Epigenetic regulation of human cancer/testis antigen gene, HAGE, in chronic myeloid leukemia. Haematologica. 2007;92:153–162. doi: 10.3324/haematol.10782. [DOI] [PubMed] [Google Scholar]
- 16.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 17.Huebner H., Strick R., Wachter D.L., Kehl S., Strissel P.L., Schneiderstock R. Hypermethylation and loss of retinoic acid receptor responder 1 expression in human choriocarcinoma. J Exp Clin Cancer Res. 2017;36:165. doi: 10.1186/s13046-017-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y.X., Wang Y., Li X., Zhang W., Zhou H.H., Yin J.Y. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics. 2017;18:901. doi: 10.1186/s12864-017-4223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilmann K., Toth R., Bossmann C., Klimo K., Plass C., Gerhauser C. Genome-wide screen for differentially methylated long noncoding RNAs identifies Esrp2 and lncRNA Esrp2-as regulated by enhancer DNA methylation with prognostic relevance for human breast cancer. Oncogene. 2017;36:6446–6461. doi: 10.1038/onc.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Lu H., Fan G., He M., Sun Y., Xu K. A novel interplay between HOTAIR and DNA methylation in osteosarcoma cells indicates a new therapeutic strategy. J Cancer Res Clin Oncol. 2017;143:2189–2200. doi: 10.1007/s00432-017-2478-3. [DOI] [PubMed] [Google Scholar]
- 21.Adams C.M., Hiebert S.W., Eischen C.M. Myc induces miRNA-mediated apoptosis in response to HDAC inhibition in hematologic malignancies. Cancer Res. 2016;76:736. doi: 10.1158/0008-5472.CAN-15-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ceccacci E., Minucci S. Inhibition of histone deacetylases in cancer therapy: lessons from leukaemia. Br J Cancer. 2016;114:605–611. doi: 10.1038/bjc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valiuliene G., Stirblyte I., Cicenaite D., Kaupinis A., Valius M., Navakauskiene R. Belinostat, a potent HDACi, exerts antileukaemic effect in human acute promyelocytic leukaemia cells via chromatin remodelling. J Cell Mol Med. 2015;19:1742–1755. doi: 10.1111/jcmm.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W., Liu H., Liu Z.G., Wang H.S., Zhang F., Wang H. Histone deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation and stabilization of Snail to promote metastasis of hepatoma cells. Cancer Lett. 2018;420:1–13. doi: 10.1016/j.canlet.2018.01.068. [DOI] [PubMed] [Google Scholar]
- 25.Gallagher S.J., Gunatilake D., Beaumont K.A., Sharp D.M., Tiffen J.C., Heinemann A. HDAC inhibitors restore BRAF-inhibitor sensitivity by altering PI3K and survival signalling in a subset of melanoma. Int J Cancer. 2017;142:1926–1937. doi: 10.1002/ijc.31199. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez G.J., Richmond P.A., Bunker E.N., Karman S.S., Azofeifa J., Garnett A.T. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res. 2018;46:1756–1776. doi: 10.1093/nar/gkx1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi N., Wagatsuma H., Wakana S., Shiroishi T., Nomura M., Aisaka K. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Rice K., Wang Y., Chen W., Zhong Y., Nakayama Y. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao Y., Suo A.L., Li Z.F., Liu L.Y., Tian T., Ni L. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 30.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han L., Dong Z., Liu N., Xie F., Wang N. Maternally expressed gene 3 (MEG3) enhances PC12 cell hypoxia injury by targeting MiR-147. Cell Physiol Biochem. 2017;43:2457–2469. doi: 10.1159/000484452. [DOI] [PubMed] [Google Scholar]
- 32.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 33.Riccio A. New endogenous regulators of class I histone deacetylases. Sci Signal. 2010;3 doi: 10.1126/scisignal.3103pe1. [DOI] [PubMed] [Google Scholar]
- 34.Feng Q., Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 36.Kleppe M., Koche R., Zou L., van Galen P., Hill C.E., Dong L. Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell. 2018;33:785–787. doi: 10.1016/j.ccell.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nairismagi M., Gerritsen M.E., Li Z.M., Wijaya G.C., Chia B.K.H., Laurensia Y. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia. 2018;32:1147–1156. doi: 10.1038/s41375-017-0004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z., Liu J., Hu S., Zhu Y., Li S. Serine/threonine kinase 35, a target gene of STAT3, regulates the proliferation and apoptosis of osteosarcoma cells. Cell Physiol Biochem. 2018;45:808–818. doi: 10.1159/000487172. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Xia M., Jin K., Wang S., Wei H., Fan C. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45. doi: 10.1186/s12943-018-0796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]