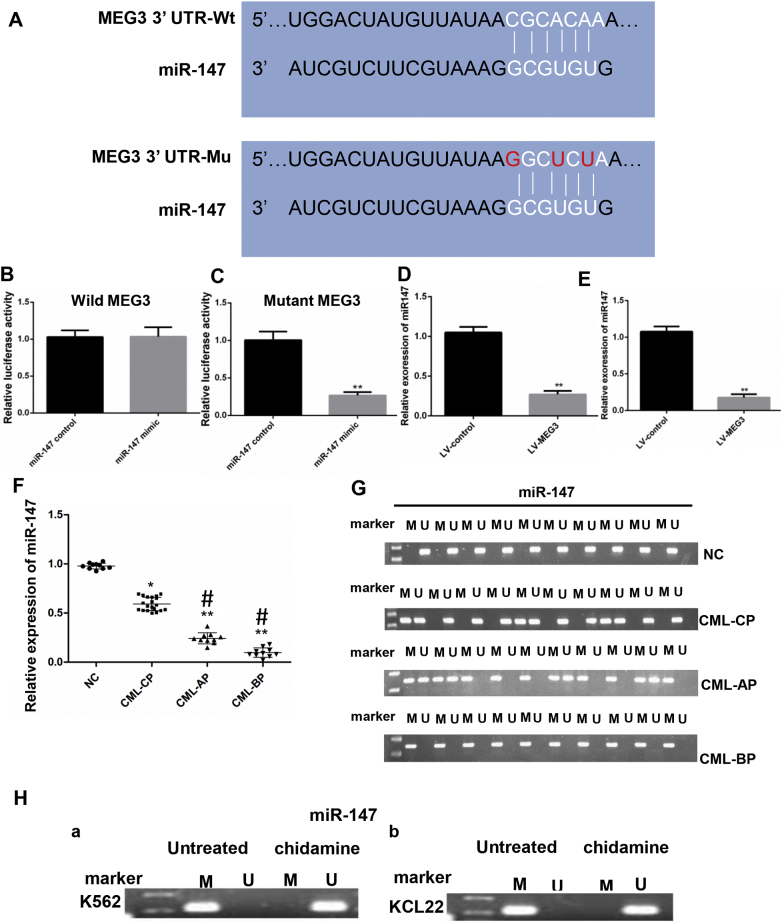
Fig. 4.
The relative luciferase activity was detected by a dual luciferase assay. A. The RNAup algorithm predicted the potential binding of miR-147 to MEG3, with a considerable sequence complementary in the indicated regions. B. The relative luciferase activity was not different between the miR-147 mimics group and the miR-147 control group. C. The relative luciferase activity was lower in the miR-147 mimic group compared to the miR-147 control group. D. In the KCL22 cells, MEG3 overexpression decreased the expression level of miR-147; E. In the K562 cells, MEG3 overexpression decreased the expression level of miR-147. F. The mRNA level of miR-147 was lower in CML-AP and CML-BP patients than that in the CML-CP patients and the healthy donors. G. The methylation of the miR-147 DNA promoter was not detected in the normal controls, and methylation can be detected in 37.5% of the CML-CP samples. All CML-AP samples showed methylation of the miR-147 promoter, and methylation can be detected in 100% of the CML-BP patients. H. After treatment with chidamide, the miR-147 was unmethylated in both the K562 (a) and KCL22 (b) cells. For 4B, C, D and E, Student's t-test was used for data analysis. *: P < 0.05 compared to NC group. **: P < 0.01 compared to NC group. For 4F, the one-way ANOVA was used for data analysis. *: P < 0.05 compared to NC group. **: P < 0.01 compared to NC group. #: P < 0.05 compared to CML-CP group. M, methylated; U, unmethylated.