Abstract
Diamond–Blackfan anemia (DBA) is a rare congenital hypoplastic anemia characterized by a block in erythropoiesis at the progenitor stage, although the exact stage at which this occurs remains to be fully defined. DBA presents primarily during infancy with macrocytic anemia and reticulocytopenia with 50% of cases associated with a variety of congenital malformations. DBA is most frequently due to a sporadic mutation (55%) in genes encoding several different ribosomal proteins, although there are many cases where there is a family history of the disease with varying phenotypes. The erythroid tropism of the disease is still a matter of debate for a disease related to a defect in global ribosome biogenesis. Assessment of biological features in conjunction with genetic testing has increased the accuracy of the diagnosis of DBA. However, in certain cases, it continues to be difficult to firmly establish a diagnosis. This review will focus on the diagnosis of DBA along with a description of new advances in our understanding of the pathophysiology and treatment recommendations for DBA.
Keywords: Diamond-Blackfan anemia, Ribosomapthy, erythropoiesis, inherited bone marrow failure
Introduction
Diamond–Blackfan anemia (DBA) was described for the first time in the 1930’s as a constitutional hypoplastic anemia 1, 2. There was a gap of almost 60 years after the first description of the disease 2, 3 before the first gene was identified in DBA, namely ribosomal protein (RP) S19 ( RPS19) in 1999 4. Surprisingly, for a disease in which the major defect is disordered erythropoiesis, the genes involved in the disease belong to both small and large subunits of the ribosome, which would be expected to have widespread consequences. The mutant RP is responsible for a defect in rRNA maturation, which is the signature feature for most DBA cases. DBA was indeed described as the first ribosomopathy in 2005 5. Since this original classification, the actual definition of DBA is evolving with further developments in the genetic basis for the disorder, with the discovery of new DBA genes that are not directly involved in ribosome biogenesis. In this review, we will update these new insights into our understanding of DBA.
Clinical features
DBA typically presents in infancy, most commonly with pallor and lethargy, at an estimated incidence of seven cases per million live births within some families who have a history of the disease. The median age at presentation is 8 weeks, with a median age at diagnosis of 12 weeks. There have been cases of hydrops fetalis 6, 7. The male-to-female ratio of cases is approximately 1:1 despite rare cases of X-linked inheritance. More than 90% of the reported cases present clinically by 1 year of age. DBA is characterized by a macrocytic moderate or severe anemia in association with aregenerative bone marrow and reticulocytopenia. The disorder is also characterized by elevated erythrocyte adenosine deaminase (eADA) activity in over 75% of cases.
The DBA Registry of North America (DBAR), a database of more than 700 patients, was established in 1991 and provides important information regarding the epidemiology and biology of DBA 8– 12. Almost half of patients exhibit physical abnormalities, except short stature, which is a known feature of DBA but could also be a result of chronic anemia, iron overload, corticosteroid administration, or a combination of all three. Included in the constellation of physical anomalies are a high number of craniofacial anomalies (50% of patients), upper limb and hand––in particular thumb (38%)––abnormalities, and genitourinary (39%) as well as cardiac (30%) abnormalities.
DBA is recognized as a cancer predisposition syndrome with an observed to expected ratio for all cancers of 5.4. The most common malignancies were MDS, AML, colon carcinoma, osteosarcoma, and genitourinary cancers 12– 15.
Of interest, there appears to be no genotype–phenotype correlation with regard to steroid responsiveness, remission, or cancer predisposition. However, RPL5 gene mutations have been associated with cleft palate malformation and are the most important rate of malformations in DBA cases 16, 17, while RPL11 mutations are associated with the classic triphalangeal thumb 17. Recently, mutations in the RPL15 gene have been identified in cases of hydrops fetalis in DBA patients 7, and RPL35a gene mutations are associated with neutropenia.
Biological features
DBA is one of the inherited bone marrow failure (IBMF) syndromes that include Fanconi anemia, Shwachman–Bodian–Diamond syndrome, dyskeratosis congenita, and cartilage hair hypoplasia 18– 24. All of these syndromes have a quantitative defect in hematopoiesis. Among the IBMF syndromes, DBA is unique in that it involves a specific intrinsic quantitative defect in erythropoiesis 25.
There is strong evidence that the erythroid blockage likely occurs between the BFU-e and CFU-e stage of erythroid development 26. It should be noted that some previous reports have suggested a general blockade upstream during hematopoiesis, since long-term culture experiments have shown a defect in megakaryocytic and granulocytic progenitors 27, 28 and there are rare cases of DBA which progress to a complete aplasia 14, 29.
The erythroid blockade is responsible for the erythroblastopenia characterized by the absence or less than 5% of erythroid progenitors in the bone marrow aspirate or an important paucity of the erythroid progenitors in the bone marrow biopsy in an otherwise normal bone marrow with no qualitative dyserythropoiesis or defects in other hematopoietic cell lineages. Neutropenia and thrombocytopenia, and in some instances thrombocytosis, have been described at diagnosis or during DBA evolution, implying that DBA diagnosis should not be ruled out when these particular blood cell anomalies are noted at DBA presentation.
Strikingly, DBA is associated with an increased eADA activity 30– 33. eADA is a critical enzyme of the purine salvage pathway, which enables the deamination of adenosine in inosine and 2'-deoxyadenosine deamination in deoxyinosine. In the French registry of over 300 DBA patients, eADA has been found to be elevated in 90% of non-transfused DBA patients as reported in a previous study 32 and in 75% of DBA patients from the American registry with a sensitivity of 84%, specificity of 95%, and positive and negative predictive values of 91% for the diagnosis of DBA compared with other IBMF syndromes 31. While an elevated eADA activity is a strong feature of DBA, it is also increased in some leukemias, lymphomas, and immune system disorders 34. The challenge in performing eADA testing is that the test is not routinely available and is currently performed in only one lab in each of the following countries: the USA, France, Germany, Italy, Poland, Israel, and Turkey 35. It should be noted that the test needs to be performed on fresh blood samples or samples stored at 4°C for less than a few days and on samples prior to red cell transfusions.
In order to eliminate the most frequent differential diagnosis, namely a parvovirus B19 infection, parvovirus B19 serology (IgM/IgG) or parvovirus B19 PCR in the blood (or in the bone marrow, which has a higher sensitivity) is mandatory.
The other biological tests that may be useful in DBA diagnosis are 1) the erythropoietin (EPO) level, which is consistently elevated in DBA as a result of a lack of effective erythropoiesis with a normal kidney response to the anemia and a quantitative deficiency of the EPO receptors that bind EPO due to the large decreases in the number of erythroid precursors, and 2) immunophenotyping and IgG/IgA agglutinin titer. A DAT test in association with an erythroid clonogenic in vitro culture assay with and without the patient’s sera may be helpful in rare cases, mostly in adults, in order to eliminate immune erythroblastopenia in doubtful DBA cases.
Molecular diagnosis
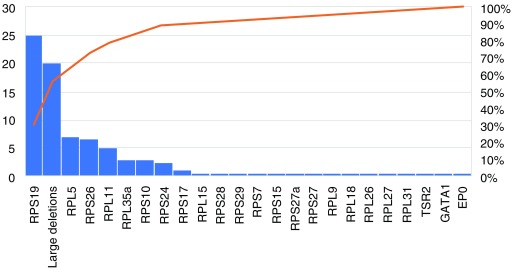
The first gene associated with DBA was identified in 1999 in a Swedish patient with DBA 4, 36 who carried a balanced translocation between the chromosomes X and 19. Since DBA exhibits a 1:1 sex ratio, it was thought unlikely to be of X-linked inheritance, and the candidate gene in 19q chromosomal breakpoint was explored and identified. Surprisingly, for a disease with an erythroid tropism, the identified gene was an RP from the small ribosome subunit gene RPS19. Subsequently, mutations or deletions in 19 other RP genes have been identified by whole exome/genome sequencing and CGH/SNP array. These include RPL5, RPL11, RPL35a, RPS10, RPS24, RPS17, RPL15, RPS28, RPS29, RPS7, RPS15, RPS27a, RPS27, RPL9, RPL18, RPL26, RPL27, and RPL31 as well as three other non-RP genes, TSR2, GATA1, and EPO ( Table 1, Figure 1, and 4, 16, 17, 35, 37– 49). It is still debated if the disease associated with non-RP genes is classical DBA or “DBA-like” disease.
Table 1. Genes involved in Diamond–Blackfan anemia (DBA) from the most mutated to reported cases, incidence, and references.
| Mutated gene | Incidence in DBA
population |
References |
|---|---|---|
| RPS19 | 25% | Draptchinskaia
et al.
4
Willig et al. 51 Ramenghi et al. 55 Cmejla et al. 56 Proust et al. 57 Campagnoli et al. 58 |
| Large deletions | 10–20% | Gustavsson
et al.
59
Quarello et al. 60 Farrar et al. 39 Quarello et al. 49 Kuramitsu et al. 48 |
| RPL5 | 7% | Gazda
et al.
17
Cmejla et al. 16 Quarello et al. 61 |
| RPS26 | 6.6% | Doherty et al. 37 |
| RPL11 | 5% | Gazda
et al.
17
Cmejla et al. 16 Quarello et al. 61 |
| RPL35a | 3% | Farrar et al. 38 |
| RPS10 | 3% | Doherty et al. 37 |
| RPS24 | 2.4% | Gazda et al. 41 |
| RPS17 | 1% | Cmejla
et al.
42
Song et al. 62 |
| RPL15 | One case
Six cases |
Landowski
et al.
40
Wlodarski et al. 7 |
| RPS28 | Two families | Gripp et al. 52 |
| RPS29 | Two families | Mirabello et al. 45 |
| RPS7 | One case | Gazda et al. 17 |
| RPS15 | One case | Gazda et al. 17 |
| RPS27a | One case | Gazda et al. 17 |
| RPS27 | One case | Wang et al. 63 |
| RPL9 | One case
Two cases |
Gazda
et al.
17
euroDBA group, in preparation |
| RPL18 | One family | Mirabello et al. 64 |
| RPL26 | One case | Gazda et al. 65 |
| RPL27 | One case | Wang et al. 63 |
| RPL31 | One case | Farrar et al. 66 |
| TSR2 (X-linked) | One family | Gripp et al. 52 |
| GATA1 (X-linked) | Five families | Sankaran
et al.
43
Klar et al. 67 Ludwig et al. 53 Parrella et al. 68 |
| EPO | One case | Kim et al. 44 |
Figure 1. Representation of the frequency of the mutated genes involved in Diamond–Blackfan anemia (DBA) from DBA-affected populations all over the world (literature data).
DBA is thus a polygenic disease with mutations in 20 of the 80 RP genes that code for the complete ribosome. Interestingly, mutations including deletions in six of 20 identified genes, namely RPS19, RPL5, RPS26, RPL11, RPL35a, and RPS24, account for 70% of all DBA cases ( Figure 1). Some RP gene mutations exhibit a phenotype–genotype relationship (see “clinical features” above). All the RP mutations identified to date are heterozygous; homozygosity is thought to be lethal, which has been confirmed in zebrafish and murine models of DBA 50. Multiple pathogenic RP mutations have not been reported in any DBA patient to date. Except for RPS19 51, no hot spot regions in the DBA genes have been reported. Various types of mutations may be associated with DBA phenotype, although most RPS19 gene mutations are missense mutations, while nonsense mutations are more frequent in RPL5- and RPL11-associated DBA. Large deletions have been reported in 20% of patients, which makes large deletions the second most frequent genetic defect after RPS19 gene mutation (25%) in DBA.
With regard to the “non-RP” genes linked to DBA, TSR2 is known to play a role in ribosome biogenesis, since it is involved in the pre-rRNA processing and binds to RPS26 52. GATA1 is the major erythroid transcription factor and plays a critical role in regulating normal erythroid differentiation by activating an array of erythroid genes. Ludwig et al. 53 have shown that GATA1 transcripts are specifically less translated compared to others in DBA owing to a higher threshold for initiation of translation of GATA1 mRNA due to defective ribosomal biogenesis. Gastou et al. 54 showed that HSP70, the chaperon of GATA1, is degraded by the proteasome following polyubiquitination during the BFU-E and CFU-E stages of erythropoiesis. Decreased HSP70 expression has been noted in all of the RP mutated-gene-tested DBA patients and in shRNA models other than RPS19, which exhibit a normal expression of HSP70. This correlates perfectly with the low level of induced apoptosis in these RPS19-mutated DBA patients compared to the RPL5- or RPL11-mutated ones. Interestingly, HSP70 degradation is responsible for caspase-3-dependent GATA1 cleavage during terminal erythroid differentiation and the resultant decreased GATA1 protein expression at late stages of erythroid differentiation 54. Finally, the EPO gene has been found to be mutated in one consanguineous Turkish family in the USA, with a homozygous missense mutation in exon 5 of EPO. The pathogenicity of this mutation has been validated in vitro by functional analysis with a well-established defect in erythroid proliferation and differentiation 44.
Following detailed mutational screening analysis using various methods, no molecular defect can be documented in 20 to 30% of DBA cases. It is likely that mutations in a regulatory region including intronic regions and promoters in one of the known RP genes may account for the DBA phenotype. It is also important to emphasize that the first step in molecular diagnosis should always be to characterize the phenotype including family history, pregnancy complications, congenital malformations, and characteristics of the anemia including an evaluation of the bone marrow, which we still believe is an essential part of the evaluation. Indeed, hypoplastic anemia can be seen as a part of some congenital dyserythropoietic anemias (CDAs) 69 and acquired minus 5q syndrome 70; therefore, the frontier between DBA and these syndromes may sometimes be difficult to ascertain without a meticulous clinical and microscopic examination by an expert hematologist and hematopathologist.
DBA pathophysiology
In terms of the history of insight into DBA as a disease, there was widespread use of erythroid cell cultures from the 1950’s to the end of the 1990’s prior to the advent of the molecular biological approaches. In the earlier stages, immune mechanisms (humoral or cell-mediated immune inhibition by T-cytotoxic or T-helper lymphocytes) were thought to play a role in the pathophysiology of DBA owing to the effectiveness of corticosteroid therapy in correcting the anemia phenotype 71– 73. However, no conclusive evidence in support of immune-meditated suppression of erythropoiesis could be documented.
A defective microenvironment as a major cause of DBA pathophysiology was also ruled out because of the effectiveness of bone marrow transplantation as a curative treatment in DBA and also from findings that documented normal erythroid proliferation of control CD34 + cells cultured in a DBA microenvironment 28, 74. DBA can thus be defined as an intrinsic defect in erythropoiesis due to defective ribosome processing. However, the exact stage at which blockade occurs during erythropoiesis still needs to be fully defined. The best documented study 26 implies a blockade between the BFU-E and the CFU-E stages or between the EPO-independent and the EPO-dependent stages of erythroid development. However, data from erythroid culture studies from DBA patients are highly heterogeneous and depend on a number of variables including the erythroid culture methods used. There is, however, some convincing evidence that different RP mutations exhibit significant differences in the erythroid proliferation phenotype 54, 75.
The major unresolved questions in DBA remain how a defect in RP is responsible for a specific defect in erythropoiesis and why there is a different penetrance of the same mutation among different individuals. Red blood cells do exhibit a high level of cell production (2 × 10 11/day) in humans, and the level of protein translation needed is indeed very high, which may explain, in part, the erythroid tropism of DBA. In this context, any factor that affects GATA1 expression would be deleterious in DBA as well. Indeed, HSP70 54, and the recently reported ribonuclease inhibitor 1 (RNH1) 76 that binds to the 40S ribosome small subunit, could be involved in the translational control of GATA1 and consequently affect erythropoiesis, resulting in the DBA phenotype. In addition, ribosome amounts have been shown to be critical for the cell lineage commitment. Recently, a global reduction in ribosome levels in DBA has been documented, while the ribosome composition was normal, which altered the translation of specific RNA transcripts 77. These exciting new discoveries are enabling the establishment of a link among a mutated RP gene, a defect in ribosome biogenesis, and the specific effect on erythropoiesis in DBA.
p53 also plays an important role in DBA pathophysiology, with p53 activation and the altered expression of its targets (p21, Bax, noxa) having been documented in erythroid cells from affected patients and in CD34 + cord blood erythroid progenitors following lentiviral transfection with various shRNAs targeting RPS19, RPL5, or RPL11 transcripts 75, 78. The relationship between p53 and GATA1 is well established, with GATA1 inhibiting p53 79. It has also been shown that wild-type HSP70 overexpression, by restoring GATA1, was also able to decrease p53 activation (phosphorylated p53) in RPL5- and RPL11-depleted erythroid cells 54. Decreased expression of GATA1 may thus account for p53 activation in DBA. Indeed, activation of p53 in DBA may be due to not only the nucleolar stress and overexpression of some RPs (RPS3, RPS7, RPS27, RPS27a, RPL5, RPL11, and RPL23), which are able to directly bind MDM2 in order to release the p53/MDM2 binding, but also the defect in GATA1 in DBA.
Another level of complexity in DBA involves the role of free excess heme in DBA pathophysiology due to imbalance between decreased globin synthesis and excess of free heme, which can generate reactive oxygen species and increased apoptosis, leading to the death of erythroid progenitors and precursors. Autophagy and cell metabolism have also been shown to be important in DBA pathophysiology 80, 81.
Thus, it appears that deciphering the complex interplay between the multiple mechanisms identified to date, and perhaps others yet to be discovered, is needed to develop detailed understanding of the documented variations in clinical severity of the highly heterogeneous DBA phenotype.
Therapeutic options
The current standard of care for DBA includes corticosteroids and/or chronic transfusions with the only definitive treatment (for the hematologic complications) being bone marrow transplantation. Approximately 80% of patients respond initially to corticosteroids with an improvement in, or complete remission of, their anemia. However, prolonged corticosteroid treatment has been problematic for many patients such that only about 40% will remain on corticosteroids for a considerable period of time. With existing treatments, the overall survival of patients, as reported by the DBAR, is 75% at 40 years of age; median overall survival is 58 years. As our understanding of the pathophysiology of the ribosomopathies increases, the goal is to be able to translate these findings into novel therapeutic options for patients with DBA.
Recently, the use of hematopoietic stem cell transplantation (HSCT) in DBA patients is increasing and has provided encouraging results. August and colleagues reported the first successful transplant for DBA in 1976 82. Alter reviewed stem cell transplantation in DBA in 1998 83; in an analysis of 35 of the 37 cases reported until that point, the actuarial survival for primarily allogeneic HLA-matched donor transplants was 66%. Recent work has proposed that this figure is approximately 90% for matched, related HSCT in young, otherwise healthy patients 84, 85. The latest analysis from the DBAR found that HLA-matched related-donor transplant resulted in an overall survival of 76.9 ± 8.4% and, for patients aged 9 years or younger, survival was 93.8 ± 6.1%. In transplants from an unrelated donor, the overall survival showed an improvement: in 1994 to 1999, it was reported to be 32.1 ± 11.7%, and in 2000 to the present, it was 85.7 ± 13.2% 11. A “How I Treat” article by Vlachos and Muir 86 provides a detailed approach to the treatment of DBA. There are also several open clinical trials (details available at clinicaltrials.gov) for patients with DBA and more in development.
Conclusion
DBA, a rare congenital hypoplastic anemia characterized by a block in erythropoiesis, is most frequently due to a sporadic mutation in genes encoding several different RPs. DBA was indeed the first identified human ribosomopathy 5. The erythroid tropism of the disease is still a matter of debate for a disease related to a defect in global ribosome biogenesis. Assessment of biological features in conjunction with genetic testing have increased the accuracy for the diagnosis of DBA. The current standard of care for DBA includes corticosteroids and/or chronic transfusions, with the only definitive treatment being bone marrow transplantation.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Steven R. Ellis, Department of Biochemistry and Molecular Genetics, University of Louisville, Louisville, Kentucky, USA
Jeffrey M. Lipton, Feinstein Institute for Medical Research, Manhasset, New York, USA; Division of Hematology/Oncology and Stem Cell Transplantation, Cohen Children's Medical Center of New York, New Hyde Park, New York, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 2 approved]
References
- 1. Diamond LK: Congenital hypoplastic anemia: Diamond-Blackfan syndrome. Historical and clinical aspects. Blood Cells. 1978;4(1–2):209–13. [PubMed] [Google Scholar]
- 2. Diamond LK, Blackfan, KD: Hypoplastic anemia. Am J Dis Child. 1938;56:464–467. [Google Scholar]
- 3. Josephs HW: Anaemia of infancy and early childhood. Medicine. 1936;15:307–451. 10.1097/00005792-193615030-00001 [DOI] [Google Scholar]
- 4. Draptchinskaia N, Gustavsson P, ersson B, et al. : The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–75. 10.1038/5951 [DOI] [PubMed] [Google Scholar]
- 5. Léger-Silvestre I, Caffrey JM, Dawaliby R, et al. : Specific Role for Yeast Homologs of the Diamond Blackfan Anemia-associated Rps19 Protein in Ribosome Synthesis. J Biol Chem. 2005;280(46):38177–85. 10.1074/jbc.M506916200 [DOI] [PubMed] [Google Scholar]
- 6. Da Costa L, Chanoz-Poulard G, Simansour M, et al. : First de novo mutation in RPS19 gene as the cause of hydrops fetalis in Diamond-Blackfan anemia. Am J Hematol. 2013;88(2):160. 10.1002/ajh.23366 [DOI] [PubMed] [Google Scholar]
- 7. Wlodarski MW, Da Costa L, O'Donohue MF, et al. : Recurring mutations in RPL15 are linked to hydrops fetalis and treatment independence in Diamond-Blackfan anemia. Haematologica. 2018;103(6):949–58. 10.3324/haematol.2017.177980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vlachos A, Osorio DS, Atsidaftos E, et al. : Increased Prevalence of Congenital Heart Disease in Children With Diamond Blackfan Anemia Suggests Unrecognized Diamond Blackfan Anemia as a Cause of Congenital Heart Disease in the General Population: A Report of the Diamond Blackfan Anemia Registry. Circ Genom Precis Med. 2018;11(5):e002044. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Vlachos A, Klein GW, Lipton JM: The Diamond Blackfan Anemia Registry: tool for investigating the epidemiology and biology of Diamond-Blackfan anemia. J Pediatr Hematol Oncol. 2001;23(6):377–82. 10.1097/00043426-200108000-00015 [DOI] [PubMed] [Google Scholar]
- 10. Vlachos A, Ball S, Dahl N, et al. : Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142(6):859–76. 10.1111/j.1365-2141.2008.07269.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vlachos A, Federman N, Reyes-Haley C, et al. : Hematopoietic stem cell transplantation for Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Bone Marrow Transplant. 2001;27(4):381–6. 10.1038/sj.bmt.1702784 [DOI] [PubMed] [Google Scholar]
- 12. Vlachos A, Rosenberg PS, Atsidaftos E, et al. : Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119(16):3815–9. 10.1182/blood-2011-08-375972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lipton JM, Federman N, Khabbaze Y, et al. : Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23(1):39–44. 10.1097/00043426-200101000-00009 [DOI] [PubMed] [Google Scholar]
- 14. Alter BP, Giri N, Savage SA, et al. : Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30–9. 10.3324/haematol.2017.178111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Lipton JM, Alter BP: Heritable cancer: Rounding up the not so usual suspects. Pediatr Blood Cancer. 2017;64(2):219–20. 10.1002/pbc.26190 [DOI] [PubMed] [Google Scholar]
- 16. Cmejla R, Cmejlova J, Handrkova H, et al. : Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat. 2009;30(3):321–7. 10.1002/humu.20874 [DOI] [PubMed] [Google Scholar]
- 17. Gazda HT, Sheen MR, Vlachos A, et al. : Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769–80. 10.1016/j.ajhg.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Dokal I: Dyskeratosis congenita. Hematology Am Soc Hematol Educ Program. 2011;2011:480–6. 10.1182/asheducation-2011.1.480 [DOI] [PubMed] [Google Scholar]
- 19. Dokal I, Vulliamy T, Mason P, et al. : Clinical utility gene card for: Dyskeratosis congenita - update 2015. Eur J Hum Genet. 2015;23(4):558. 10.1038/ejhg.2014.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Donadieu J, Fenneteau O, Beaupain B, et al. : Classification of and risk factors for hematologic complications in a French national cohort of 102 patients with Shwachman-Diamond syndrome. Haematologica. 2012;97(9):1312–9. 10.3324/haematol.2011.057489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dror Y, Donadieu J, Koglmeier J, et al. : Draft consensus guidelines for diagnosis and treatment of Shwachman-Diamond syndrome. Ann N Y Acad Sci. 2011;1242(1):40–55. 10.1111/j.1749-6632.2011.06349.x [DOI] [PubMed] [Google Scholar]
- 22. Malric A, Defachelles AS, Leblanc T, et al. : Fanconi anemia and solid malignancies in childhood: a national retrospective study. Pediatr Blood Cancer. 2015;62(3):463–70. 10.1002/pbc.25303 [DOI] [PubMed] [Google Scholar]
- 23. Pinto FO, Leblanc T, Chamousset D, et al. : Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica. 2009;94(4):487–95. 10.3324/haematol.13592 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Soulier J: Fanconi anemia. Hematology Am Soc Hematol Educ Program. 2011;2011(1):492–7. 10.1182/asheducation-2011.1.492 [DOI] [PubMed] [Google Scholar]
- 25. Tsai PH, Arkin S, Lipton JM: An intrinsic progenitor defect in Diamond-Blackfan anaemia. Br J Haematol. 1989;73(1):112–20. 10.1111/j.1365-2141.1989.tb00229.x [DOI] [PubMed] [Google Scholar]
- 26. Ohene-Abuakwa Y, Orfali KA, Marius C, et al. : Two-phase culture in Diamond Blackfan anemia: localization of erythroid defect. Blood. 2005;105(2):838–46. 10.1182/blood-2004-03-1016 [DOI] [PubMed] [Google Scholar]
- 27. Casadevall N, Croisille L, Auffray I, et al. : Age-related alterations in erythroid and granulopoietic progenitors in Diamond-Blackfan anaemia. Br J Haematol. 1994;87(2):369–75. 10.1111/j.1365-2141.1994.tb04924.x [DOI] [PubMed] [Google Scholar]
- 28. Santucci MA, Bagnara GP, Strippoli P, et al. : Long-term bone marrow cultures in Diamond-Blackfan anemia reveal a defect of both granulomacrophage and erythroid progenitors. Exp Hematol. 1999;27(1):9–18. 10.1016/S0301-472X(98)00068-X [DOI] [PubMed] [Google Scholar]
- 29. Alter BP: Inherited bone marrow failure syndromes: considerations pre- and posttransplant. Hematology Am Soc Hematol Educ Program. 2017;2017(1):88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Glader BE, Backer K, Diamond LK: Elevated erythrocyte adenosine deaminase activity in congenital hypoplastic anemia. N Engl J Med. 1983;309(24):1486–90. 10.1056/NEJM198312153092404 [DOI] [PubMed] [Google Scholar]
- 31. Narla A, Davis NL, Lavasseur C, et al. : Erythrocyte adenosine deaminase levels are elevated in Diamond Blackfan anemia but not in the 5q- syndrome. Am J Hematol. 2016;91(12):E501–E502. 10.1002/ajh.24541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Willig TN, Pérignon JL, Gustavsson P, et al. : High adenosine deaminase level among healthy probands of Diamond Blackfan anemia (DBA) cosegregates with the DBA gene region on chromosome 19q13. The DBA Working Group of Société d'Immunologie Pédiatrique (SHIP). Blood. 1998;92(11):4422–7. [PubMed] [Google Scholar]
- 33. Fargo JH, Kratz CP, Giri N, et al. : Erythrocyte adenosine deaminase: diagnostic value for Diamond-Blackfan anaemia. Br J Haematol. 2013;160(4):547–54. 10.1111/bjh.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Glader BE, Backer K: Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases. Br J Haematol. 1988;68(2):165–8. 10.1111/j.1365-2141.1988.tb06184.x [DOI] [PubMed] [Google Scholar]
- 35. Da Costa L, O'Donohue MF, van Dooijeweert B, et al. : Molecular approaches to diagnose Diamond-Blackfan anemia: The EuroDBA experience. Eur J Med Genet. 2017; pii: S1769-7212(17)30505-0. 10.1016/j.ejmg.2017.10.017 [DOI] [PubMed] [Google Scholar]
- 36. Gustavsson P, Skeppner G, Johansson B, et al. : Diamond-Blackfan anaemia in a girl with a de novo balanced reciprocal X;19 translocation. J Med Genet. 1997;34(9):779–82. 10.1136/jmg.34.9.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doherty L, Sheen MR, Vlachos A, et al. : Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86(2):222–8. 10.1016/j.ajhg.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Farrar JE, Nater M, Caywood E, et al. : Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582–92. 10.1182/blood-2008-02-140012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farrar JE, Vlachos A, Atsidaftos E, et al. : Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118(26):6943–51. 10.1182/blood-2011-08-375170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Landowski M, O'Donohue MF, Buros C, et al. : Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum Genet. 2013;132(11):1265–74. 10.1007/s00439-013-1326-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gazda HT, Grabowska A, Merida-Long LB, et al. : Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79(6):1110–8. 10.1086/510020 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Cmejla R, Cmejlova J, Handrkova H, et al. : Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28(12):1178–82. 10.1002/humu.20608 [DOI] [PubMed] [Google Scholar]
- 43. Sankaran VG, Ghazvinian R, Do R, et al. : Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439–43. 10.1172/JCI63597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim AR, Ulirsch JC, Wilmes S, et al. : Functional Selectivity in Cytokine Signaling Revealed Through a Pathogenic EPO Mutation. Cell. 2017;168(6):1053–1064.e15. 10.1016/j.cell.2017.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Mirabello L, Macari ER, Jessop L, et al. : Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood. 2014;124(1):24–32. 10.1182/blood-2013-11-540278 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Boria I, Garelli E, Gazda HT, et al. : The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum Mutat. 2010;31(12):1269–79. 10.1002/humu.21383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boria I, Quarello P, Avondo F, et al. : A new database for ribosomal protein genes which are mutated in Diamond-Blackfan Anemia. Hum Mutat. 2008;29(11):E263–70. 10.1002/humu.20864 [DOI] [PubMed] [Google Scholar]
- 48. Kuramitsu M, Sato-Otsubo A, Morio T, et al. : Extensive gene deletions in Japanese patients with Diamond-Blackfan anemia. Blood. 2012;119(10):2376–84. 10.1182/blood-2011-07-368662 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Quarello P, Garelli E, Brusco A, et al. : High frequency of ribosomal protein gene deletions in Italian Diamond-Blackfan anemia patients detected by multiplex ligation-dependent probe amplification assay. Haematologica. 2012;97(12):1813–7. 10.3324/haematol.2012.062281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsson H, Davey EJ, Fröjmark AS, et al. : Erythropoiesis in the Rps19 disrupted mouse: Analysis of erythropoietin response and biochemical markers for Diamond-Blackfan anemia. Blood Cells Mol Dis. 2006;36(2):259–64. 10.1016/j.bcmd.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 51. Willig TN, Draptchinskaia N, Dianzani I, et al. : Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94(12):4294–306. [PubMed] [Google Scholar]
- 52. Gripp KW, Curry C, Olney AH, et al. : Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A(9):2240–9. 10.1002/ajmg.a.36633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ludwig LS, Gazda HT, Eng JC, et al. : Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748–53. 10.1038/nm.3557 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Gastou M, Rio S, Dussiot M, et al. : The severe phenotype of Diamond-Blackfan anemia is modulated by heat shock protein 70. Blood Adv. 2017;1(22):1959–76. 10.1182/bloodadvances.2017008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ramenghi U, Campagnoli MF, Garelli E, et al. : Diamond-Blackfan anemia: report of seven further mutations in the RPS19 gene and evidence of mutation heterogeneity in the Italian population. Blood Cells Mol Dis. 2000;26(5):417–22. 10.1006/bcmd.2000.0324 [DOI] [PubMed] [Google Scholar]
- 56. Cmejla R, Blafkova J, Stopka T, et al. : Ribosomal protein S19 gene mutations in patients with diamond-blackfan anemia and identification of ribosomal protein S19 pseudogenes. Blood Cells Mol Dis. 2000;26(2):124–132. 10.1006/bcmd.2000.0286 [DOI] [PubMed] [Google Scholar]
- 57. Proust A, Da Costa L, Rince P, et al. : Ten novel Diamond-Blackfan anemia mutations and three polymorphisms within the rps19 gene. Hematol J. 2003;4(2):132–136. [DOI] [PubMed] [Google Scholar]
- 58. Campagnoli MF, Ramenghi U, Armiraglio M, et al. : RPS19 mutations in patients with Diamond-Blackfan anemia. Hum Mutat. 2008;29(7):911–920. 10.1002/humu.20752 [DOI] [PubMed] [Google Scholar]
- 59. Gustavsson P, Garelli E, Draptchinskaia N, et al. : Identification of microdeletions spanning the Diamond-Blackfan anemia locus on 19q13 and evidence for genetic heterogeneity. Am J Hum Genet. 1998;63(5):1388–1395. 10.1086/302100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Quarello P, Garelli E, Brusco A, et al. : Multiplex ligation-dependent probe amplification enhances molecular diagnosis of Diamond-Blackfan anemia due to RPS19 deficiency. Haematologica. 2008;93(11):1748–1750. 10.3324/haematol.13423 [DOI] [PubMed] [Google Scholar]
- 61. Quarello P, Garelli E, Carando A, et al. : Diamond-Blackfan anemia: genotype-phenotype correlations in Italian patients with RPL5 and RPL11 mutations. Haematologica. 2010;95(2):206–213. 10.3324/haematol.2009.011783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song MJ, Yoo EH, Lee KO, et al. : A novel initiation codon mutation in the ribosomal protein S17 gene ( RPS17) in a patient with Diamond-Blackfan anemia. Pediatr Blood Cancer. 2010;54(4):629–631. 10.1002/pbc.22316 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Wang R, Yoshida K, Toki T, et al. : Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br J Haematol. 2015;168(6):854–864. 10.1111/bjh.13229 [DOI] [PubMed] [Google Scholar]
- 64. Mirabello L, Khincha PP, Ellis SR, et al. : Novel and known ribosomal causes of Diamond-Blackfan anaemia identified through comprehensive genomic characterisation. J Med Genet. 2017;54(6):417–425. 10.1136/jmedgenet-2016-104346 [DOI] [PubMed] [Google Scholar]
- 65. Gazda HT, Preti M, Sheen MR, et al. : Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum Mutat. 2012;33(7):1037–1044. 10.1002/humu.22081 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Farrar JE, Quarello P, Fisher R, et al. : Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. Am J Hematol. 2014;89(10):985–991. 10.1002/ajh.23807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Klar J, Khalfallah A, Arzoo PS, et al. : Recurrent GATA1 mutations in Diamond-Blackfan anaemia. Br J Haematol. 2014;166(6):949–951. 10.1111/bjh.12919 [DOI] [PubMed] [Google Scholar]
- 68. Parrella S, Aspesi A, Quarello P, et al. : Loss of GATA-1 full length as a cause of Diamond-Blackfan anemia phenotype. Pediatr Blood Cancer. 2014;61(7):1319–1321. 10.1002/pbc.24944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hollanda LM, Lima CS, Cunha AF, et al. : An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38(7):807–12. 10.1038/ng1825 [DOI] [PubMed] [Google Scholar]
- 70. Vlachos A, Farrar JE, Atsidaftos E, et al. : Diminutive somatic deletions in the 5q region lead to a phenotype atypical of classical 5q- syndrome. Blood. 2013;122(14):2487–90. 10.1182/blood-2013-06-509935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hoffman R, Zanjani ED, Vila J, et al. : Diamond-Blackfan syndrome: lymphocyte-mediated suppression of erythropoiesis. Science. 1976;193(4256):899–900. 10.1126/science.986086 [DOI] [PubMed] [Google Scholar]
- 72. Ortega JA, Shore NA, Dukes PP, et al. : Congenital hypoplastic anemia inhibition of erythropoiesis by sera from patients with congenital hypoplastic anemia. Blood. 1975;45(1):83–9. [PubMed] [Google Scholar]
- 73. Zanjani ED, Rinehart JJ: Role of cell--cell interaction in normal and abnormal erythropoiesis. Am J Pediatr Hematol Oncol. 1980;2(3):233–44. [PubMed] [Google Scholar]
- 74. Bonsi L, Bagnara GP, Strippoli P, et al. : M-07e cell bioassay detects stromal cell production of granulocyte-macrophage colony stimulating factor and stem cell factor in normal and in Diamond-Blackfan anemia bone marrow. Stem Cells. 1993;11 Suppl 2:131–4. 10.1002/stem.5530110821 [DOI] [PubMed] [Google Scholar]
- 75. Moniz H, Gastou M, Leblanc T, et al. : Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3(7):e356. 10.1038/cddis.2012.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chennupati V, Veiga DF, Maslowski KM, et al. : Ribonuclease inhibitor 1 regulates erythropoiesis by controlling GATA1 translation. J Clin Invest. 2018;128(4):1597–614. 10.1172/JCI94956 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Khajuria RK, Munschauer M, Ulirsch JC, et al. : Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell. 2018;173(1):90–103.e19. 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Dutt S, Narla A, Lin K, et al. : Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–76. 10.1182/blood-2010-07-295238 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Trainor CD, Mas C, Archambault P, et al. : GATA-1 associates with and inhibits p53. Blood. 2009;114(1):165–73. 10.1182/blood-2008-10-180489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Doulatov S, Vo LT, Macari ER, et al. : Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med. 2017;9(376): pii: eaah5645. 10.1126/scitranslmed.aah5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Heijnen HF, van Wijk R, Pereboom TC, et al. : Ribosomal protein mutations induce autophagy through S6 kinase inhibition of the insulin pathway. PLoS Genet. 2014;10(5):e1004371. 10.1371/journal.pgen.1004371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. August CS, King E, Githens JH, et al. : Establishment of erythropoiesis following bone marrow transplantation in a patient with congenital hypoplastic anemia (Diamond-Blackfan syndrome). Blood. 1976;48(4):491–8. [PubMed] [Google Scholar]
- 83. Alter BP: Bone marrow transplant in Diamond-Blackfan anemia. Bone Marrow Transplant. 1998;21(9):965–6. 10.1038/sj.bmt.1701243 [DOI] [PubMed] [Google Scholar]
- 84. Dietz AC, Duncan CN, Alter BP, et al. : The Second Pediatric Blood and Marrow Transplant Consortium International Consensus Conference on Late Effects after Pediatric Hematopoietic Cell Transplantation: Defining the Unique Late Effects of Children Undergoing Hematopoietic Cell Transplantation for Immune Deficiencies, Inherited Marrow Failure Disorders, and Hemoglobinopathies. Biol Blood Marrow Transplant. 2017;23(1):24–9. 10.1016/j.bbmt.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Peffault de Latour R, Peters C, Gibson B, et al. : Recommendations on hematopoietic stem cell transplantation for inherited bone marrow failure syndromes. Bone Marrow Transplant. 2015;50(9):1168–72. 10.1038/bmt.2015.117 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Vlachos A, Muir E: How I treat Diamond-Blackfan anemia. Blood. 2010;116(19):3715–23. 10.1182/blood-2010-02-251090 [DOI] [PMC free article] [PubMed] [Google Scholar]