Abstract
Background
Salmonella is a major zoonotic food-borne pathogen that persists on poultry farms, and animals undergo reinfection with endemic strains. The present study aimed to investigate the characteristics and dissemination of antimicrobial-resistant Salmonella within and between broiler farms that used enrofloxacin and those that did not.
Results
Cloacal and environmental (litter, feed, and water) samples from two selected flocks in each of 12 farms owned by the same company were collected three times over a 30-day period of two production cycles during 2015–2016. The rate of Salmonella isolation was 7.8% (123/1584). Nine Salmonella serotypes (116 isolates) and seven untypable isolates were identified, and Salmonella Montevideo was the most prevalent serotype. Azithromycin-resistant (17.9%) and colistin-resistant (3.3%) isolates were detected, and multidrug-resistant isolates (43.1%) were also observed. No isolate was resistant to enrofloxacin or ciprofloxacin; however, intermediate resistance to enrofloxacin was significantly higher (P < 0.05) in farms that used enrofloxacin than in those that did not. The rate of multi-drug resistance among litter isolates (25/44, 56.8%) was significantly higher (P < 0.05) than that among cloacal swab (24/67, 35.8%) and feed (4/12, 33.3%) isolates. Pulsed-field gel electrophoresis (PFGE) analysis of strains of the same serotype was conducted to determine their epidemiological relationship. The PFGE types were classified into 31 groups with a 100% correlation cutoff in dendrograms for Salmonella Montevideo isolates, which showed 100% genomic identity based on age, sample type, flock, and production cycle within and between farms.
Conclusion
The present study highlights the occurrence of horizontal transmission and cyclic contamination with antimicrobial-resistant Salmonella in broiler farms owned by the same company. Litter may be a good indicator of indoor environmental contamination with antimicrobial-resistant Salmonella on farms. Additionally, enrofloxacin use may be one of the factors promoting resistance towards it in Salmonella.
Electronic supplementary material
The online version of this article (10.1186/s12917-018-1590-1) contains supplementary material, which is available to authorized users.
Keywords: Salmonella, Antimicrobial resistance, Circulating contamination, Enrofloxacin, Broiler farm, Litter, PFGE
Background
Salmonella is the leading cause of food-borne illness worldwide, and dissemination of antimicrobial-resistant Salmonella through the food chain, especially through chicken, has important implications for the failure of salmonellosis treatment. There is increasing risk of chicken-mediated spread of antimicrobial-resistant Salmonella to public health [1]. The prevalence of Salmonella on farms is linked to the prevalence of Salmonella in the derived meat products [2]. Prevention of Salmonella contamination in chicken products requires detailed knowledge of the major sources of contamination. Although measures for eliminating Salmonella on breeder farms through vaccination, use of all-in/all-out replacement systems on broiler farms, and “antimicrobial-free” strategies have been implemented, high Salmonella prevalence rates and antimicrobial-resistance rates are still observed in broiler farms [3, 4]. Interventions performed at poultry farms, including enhanced biosecurity, rodent control, routine surveillance of the farm environment, feed decontamination, and use of autogenous poultry vaccine, can reduce, but cannot eliminate Salmonella from live poultry [5]. Environmental samples, especially poultry litter, have been reported to be a good indicator for the presence of Salmonella in poultry farms [6, 7]. We speculated that the transmission of antimicrobial-resistant Salmonella among farms might occur through indoor environmental contamination in farms.
Salmonella strains can develop antimicrobial resistance (AMR), initially to the traditional first-line drugs chloramphenicol, ampicillin, and trimethoprim/sulfamethoxazole. Because of such resistance, fluoroquinolones (FQs), third-generation cephalosporins, and macrolides (azithromycin) have become critically important for treating salmonellosis in humans [8]. Prophylactic treatment of poultry with enrofloxacin (ENR), a fluoroquinolone antibiotic used to treat animal infections, has been implicated in the increasing resistance to ciprofloxacin, posing a risk to human health [9–11]. In Korea, ENR was licensed for veterinary use in 1987, and the quantity of ENR used has increased since its introduction [12]. In fact, the quantity of ENR sold was the highest of all antimicrobials used to treat chicken in Korea [13]. Concerns over the entry of antimicrobial-resistant zoonotic bacteria into the food chain and the consequent human infections led the Food and Drug Administration (FDA) to ban the use of ENR in poultry in the USA in September 2005 [14]. The use of antimicrobials in poultry farming and the accompanying selection pressure for resistant Salmonella have been the subjects of numerous studies [15, 16]. Antimicrobial use in farming has led to the widespread dissemination of antimicrobial-resistant Salmonella in broiler farms [4]. Therefore, to institute effective measures for reducing the infection of chicken with antimicrobial-resistant Salmonella, Salmonella contamination on broiler farms needs to be well-understood.
To explore production cycle-contamination with antimicrobial-resistant Salmonella in broiler farms that use antimicrobials and in those that do not, samples were collected from 12 farms (which either used or did not use ENR) owned by a single company during two production cycles. Trace-back investigations of the relationship between antimicrobial-resistant Salmonella and ENR use in broiler farms have not been previously conducted. The present study was aimed at determining the distribution and spread of indoor contamination with antimicrobial-resistant Salmonella through a comprehensive investigation of its prevalence within and between broiler farms. We also determined the epidemiological relationships among strains of the same serotype using electrophoresis-based DNA fingerprinting.
Methods
Sampling
The samples used in the present study were collected specifically for the purposes of the study. In total, 1584 fresh samples including 660 cloacal samples and 924 environmental samples (396 litter samples, 264 feed samples, and 264 water samples), were collected during 2015 and 2016 from 12 farms belonging to one of the largest integrated poultry companies in South Korea. In seven farms, ENR (50 mg/L) has been administered to young chicks via addition to drinking water for three consecutive days, while on the other five farms; no ENR was used [17]. The same ENR administration protocol was followed in all seven farms. The sampling farms contained an average of 70–100 thousands broilers, and 3–5 separate flocks. Two flocks per farm were sampled three times over a thirty-day period (chickens at 1, 15, and 30 days of age) during one production cycle. This sampling was then repeated during a second, separate production cycle. The cloacal samples were randomly collected from 25 broilers in entire area of the flock, and samples from five broilers were pooled into one test sample. The environmental samples, including litter (n = 15), feed (n = 10), and water (n = 10) were uniformly collected from equal areas of the flock, and five samples obtained from same area were pooled into one test sample. Finally cloacal swabs (S, n = 5), litter (L, n = 3), feed (F, n = 2) and water (W, n = 2) were collected from each flock.
Salmonella isolation
Samples were collected in sterile plastic conical tubes (50 mL; SPL Life Sciences Co., Ltd., Seoul, Korea) and were stored under refrigeration in the laboratory until analysis, which was performed within 48 h of arrival. Fresh samples [1 g (or mL)] were separately mixed with 9 mL (1:9 dilutions) of buffered peptone water (BPW; BD Difco, Sparks, MD, USA) and incubated at 37 °C for 24 h for enrichment. An aliquot of the enriched BPW culture (100 μL) was transferred to 10 mL of Rappaport Vassiliadis (RV) broth (Thermo Fisher Scientific, Oxoid Ltd., Basingstoke, UK), and incubated at 42 °C for 24 h for selective enrichment [18]. A loopful of each RV culture was streaked onto a xylose-lysine-deoxycholate (XLD) agar plate (BD Difco™ XLD agar, USA), and the plate was incubated overnight at 37 °C. Presumptive Salmonella colonies were then tested with a Salmonella latex test kit (Thermo Fisher Scientific, Oxoid Ltd., Basingstoke, UK).
Salmonella serotyping
The serogroup and serovar of each Salmonella isolate were identified following Edwards and Ewing’s procedure for the identification of Enterobacteriaceae using an antisera kit (BD Difco, Sparks, MD, USA) [19]. Somatic O antigen (BD Difco) was identified using the slide agglutination test with a commercially available antiserum. Flagellar (H) antigens (phases 1 and 2) were identified via successive inoculation onto 0.3% brain heart infusion (BHI) agar (BD) to activate flagella, followed by inoculation into BHI broth. The broth was cultured overnight, fixed with 0.6% formalin, and then analyzed using a tube agglutination test [20].
Antimicrobial susceptibility
The minimum inhibitory concentrations (MICs) of the test antimicrobials nalidixic acid (NAL), ciprofloxacin (CIP), neomycin (NEO), gentamicin (GEN), streptomycin (STR), tetracycline (TET), azithromycin (AZM), amoxicillin/clavulanic acid (AMC), cefoxitin (FOX), ceftiofur (XNL), ampicillin (AMP), trimethoprim/sulfamethoxazole (SXT), colistin (COL), florfenicol (FFN), and chloramphenicol (CHL) were determined using the Sensititre panel KRNV4F (TREK Diagnostic Systems, Korea), while the MICs of enrofloxacin (ENR), tigecycline (TIG), and fosfomycin (FOS) were determined using the agar dilution method. Escherichia coli (ATCC 25922) were used as the quality-control strain. The susceptibility breakpoints of most tested antimicrobials were interpreted based on CLSI guidelines [21], while those of XNL, ENR, and FFN were interpreted based on CLSI BM31-A3 standards [22]. No CLSI interpretation criteria were available for STR, TIG, COL, and NEO; therefore, the following MIC values were considered to indicate resistance: STR, ≥ 64 μg/mL [23]; TIG, ≥ 8 μg/mL [24]; COL, ≥ 4 μg/mL [25]; and NEO, ≥ 16 μg/mL [26] (Table 1). Salmonella isolates resistant to at least three antimicrobial classes were designated multidrug resistant (MDR).
Table 1.
Antimicrobials used in the study and the tested concentration ranges
| Antimicrobialsa | Abbreviation | Breakpoints (μg/mL) | Concentration ranges (μg/mL) | ||
|---|---|---|---|---|---|
| Sb | I | R | |||
| Nalidixic acid | NAL | 16 | -c | 32 | 2–128 |
| Ciprofloxacin | CIP | 0.06 | 0.12–0.5 | 1 | 0.12–16 |
| Enrofloxacin | ENR | 0.25 | 0.5–1 | 2 | 0.12–64 |
| Neomycin | NEO | – | – | 16 | 2–32 |
| Gentamicin | GEN | 4 | 8 | 16 | 1–64 |
| Streptomycin | STR | 32 | – | 64 | 2–128 |
| Tetracycline | TET | 4 | 8 | 16 | 2–128 |
| Azithromycin | AZM | 16 | – | 32 | 0.25–64 |
| Amoxicillin/clavulanic acid | AMC | 8/4 | 16/8 | 32/16 | 2/1–64/32 |
| Cefoxitin | FOX | 8 | 16 | 32 | 1–32 |
| Ceftiofur | XNL | 2 | 4 | 8 | 0.5–8 |
| Ampicillin | AMP | 8 | 16 | 32 | 2–32 |
| Trimethoprim/sulfamethoxazole | SXT | 2/38 | – | 4/76 | 0.12/2.38–4/76 |
| Colistin | COL | 2 | – | 4 | 2–32 |
| Florfenicol | FFN | – | – | 32 | 2–64 |
| Chloramphenicol | CHL | 8 | 16 | 32 | 2–64 |
| Tigecycline | TIG | 2 | 4 | 8 | 0.12–32 |
| Fosfomycin | FOS | 64 | 128 | 256 | 0.25–256 |
aSorted based on category of antimicrobials
bS, sensitivity; I, intermediate resistance; R, resistance
c-, no standard breakpoint value in related references
PFGE and BioNumerics analysis
Isolates of Salmonella Montevideo (n = 75) and Salmonella Senftenberg (n = 16) were genotyped using PFGE following protocols of the Centers for Disease Control and Prevention available on PulseNet, with some modifications. In brief, the Salmonella isolates were streaked onto MacConkey agar plates and incubated overnight at 37 °C. Then, the bacteria were suspended in PBS at an OD value of 0.6–0.8. Genomic DNA (extraction using 1% SDS and 1 mg/mL proteinase K, Biosesang, Seoul, Korea) samples were digested with 50 U of XbaI (Thermo Fisher Scientific, Inchon, Korea) at 37 °C for 3 h. The digested DNA was separated by electrophoresis in 0.5 x TBE buffer at 14 °C for 18 h using a CHEF-DR@ electrophoresis system (Bio-Rad, Hercules, CA, USA). The pulse time was ramped from 2.16 to 63.8 s. Salmonella Braenderup H9812, which was included as a molecular weight standard, was processed with each batch of isolate. The gels were stained with ethidium bromide, and the DNA patterns were visualized on a UV transilluminator (Bio Doc-It Imaging System, Upland, CA, USA). The DNA fingerprints obtained with PFGE were analyzed using BioNumerics (version 5.10 for Windows). Dice similarity coefficients were calculated based on pairwise comparison of the PFGE types of the isolates. The isolates were considered to have closely related band patterns based on molecular typing when their PFGE types had dice similarity coefficients of 100%, and were clustered at the 90% similarity level. Band-matching settings, with an optimization of 1.0% and a position tolerance of 1.0%, were applied.
Statistical analysis
The chi-square test was used to test for significant differences in the rates of Salmonella isolation, MDR prevalence, and AMR rates between farms that used ENR and those that did not. P values less than 0.05 were considered statistically significant. Analyses were performed using SPSS version 19.0 (IBM Co., Armonk, NY, USA).
Results
Prevalence and serovars of Salmonella
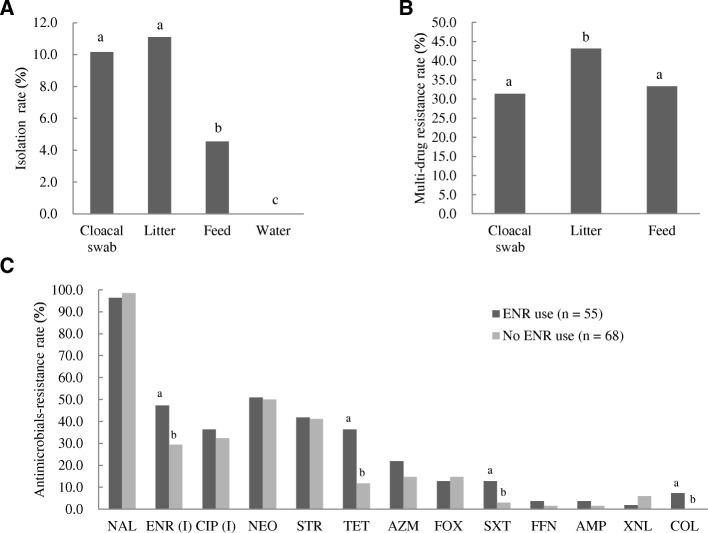
The cloacal and environmental samples (n = 1584) collected from the broiler farms were analyzed for the presence of Salmonella. In total, 123 (7.8%) samples were positive for Salmonella. There were no significant differences in isolation rates between the litter samples (44/396, 11.1%) and the cloacal swabs (67/660, 10.2%); however, significantly higher isolation rates were found in litter samples and cloacal swabs compared to feed (12/264, 4.5%), and water samples (0/264, 0.0%) (Fig. 1a). The Salmonella isolation rate from all types of samples was significantly lower (P < 0.05) in farms that used ENR (55/924, 6.0%) than on farms that did not (68/660, 10.3%; Table 2).
Fig. 1.
Rates of Salmonella isolation from samples of cloacal swabs, litter, feed, and water in broiler farms (a); the incidence of multi-drug resistance in isolates from cloacal swabs, litter, feed, and water in broiler farms (b); rates of antimicrobial-resistance in isolates from farms that used ENR and in isolates from farms that did not (c). The chi-square test was used to assess the significance of differences. P values less than 0.05 were considered statistically significant, and were marked with lowercase letters (a/b/c)
Table 2.
Salmonella isolation from different sample types in broiler farms with or without enrofloxacin (ENR) use *
| ENR use | Sample no. | % (no. positive/total no. samples) | ||||
|---|---|---|---|---|---|---|
| Cloacal swab | Litter | Feed | Water | Total | ||
| Yes | 924 | 7.5 (29/385) a | 8.2 (19/231) a | 4.5 (7/154) | 0.0 (0/154) | 6.0 (55/924) a |
| No | 660 | 13.8 (38/275) b | 15.2 (25/165) b | 4.5 (5/110) | 0.0 (0/110) | 10.3 (68/660) b |
| Total | 1584 | 10.2 (67/660) | 11.1 (44/396) | 4.5 (12/264) | 0.0 (0/264) | 7.8 (123/1584) |
*Lowercase (a/b) were used to indicate significant difference in isolation rates between farms with enrofloxacin use and those without; different letters indicate significant differences (P < 0.05)
Isolates were assigned to nine serovars, most of which belonged to serogroups C1 (65.0%) and E1 (17.1%). S. Montevideo (75 isolates, 61.0%) was the dominant serovar, and S. Senftenberg (21 isolates, 17.1%) was a distant second, followed by S. Emek (9 isolates, 7.3%), S. Macclesfield (4 isolate, 3.3%), and S. Virchow (3 isolates, 2.4%). Only one isolate was detected for the four serotypes, S. Infantis, S. Edinburg, S. Hato, and S. Vellore, and seven isolates (5.7%) marked as S. spp. could not be assigned to specific serotypes (Table 3).
Table 3.
Antimicrobial resistance in Salmonella isolates from broiler farms a
| Serovar (Serogroup) | N (%) | NAL | NEO | STR | TET | AZM | XNL | FOX | AMP | SXT | COL | FFN | > 1 agent | ≥3 agents | ≥5 classes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | MIC50/90 | n (%) | n (%) | n (%) | n (%) | ||
| Hato (B) | 1 (0.8) | – | 1 (100) | – | 1 (100) | – | 1 (100) | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | 1 (100) | 1 (100) | 0 |
| Vellore (B) | 1 (0.8) | – | 0 | – | 1 (100) | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | 1 (100) | 0 | 0 |
| Montevideo (C1) | 75 (61.0) | 128/128 | 75 (100) | 4/32 | 35 (46.7) | 32/128 | 37 (49.3) | 4/16 | 24 (32) | 16/32 | 12 (16) | 1/1 | 3 (4.0) | 4/16 | 9 (12.0) | 4/8 | 2 (2.7) | 0.5/1 | 8 (10.7) | 2/2 | 2 (2.7) | 8/8 | 3 (4.0) | 75 (100) | 37 (49.3) | 4 (5.3) |
| Virchow (C1) | 3 (2.4) | 128/128 | 3 (100) | 16/32 | 3 (100) | 64/64 | 2 (66.7) | 8/8 | 0 | 64/64 | 2 (66.7) | 1/1 | 0 | 16/32 | 1 (33.3) | 8/8 | 0 | 0.25/0.25 | 0 | 2/2 | 0 | 8/8 | 0 | 3 (100) | 2 (66.7) | 0 |
| Infantis (C1) | 1 (0.8) | – | 1 (100) | – | 1 (100) | – | 1 (100) | – | 1 (100) | – | 1 (100) | – | 1 (100) | – | 0 | – | 0 | – | 0 | – | 1 (100) | – | 0 | 1 (100) | 1 (100) | 1 (100) |
| Edinburg (C1) | 1 (0.8) | – | 1 (100) | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 1 (100) | – | 0 | – | 0 | – | 0 | – | 0 | 1 (100) | 0 | 0 |
| Emek (C2-C3) | 9 (7.3) | 128/128 | 9 (100) | 4/16 | 3 (33.3) | 8/32 | 1 (11.1) | 1/4 | 0 | 16/32 | 3 (33.3) | 0.5/1 | 0 | 4/16 | 0 | 2/4 | 0 | 0.5/0.5 | 0 | 2/2 | 0 | 4/8 | 0 | 9 (100) | 3 (33.3) | 0 |
| Macclesfield (D2) | 4 (3.3) | 64/128 | 4 (100) | 2/16 | 1 (25) | 8/128 | 1 (25) | 1/16 | 1 (6.3) | 16/64 | 1 (25) | 0.5/1 | 0 | 4/32 | 0 | 2/8 | 0 | 0.5/4 | 1 (25) | 2/2 | 0 | 2/8 | 0 | 4 (100) | 1 (25) | 0 |
| Senftenberg (E4) | 21 (17.1) | 128/128 | 21 (100) | 4/32 | 10 (47.6) | 32/128 | 4 (19) | 1/4 | 0 | 16/16 | 2 (9.5) | 0.5/1 | 0 | 4/16 | 2 (9.5) | 2/4 | 0 | 0.5/0.5 | 0 | 2/2 | 0 | 4/8 | 0 | 21 (100) | 2 (9.5) | 0 |
| spp. (C2-C3, B, D1) | 7 (5.7) | – | 6 (71.4) | – | 7 (100) | – | 4 (57.1) | – | 2 (28.6) | – | 1 (14.3) | – | 1 (14.3) | – | 4 (57.1) | – | 1 (14.3) | – | 0 | – | 1 (14.3) | – | 0 | 7 (100) | 4 (57.1) | 2 (28.6) |
| Total | 123 | 128/128 | 120 (97.6) | 8/32 | 62 (50.4) | 8/32 | 51 (41.5) | 1/16 | 28 (22.8) | 16/32 | 22 (17.9) | 1/1 | 5 (4.1) | 4/32 | 17 (13.8) | 4/8 | 3 (2.4) | 0.5/0.5 | 9 (7.3) | 2/2 | 4 (3.3) | 8/8 | 3 (2.4) | 123 (100) | 53 (43.1) | 7 (5.7) |
a-, not analyzed; the unit of MIC50/90 was μg/mL
Antimicrobial susceptibility analysis
All the isolates analyzed in this study showed resistance to at least one tested antimicrobial (Table 3). Resistance to NAL was the most prevalent (120/123, 97.6%), followed by that to NEO (62/123, 50.4%), STR (51/123, 41.5%), TET (28/123, 22.8%), AZM (22/123, 17.9%), FOX (17/123, 13.8%), XNL (5/123, 4.1%), AMP (3/123, 2.4%), SXT (9/123, 7.3%), COL (4/123, 3.3%), and FFN (3/123, 2.4%). several isolates showed intermediate resistance (IR) to ENR (46/123, 37.4%) and CIP (42/123, 31.1%). All isolates were sensitive to the five antimicrobials CHL, GEN, TIG, FOS, and AMC.
Thirty-one AMR phenotypes were observed among the Salmonella isolates from the 12 broiler farms; the AMR profile NAL + NEO + STR + TET was the most prevalent in this study (10 isolates, 8.1%) (Table 4). Further, seven MDR isolates comprising four S. Montevideo, two S. spp., and one S. Infantis isolates, were resistant to ≥5 antimicrobial classes, all of which were isolated from the farms that used ENR.
Table 4.
Antimicrobial resistance profiles of Salmonella isolates from broiler farms (n = 123)
| No. | Antimicrobial resistance profile | na (%) | Serovars (n) |
|---|---|---|---|
| – | Susceptible | 0 | – |
| 1 | NAL | 27 (22.0) | Montevideo (13), Senftenberg (8), Emek (5), Macclesfield (1) |
| 2 | NEO | 2 (1.6) | Vellore (1), spp. (1) |
| 3 | NAL + NEO | 18 (14.6) | Montevideo (9), Senftenberg (6), Virchow (1), spp. (2) |
| 4 | NAL + STR | 6 (4.9) | Montevideo (5), Senftenberg (1) |
| 5 | NAL + AZM | 4 (3.3) | Montevideo (1), Senftenberg (1), Emek (1), Macclesfield (1) |
| 6 | NAL + FOX | 3 (2.4) | Montevideo (1), Senftenberg (1), Edinburg (1) |
| 7 | NAL + SXT | 9 (7.3) | Montevideo (8), Macclesfield (1) |
| 8 | NAL + COL | 1 (0.8) | Montevideo (1) |
| 9 | NAL + NEO + STR | 7 (5.7) | Montevideo (3), Senftenberg (2), Emek (1), Hato (1) |
| 10 | NAL + NEO + FFN | 2 (1.6) | Montevideo (2) |
| 11 | NAL + NEO + AZM | 4 (3.3) | Montevideo (1), Senftenberg (1), Emek (2) |
| 12 | NAL + NEO + COL | 1 (0.8) | Montevideo (1) |
| 13 | NEO + STR + FOX | 3 (2.4) | Montevideo (3) |
| 14 | NEO + STR + TET | 4 (3.3) | Montevideo (4) |
| 15 | NAL + STR + AZM | 2 (1.6) | Montevideo (2) |
| 16 | NEO + STR + TET + FOX | 1 (0.8) | spp. (1) |
| 17 | NAL + STR + FOX + AZM | 1 (0.8) | Montevideo (1) |
| 18 | NAL + NEO + STR + TET | 10 (8.1) | Montevideo (10) |
| 19 | NAL + NEO + STR + FOX | 2 (1.6) | Senftenberg (1), spp. (1) |
| 20 | NAL + NEO + STR + AZM | 1 (0.8) | Virchow (1) |
| 21 | NAL + NEO + FFN + AZM | 1 (0.8) | Montevideo (1) |
| 22 | NAL + NEO + STR + TET + XNL | 2 (1.6) | Montevideo (2) |
| 23 | NAL + NEO + STR + TET + FOX | 2 (1.6) | Montevideo (1), Macclesfield (1) |
| 24 | NAL + NEO + STR + TET + AZM | 2 (1.6) | Montevideo (2) |
| 25 | NAL + NEO + STR + FOX + AZM | 1 (0.8) | Virchow (1) |
| 26 | NAL + NEO + STR + FOX + COL + AZM | 1 (0.8) | spp. (1) |
| 27 | NAL + NEO + STR + TET + FOX + AZM | 2 (1.6) | Montevideo (2) |
| 28 | NAL + STR + TET + FFN + AMP + AZM | 1 (0.8) | Montevideo (1) |
| 29 | NAL + NEO + STR + TET + FOX + XNL + AMP | 1 (0.8) | spp. (1) |
| 30 | NAL + NEO + STR + TET + XNL + COL + AZM | 1 (0.8) | Infantis (1) |
| 31 | NAL + NEO + STR + TET + FOX + XNL + AMP + AZM | 1 (0.8) | Montevideo (1) |
an, number of isolates
The MDR rate among litter isolates (25/44, 56.8%) was significantly higher (P < 0.05) than that among the cloacal swab (24/67, 35.8%) and feed (4/12, 33.3%) isolates (Fig. 1b). The ratio of IR to ENR was significantly higher (P < 0.05) in isolates from farms that used ENR (26/55, 47.3%) than in those from farms that did not (20/68, 29.4%; Fig. 1c).
Serotype distribution and genetic analysis of isolates from broiler farms
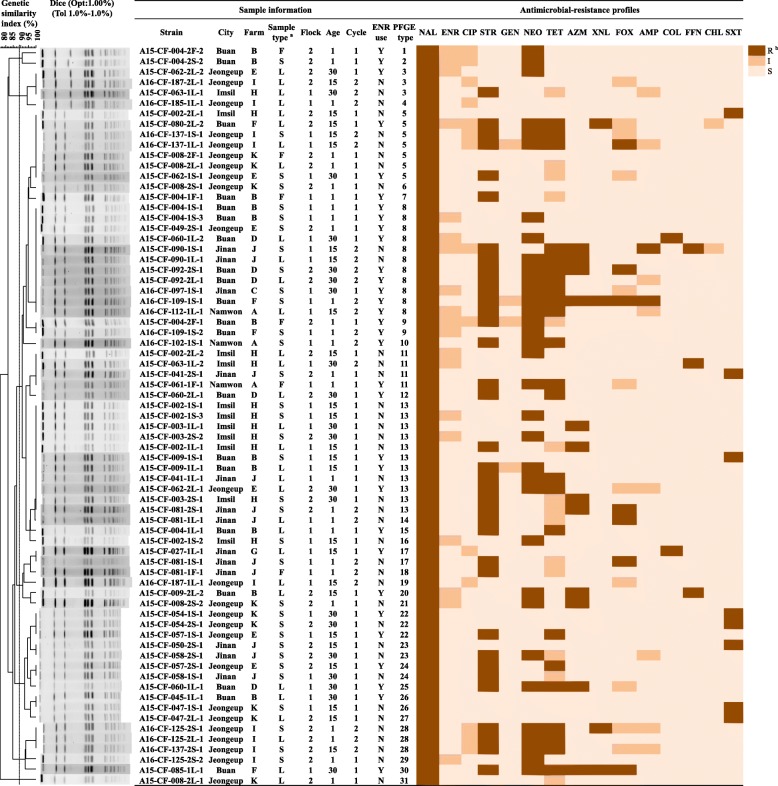
Serotype diversity was observed in the broiler farms sampled in the present study with 75% (9/12) of them showing contamination with at least two Salmonella serovars (Table 5). S. Montevideo and S. Senftenberg were isolated from 11 and 8 of the 12 farms, respectively. In farms B, H, I, and J, there were at least four different serotypes observed in a single flock. In one farm (farm H), isolates from the same flock at different ages had the same PFGE type (type 13), with a similarity index of 100% (isolates A15-CF-002-1S-3 and A15-CF-003-1 L-1); isolates from different flocks were of the same type (type 13; isolates A15-CF-003-1 L-1 and A15-CF-003-2S-2); and two isolates, A15-CF-002-2 L-2 and A15-CF-063-1 L-2, which were of the same PFGE type (type 11) and same AMR profile (including intermediate ENR resistance), were obtained from litter during different production cycles. Isolates of the same PFGE type were also observed in different farms (type 5 in farms H, F, I, K, and E; type 8 in farms B, E, D, J, C, F, and A; type 11 in farms A, H, and J; type 13 in farms B, E, H, and J; type 17 in farms G and J; type 22 in farms K and E; and type 26 in farms B and K; Fig. 2). In S. Senftenberg isolates, the same PFGE type was observed in two different farms in different cities, that neither of which used ENR (type 4 in farms H and J; type 6 in farms J and K; Additional file 1).
Table 5.
Distribution of Salmonella serotypes and genotypes in broiler farms
| Farm | ENR use a | Flock | Production cycle 1 | Production cycle 2 | ||||
|---|---|---|---|---|---|---|---|---|
| 1 day | 15 days | 30 days | 1 day | 15 days | 30 days | |||
| A | Y | 1 |
S. Montevideo (11)b S. Senftenberg (5) |
-c | – |
S. Montevideo (10) S. Macclesfield (NCd) |
S. Montevideo (8) | – |
| 2 | – | – | – | – | – |
S. spp. (NC) S. Vellore (NC) |
||
| B | 1 | S. Montevideo (7, 8, 15) | S. Montevideo (13) | S. Montevideo (26) | S. Senftenberg (7) | – | – | |
| 2 |
S. Montevideo (2, 4) S. Senftenberg (10) |
S. Montevideo (20) S. Emek (3) |
S. Macclesfield (NC) | – | – | – | ||
| C | 1 | – | – | S. Montevideo (8) |
S. Montevideo (28) S. Senftenberg (9) S. Emek (4) |
– | – | |
| 2 | – | – | – | S. spp. (NC) | – | – | ||
| D | 1 | S. Senftenberg (2) | S. Montevideo (8) | S. Montevideo (8, 25) | – | – | – | |
| 2 | S. Macclesfield (NC) | S. Montevideo (8) | S. Montevideo (12) | – | – | S. Montevideo (8) | ||
| E | 1 | – | S. Montevideo (22) | S. Montevideo (5) | – | – | – | |
| 2 | S. Montevideo (8) | S. Montevideo (24) | S. Montevideo (3, 13) | – | – | – | ||
| F | 1 | – | S. Virchow (NC) |
S. Montevideo (30) S. spp. (NC) |
S. Montevideo (8, 9) | – | – | |
| 2 | – |
S. Montevideo (5) S. Virchow (NC) |
S. Infantis (NC) | – | – | – | ||
| G | 1 | – | S. Montevideo (17) | – | Ne | N | N | |
| 2 | – | – | – | N | N | N | ||
| H | N | 1 |
S. Senftenberg (11) S. Virchow (NC) |
S. Montevideo (13, 16) S. Senftenberg (4) |
S. Montevideo (13) | S. Macclesfield (NC) | S. Montevideo (13) | S. Montevideo (3, 11) |
| 2 | S. Senftenberg (8) | S. Montevideo (5, 11) | S. Montevideo (13) | – | – | – | ||
| I | 1 | – | S. Montevideo (19) | – |
S. Montevideo (4) S. Senftenberg (8) S. Emek (2) |
S. Montevideo (5) S. Edinburg (NC) |
– | |
| 2 | S. Montevideo (29) |
S. Montevideo (28) S. Hato (NC) |
– |
S. Montevideo (28) S. Senftenberg (2) |
S. Montevideo (16, 28) S. Senftenberg (2) |
S. Montevideo (28) | ||
| J | 1 |
S. Montevideo (13) S. Emek (1,2) |
S. spp. (NC) | S. Montevideo (24) |
S. Montevideo (14, 17, 18) S. Senftenberg (4) |
S. Montevideo (8) | – | |
| 2 | S. Montevideo (11) | S. Montevideo (23) | S. Montevideo (23) |
S. Montevideo (13) S. Senftenberg (4) |
S. Montevideo (14) S. Senftenberg (6) |
– | ||
| K | 1 | S. Senftenberg (6) | S. Montevideo (26) | S. Montevideo (22) | – | – | – | |
| 2 | S. Montevideo (5, 6, 21, 31) | S. Montevideo (27) | S. Montevideo (22) | – | – | S. spp. (NC) | ||
| L | 1 | S. Senftenberg (NC) | S. Senftenberg (NC) | – | N | N | N | |
| 2 | – | – | – | N | N | N | ||
aENR use, Y implies yes; N implies no.
bPFGE types are showed within parenthesis
cThe dash (−) indicates that the farm was negative for Salmonella
dNC implies that PFGE was not conducted
eN denotes no sampling
Fig. 2.
Dendrograms showing pattern analysis on the basis of Xba I-PFGE of the 75 Salmonella Montevideo isolates obtained from broiler farms, along with related sampling information, and antimicrobial-resistance (AMR) profiles. The Dice coefficient was used to perform similarity analysis. aS, cloacal swabs; L, litter; F, feed. bR, resistance (dark pink); I, intermediate resistance (pink), S, sensitivity (light pink). Dotted lines indicate 90% similarity
Discussion
In the present study, to explore the distribution and dissemination of antimicrobial-resistant Salmonella in broiler farms, we obtained samples during two production cycles from farms that used antimicrobials and those that did not (Table 5).
S. Montevideo, one of the serotypes prevalent in human infections [27], showed high resistance to ≥3 antimicrobials (49.3%; Table 3). In recent years, there has been an increase in S. Montevideo human infections and outbreaks worldwide, including in the USA, Europe, Australia, and Asian nations, such as South Korea [5, 28–30]. In the USA, the number of human illnesses caused by S. Montevideo increased from 728 cases in 2002 to 1203 cases in 2012 [31]. In South Korea, S. Montevideo is one of the most common serotypes affecting the poultry industry [32, 33]. Additionally, the most serious case of resistance found in the present study was an S. Montevideo isolate with resistance to eight antimicrobials (NAL + NEO + STR + TET + FOX + XNL + AMP + AZM; Table 4). In addition, there was an MDR S. Infantis isolate resistant to ≥5 antimicrobial classes (Table 3); and S. Infantis is one of the most prevalent serotypes in South Korea and second in frequency among serovars [34]. In countries including Germany [35], Hungary [36], Italy [37], and Japan [38], emergence and clonal dissemination of MDR S. Infantis strains in chicken has been recent reported, and has been related to an increased incidence of human infections [31]. Given these findings, further investigations of the antimicrobial resistance and distribution of S. Infantis in slaughterhouses and retail chicken meat are required.
All isolates showed resistance to at least one antimicrobial, suggesting that AMR Salmonella is widespread in broiler farms, as was reported previously in South Korea [33]. Contrary to the increasing prevalence of FQ-resistant Salmonella reported in other countries [39, 40], we did not observe any FQ-resistant isolates in this study; however, 31.1% of the isolates showed reduced susceptibility to CIP (with a MIC of 0.12 to 0.25 or 0.5 μg/mL), and 37.4% of the isolates showed IR to ENR [41, 42]. The rate of IR to CIP was higher in farms that used ENR than in farms that did not, although the difference was not statistically significant (p > 0.05). The ratio of IR to ENR was significantly higher (P < 0.05) in farms that used ENR than in those that did not (Fig. 1c). Resistance to SXT, TET, and COL was significantly higher in farms that used ENR than in those that did not (P < 0.05; Fig. 1c). Resistance to the three antimicrobials ENR, TET, and SXT was most commonly observed in isolates from chicken, in a previous study [43]. One plausible explanation is that the selective pressure exerted by the antimicrobials contributed to the co-selection of this antimicrobial resistance pattern. The co-selection of resistance to more than one antimicrobial, owing to the genetic linkage of resistance genes, is a common feature of resistance acquired by horizontal gene transfer [44, 45].
In the present study, 17.9% of isolates showed resistance to AZM (Table 3), a macrolide antimicrobial used for the limited treatment of MDR Salmonella infections [8, 46]. In recent years, AZM has been used for typhoid treatment due to the increased prevalence of MDR Salmonella isolates, which has caused serious problems in antimicrobial selection. The sporadic occurrence of AZM-resistant Salmonella isolates has also caused problems in the selection of suitable antimicrobials for typhoid treatment [47, 48]. In Europe, the first case of failure of AZM treatment of Salmonella infection was reported [49]. Similarly, emergence of clinical AZM-resistant Salmonella isolates was recently reported in Asia [50, 51]; 15.24% of Salmonella isolates from chicken showed resistance to AZM (with an MIC ≥16 μg/mL) in another study [52]. However, reports of AZM resistance in Salmonella isolates from humans or animals in South Korea are rare. Considering the fact that, AZM has not been used on these farms to the best of our knowledge, the prevalence of AZM resistance in Salmonella is probably either due to prolonged use of antimicrobials in the same class as AZM such as erythromycin and tylosin for treating poultry diseases [53], or due to co-resistance to AZM and other antimicrobials [54]. Further, the MIC50 of AZM was 16 μg/mL against all isolates and 64 μg/mL against S. Virchow (Table 3). However, the exact reason for the emergence of AZM resistance in Salmonella and its mechanism require further studies. Additionally, because COL is a last-line drug used to treat MDR Enterobacteriaceae infections, the four COL-resistant isolates identified in the present study warrant more attention; due to the prevalence of COL-resistant Salmonella, mcr family genes can be easily and quickly transmitted [8, 55, 56]. In Europe, plasmid-mediated COL resistance in Enterobacteriaceae has already spread widely in avian and pig farms, and this has necessitated prompt international action to restrict or ban COL use in agriculture to avoid further spread of resistance, similar to the solution involving NDM-1 (New Delhi metallo-β-lactamase-1) several years ago [56]. COL resistance in Salmonella isolates from humans had not been reported until recently, when the first COL-resistant isolates from humans were reported in the Arabian Peninsula [57], followed by two COL-resistant Salmonella strains in Italy [58]. The observation of co-resistance to AZM and COL in two MDR isolates (AMR profiles: NAL + NEO + STR + FOX + COL + AZM and NAL + NEO + STR + TET + XNL + COL + AZM), both of which exhibited intermediate resistance to CIP and ENR (data not shown), is of high importance. This might pose a considerable challenge when selecting drugs to treat human Salmonella infections. The most important finding of the present study was that the AMR profile XNL + COL + AZM, conferring resistance to the critically important antimicrobials used to treat salmonellosis or MDR Salmonella infections in humans, has never been reported previously in Salmonella strains of animal origin.
Based on the results of distribution of Salmonella serotypes and genotypes, a considerable cross-contamination among the farms could be inferred. For example, isolates with the same PFGE type shared between flocks, production cycles, sample types, and among chickens of different ages within the farms were frequently identified (Fig. 2). This could be because the farms shared resources including breeders, trucks for transport, veterinarians, chicks, and feed [59]. Chicks in five farms (farms A, B, D, E, and J) were from the same breeder farm which was negative for Salmonella (data not shown). Moreover, there was contamination between flocks, production cycles, sample types, and among chicken of different ages within the farms. Antimicrobial-resistant Salmonella can be circularly transmitted between continuous production cycles. Although all-in/all-out replacement systems have been applied in the commercial poultry industry, Salmonella contamination still occurs, especially in the farm environment. Plausible explanations include insufficient disinfection, development of resistance to disinfectants in the first production cycle and subsequent survival, and inherent resistance in Salmonella [1, 60]. Another explanation is that other factors including air, unclean facilities, and vectors such as insects, wild birds, farmers, and rodents, might contribute to Salmonella transmission in poultry farms [61].
The dissemination of antimicrobial-resistant Salmonella in the farms might also have occurred via litter contamination (farm H; Fig. 2), because the litter samples had higher Salmonella isolation and MDR rates than the cloacal swab and feed samples (Fig. 1a, b), with no significant difference in isolation rates from litter samples between production cycles (Additional file 2). The spread of indoor Salmonella contamination in the broiler farms was apparently caused by litter from the broilers. Salmonella-positive litter samples detected in our study may have important public health implications. A recent study reported a positive correlation between prevalence of Salmonella in litter samples and Salmonella isolation from broiler carcasses [6]. Antimicrobial-resistant Salmonella could re-circulate in the farms because of litter contamination during different production cycles. Therefore, litter in broiler farms may be an important reservoir of Salmonella, consistent with a speculation in a report from the USA [62]. In summary, we concluded that serious Salmonella contamination occurs in farms during production cycles, as does cross-contamination among farms owned by the same company. Because of dissemination and cross-contamination between the farms that used antimicrobials and those that did not, close attention should be paid to farm-level hygiene management.
Conclusion
In conclusion, we detected considerable contamination with antimicrobial-resistant Salmonella in broiler farms. The litter in the farm was one of the important reservoirs for Salmonella showing high Salmonella prevalence and MDR rates. Other reservoirs of Salmonella, such as feed, air, fans, and vectors such as insects, wild birds, farmers, and rodents might also contribute to its transmission in chicken farms [61]. Additional measures for litter and feed management might be required to prevent the transmission of antimicrobial-resistant Salmonella in such farms. Moreover, ENR use may be an important factor causing ENR resistance among Salmonella in the farms. Our results provide useful information regarding the distribution of AMR phenotypes among Salmonella isolates from broiler farms that use ENR and those that do not, highlighting the need for improved farming practices and more cautious use of antimicrobial agents. Further studies are required to develop protocols to prevent the contamination of litter and feed; this, together with instructions for strict all-in/all-out replacement and biosecurity systems, may markedly reduce the occurrence of antimicrobial-resistant Salmonella species in broiler farms.
Additional files
Dendrograms showing pattern analysis on the basis of Xba I-PFGE of the 16 Salmonella Senftenberg isolates obtained from broiler farms and their association with antimicrobial-resistance. The Dice coefficient was used to perform similarity analysis. aS, cloacal swabs; L, litter; F, feed. bR, resistance (dark pink); I, intermediate resistance (pink), S, sensitivity (light pink). Dotted lines indicate 90% similarity. (DOCX 271 kb)
Isolation rates and multidrug resistant (MDR) rates in different types of samples obtained during production cycles 1 and 2. Different lowercase (a/b) and capital letters (A/B) in the same row were used to indicate significant (P < 0.05) differences in isolation rates and MDR rates between production cycles 1 and 2, respectively. (DOCX 36 kb)
Acknowledgements
The authors wish to thank the veterinary practitioners and farmers for their help with sample collection, and lab members for their help with internal review of the paper.
Funding
This work was supported by Korea Institute of Planning and Evaluation for technology in Food, Agriculture, Forestry and Fisheries (716002–7, 315035–5), Basic Science Research Program through the NRF funded by the Ministry of Education (2017R1D1A1B03030883), and Research of Animal and Plant Quarantine Agency (Z-1543073-2015-16-01), South Korea.
Availability of data and materials
The datasets analyzed in the present study are available from the corresponding author on reasonable request. Additional supporting files can be found in the supplementary material section.
Abbreviations
- AMC
Amoxicillin/clavulanic acid
- AMP
Ampicillin
- AMR
Antimicrobial resistance
- AZM
Azithromycin
- BHI
Brain heart infusion
- CHL
Chloramphenicol
- CIP
Ciprofloxacin
- CLSI
Clinical Laboratory Standardization Institute
- COL
Colistin
- ENR
Enrofloxacin
- FDA
Food and Drug Administration
- FFN
Florfenicol
- FOS
Fosfomycin
- FOX
Cefoxitin
- FQs
Fluoroquinolones
- GEN
Gentamicin
- MDR
Multidrug resistant
- MICs
Minimum inhibitory concentrations
- NAL
Nalidixic acid
- NDM-1
New Delhi metallo-β-lactamase-1
- NEO
Neomycin
- PFGE
A pulsed-field gel electrophoresis
- RV
Rappaport Vassiliadis
- S. Edinburg
Salmonella Edinburg
- S. Emek
Salmonella Emek
- S. Hato
Salmonella Hato
- S. Infantis
Salmonella Infantis
- S. Macclesfield
Salmonella Macclesfield
- S. Montevideo
Salmonella Montevideo
- S. Senftenberg
Salmonella Senftenberg
- S. spp.
Salmonella species
- S. Vellore
Salmonella Vellore
- S. Virchow
Salmonella Virchow
- STR
Streptomycin
- SXT
Trimethoprim/sulfamethoxazole
- TET
Tetracycline
- TIG
Tigecycline
- USA
United States of America
- XLD
Xylose-lysine-deoxycholate
- XNL
Ceftiofur
Authors’ contributions
KS and BW performed experiments, analyzed the data and wrote manuscript; MK designed the experiments, analyzed the data, edited the manuscript and coordinated the whole project. All authors critically read and approved the final manuscript.
Ethics approval and consent to participate
The current work was carried out according to the ethical guide lines of Chonbuk National University (CBNU). Prior to the initiation of this study, proper approval was obtained from Ethics Committee for Animal Experiments of the Chonbuk National University. There are no vulnerable populations involved, and no endangered species was used in the experiments. Farm managers provided verbal consent to collect the cloacal swab samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ke Shang, Email: shangke0624@gmail.com.
Bai Wei, Email: weibai116@hotmail.com.
Min Kang, Email: vet.minkang@gmail.com.
References
- 1.Youn SY, Jeong OM, Choi BK, Jung SC, Kang MS. Comparison of the antimicrobial and sanitizer resistance of Salmonella isolates from chicken slaughter processes in Korea. J Food Sci. 2017;82(3):711–717. doi: 10.1111/1750-3841.13630. [DOI] [PubMed] [Google Scholar]
- 2.Choi SW, Ha JS, Kim BY, Lee DH, Park JK, Youn HN, Hong YH, Lee SB, Lee JB, Park SY, et al. Prevalence and characterization of Salmonella species in entire steps of a single integrated broiler supply chain in Korea. Poult Sci. 2014;93(5):1251–1257. doi: 10.3382/ps.2013-03558. [DOI] [PubMed] [Google Scholar]
- 3.Ishihara K, Takahashi T, Morioka A, Kojima A, Kijima M, Asai T, Tamura Y. National surveillance of Salmonella enterica in food-producing animals in Japan. Acta Vet Scand. 2009;51(1):35. doi: 10.1186/1751-0147-51-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayamajhi N, Jung BY, Cha SB, Shin MK, Kim A, Kang MS, Lee KM, Yoo HS. Antibiotic resistance patterns and detection of blaDHA-1 in Salmonella species isolates from chicken farms in South Korea. Appl Environ Microbiol. 2010;76(14):4760–4764. doi: 10.1128/AEM.02536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaffga NH, Barton Behravesh C, Ettestad PJ, Smelser CB, Rhorer AR, Cronquist AB, Comstock NA, Bidol SA, Patel NJ, Gerner-Smidt P, et al. Outbreak of salmonellosis linked to live poultry from a mail-order hatchery. N Engl J Med. 2012;366(22):2065–2073. doi: 10.1056/NEJMoa1111818. [DOI] [PubMed] [Google Scholar]
- 6.Volkova VV, Bailey RH, Rybolt ML, Dazo-Galarneau K, Hubbard SA, Magee D, Byrd JA, Wills RW. Inter-relationships of Salmonella status of flock and grow-out environment at sequential segments in broiler production and processing. Zoonoses Public Health. 2010;57(7–8):463–475. doi: 10.1111/j.1863-2378.2009.01263.x. [DOI] [PubMed] [Google Scholar]
- 7.Berghaus RD, Thayer SG, Law BF, Mild RM, Hofacre CL, Singer RS. Enumeration of Salmonella and Campylobacter spp. in environmental farm samples and processing plant carcass rinses from commercial broiler chicken flocks. Appl Environ Microbiol. 2013;79(13):4106–4114. doi: 10.1128/AEM.00836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. Critically important antimicrobials for human medicine: ranking of antimicrobial agents for risk management of antimicrobial resistance due to non-human use. Geneva: World Health Organization; 2017.
- 9.Angulo FJ, Johnson KR, Tauxe RV, Cohen ML. Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb Drug Resist. 2000;6(1):77–83. doi: 10.1089/mdr.2000.6.77. [DOI] [PubMed] [Google Scholar]
- 10.Randall LP, Eaves DJ, Cooles SW, Ricci V, Buckley A, Woodward MJ, Piddock LJV. Fluoroquinolone treatment of experimental Salmonella enterica serovar typhimurium DT104 infections in chickens selects for both gyrA mutations and changes in efflux pump gene expression. J Antimicrob Chemother. 2005;56(2):297–306. doi: 10.1093/jac/dki189. [DOI] [PubMed] [Google Scholar]
- 11.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease 'Burden of Illness S The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50(6):882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 12.Hong J, Kim JM, Jung WK, Kim SH, Bae W, Koo HC, Gil J, Kim M, Ser J, Park YH. Prevalence and antibiotic resistance of Campylobacter spp. isolated from chicken meat, pork, and beef in Korea, from 2001 to 2006. J Food Prot. 2007;70(4):860–866. doi: 10.4315/0362-028X-70.4.860. [DOI] [PubMed] [Google Scholar]
- 13.Park H, Kang J, Jang Y, Song JY, Lee KJ. Studies on pharmacokinetics/Pharmacodanymics of Enrofloxacin in Broiler chicken. FASEB J. 2016;30:539. [Google Scholar]
- 14.Han FF, Lestari SI, Pu SH, Ge BL. Prevalence and antimicrobial resistance among Campylobacter spp. in Louisiana retail chickens after the enrofloxacin ban. Foodborne Pathog Dis. 2009;6(2):163–171. doi: 10.1089/fpd.2008.0171. [DOI] [PubMed] [Google Scholar]
- 15.Asai T, Kojima A, Harada K, Ishihara K, Takahashi T, Tamura Y. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn J Infect Dis. 2005;58(6):369–372. [PubMed] [Google Scholar]
- 16.Diarra MS, Silversides FG, Diarrassouba F, Pritchard J, Masson L, Brousseau R, Bonnet C, Delaquis P, Bach S, Skura BJ, et al. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolates. Appl Environ Microbiol. 2007;73(20):6566–6576. doi: 10.1128/AEM.01086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Da Costa PM, Belo A, Gonçalves J, Bernardo F. Field trial evaluating changes in prevalence and patterns of antimicrobial resistance among Escherichia coli and Enterococcus spp. isolated from growing broilers medicated with enrofloxacin, apramycin and amoxicillin. Vet Microbiol. 2009;139(3–4):284–292. doi: 10.1016/j.vetmic.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 18.USDA-FSIS . Microbiology laboratory guidebook. 2014. Isolation and identification of Salmonella from meat, poultry, pasteurized egg, and catfish products and carcass and environmental sponges; p. 4. [Google Scholar]
- 19.Ewing W. Edwards and Ewing’s identification of Enterobacteriaceae. New York: Elsevier Science Publishing Co. Inc.; 1986.
- 20.Kim YJ, Song BR, Lim JS, Heo EJ, Park HJ, Wee SH, Oh SM, Moon JS. First report on isolation of Salmonella enteritidis from eggs at grocery stores in Korea. Korean J Food Sci An. 2013;33(2):239–243. doi: 10.5851/kosfa.2013.33.2.239. [DOI] [Google Scholar]
- 21.CLSI . Performance standards for antimicrobial susceptibility testing-26th. Wayne: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 22.CLSI . Performance standards for antimicrobial susceptibility testing. Wayne: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 23.NARMS. The National Antimicrobial Resistance Monitoring System: enteric Bacteria. In: NARMS Integrated Report: 2012–2013: National Antimicrobial Resistance Monitoring System; 2013.
- 24.Chen Y, Hu D, Zhang Q, Liao XP, Liu YH, Sun J. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar typhimurium. Front Cell Infect Microbiol. 2017;7:37. doi: 10.3389/fcimb.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti-Infect Ther. 2012;10(8):917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 26.Ojo OE, Ogunyinka OG, Agbaje M, Okuboye JO, Kehinde OO, Oyekunle MA. Antibiogram of Enterobacteriaceae isolated from free-range chickens in Abeokuta, Nigeria. Veterinarski Arhiv. 2012;82(6):577–589. [Google Scholar]
- 27.Foley S, Lynne A, Nayak R. Challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J Anim Sci. 2008;86(14 suppl):E149–E162. doi: 10.2527/jas.2007-0464. [DOI] [PubMed] [Google Scholar]
- 28.Kim J-Y, Park Y-J, Lee S-O, Song W, Jeong SH, Yoo YA, Lee K-Y. Bacteremia due to Salmonella enterica serotype Montevideo producing plasmid-mediated AmpC β-lactamase (DHA-1) Ann Clin Lab Sci. 2004;34(2):214–217. [PubMed] [Google Scholar]
- 29.Harada T, Sakata J, Kanki M, Seto K, Taguchi M, Kumeda Y. Molecular epidemiological investigation of a diffuse outbreak caused by Salmonella enterica serotype Montevideo isolates in Osaka Prefecture, Japan. Foodborne Pathog Dis. 2011;8(10):1083–1088. doi: 10.1089/fpd.2011.0862. [DOI] [PubMed] [Google Scholar]
- 30.Lalsiamthara J, Lee JH. Pathogenic traits of Salmonella Montevideo in experimental infections in vivo and in vitro. Sci Rep. 2017;7:46786. doi: 10.1038/srep46786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci. 2017;96(3):687–702. doi: 10.3382/ps/pew342. [DOI] [PubMed] [Google Scholar]
- 32.Yoon RH, Cha SY, Wei B, Roh JH, Seo HS, Oh JY, Jang HK. Prevalence of Salmonella isolates and antimicrobial resistance in poultry meat from South Korea. J Food Prot. 2014;77(9):1579–1582. doi: 10.4315/0362-028X.JFP-14-018. [DOI] [PubMed] [Google Scholar]
- 33.Kim MS, Lim TH, Jang JH, Lee DH, Kim BY, Kwon JH, Choi SW, Noh JY, Hong YH, Lee SB, et al. Prevalence and antimicrobial resistance of Salmonella species isolated from chicken meats produced by different integrated broiler operations in Korea. Poult Sci. 2012;91(9):2370–2375. doi: 10.3382/ps.2012-02357. [DOI] [PubMed] [Google Scholar]
- 34.Kim S. Salmonella serovars from foodborne and waterborne diseases in Korea, 1998-2007: total isolates decreasing versus rare serovars emerging. J Korean Med Sci. 2010;25(12):1693–1699. doi: 10.3346/jkms.2010.25.12.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauser E, Tietze E, Helmuth R, Junker E, Prager R, Schroeter A, Rabsch W, Fruth A, Toboldt A, Malorny B. Clonal dissemination of Salmonella enterica serovar Infantis in Germany. Foodborne Pathog Dis. 2012;9(4):352–360. doi: 10.1089/fpd.2011.1038. [DOI] [PubMed] [Google Scholar]
- 36.Nogrady N, Toth A, Kostyak A, Paszti J, Nagy B. Emergence of multidrug-resistant clones of Salmonella Infantis in broiler chickens and humans in Hungary. J Antimicrob Chemother. 2007;60(3):645–648. doi: 10.1093/jac/dkm249. [DOI] [PubMed] [Google Scholar]
- 37.Dionisi AM, Lucarelli C, Benedetti I, Owczarek S, Luzzi I. Molecular characterisation of multidrug-resistant Salmonella enterica serotype Infantis from humans, animals and the environment in Italy. Int J Antimicrob Ag. 2011;38(5):384–389. doi: 10.1016/j.ijantimicag.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Noda T, Murakami K, Ishiguro Y, Asai T. Chicken meat is an infection source of Salmonella serovar Infantis for humans in Japan. Foodborne Pathog Dis. 2010;7(6):727–735. doi: 10.1089/fpd.2009.0438. [DOI] [PubMed] [Google Scholar]
- 39.Medalla F, Sjolund-Karlsson M, Shin S, Harvey E, Joyce K, Theobald L, Nygren BL, Pecic G, Gay K, Austin J, et al. Ciprofloxacin-resistant Salmonella enterica serotype Typhi, United States, 1999-2008. Emerg Infect Dis. 2011;17(6):1095–1098. doi: 10.3201/eid/1706.100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattiello SP, Drescher G, Barth VC, Jr, Ferreira CA, Oliveira SD. Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Anton Leeuw Int J G. 2015;108(5):1227–1238. doi: 10.1007/s10482-015-0577-1. [DOI] [PubMed] [Google Scholar]
- 41.Tamang MD, Nam HM, Kim TS, Jang GC, Jung SC, Lim SK. Emergence of extended-spectrum β-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J Clin Microbiol. 2011;49(7):2671–2675. doi: 10.1128/JCM.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JE, Lee YJ. Molecular characterization of antimicrobial resistant non-typhoidal Salmonella from poultry industries in Korea. Ir Vet J. 2017;70(1):20. doi: 10.1186/s13620-017-0095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarquin C, Alvarez D, Morales O, Morales AJ, Lopez B, Donado P, Valencia MF, Arevalo A, Munoz F, Walls I, et al. Salmonella on raw poultry in retail markets in Guatemala: levels, antibiotic susceptibility, and serovar distribution. J Food Prot. 2015;78(9):1642–1650. doi: 10.4315/0362-028X.JFP-15-117. [DOI] [PubMed] [Google Scholar]
- 44.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8(4):260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 45.Enne VI, Bennett PM, Livermore DM, Hall LM. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemother. 2004;53(6):958–963. doi: 10.1093/jac/dkh217. [DOI] [PubMed] [Google Scholar]
- 46.Chande CA, Chopdekar KA, Pradnya V, Unnati R, Jyoti B, Ritesh S, Shazia C, Joshi SG, Abhay C. Current trend of antibiotic sensitivity of Salmonella typhi and other Salmonellae in Mumbai: a 5 years study. Indian J Med Microbiol. 2016;34(1):115–116. doi: 10.4103/0255-0857.174100. [DOI] [PubMed] [Google Scholar]
- 47.Rai S, Jain S, Prasad KN, Ghoshal U, Dhole TN. Rationale of azithromycin prescribing practices for enteric fever in India. Indian J Med Microbiol. 2012;30(1):30–33. doi: 10.4103/0255-0857.93017. [DOI] [PubMed] [Google Scholar]
- 48.Hassing RJ, Goessens WH, van Pelt W, Mevius DJ, Stricker BH, Molhoek N, Verbon A, van Genderen PJ. Salmonella subtypes with increased MICs for azithromycin in travelers returned to the Netherlands. Emerg Infect Dis. 2014;20(4):705–708. doi: 10.3201/eid2004.131536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molloy A, Nair S, Cooke FJ, Wain J, Farrington M, Lehner PJ, Torok ME. First report of Salmonella enterica serotype paratyphi a azithromycin resistance leading to treatment failure. J Clin Microbiol. 2010;48(12):4655–4657. doi: 10.1128/JCM.00648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong MH, Yan M, Chan EW, Biao K, Chen S. Emergence of clinical Salmonella enterica serovar typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother. 2014;58(7):3752–3756. doi: 10.1128/AAC.02770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Li Y, Xu X, Liang B, Wu F, Yang X, Ma Q, Yang C, Hu X, Liu H, et al. Antimicrobial resistance of Salmonella enterica serovar Typhimurium in Shanghai, China. Front Microbiol. 2017;8:510. doi: 10.3389/fmicb.2017.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuang X, Hao H, Dai M, Wang Y, Ahmad I, Liu Z, Zonghui Y. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol. 2015;6:602. doi: 10.3389/fmicb.2015.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brennan J, Moore G, Poe SE, Zimmermann A, Vessie G, Barnum DA, Wilson J. Efficacy of in-feed tylosin phosphate for the treatment of necrotic enteritis in broiler chickens. Poult Sci. 2001;80(10):1451–1454. doi: 10.1093/ps/80.10.1451. [DOI] [PubMed] [Google Scholar]
- 54.Hoge CW, Gambel JM, Srijan A, Pitarangsi C, Echeverria P. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis. 1998;26(2):341–345. doi: 10.1086/516303. [DOI] [PubMed] [Google Scholar]
- 55.Hu Y, Liu F, Lin IYC, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):146–147. doi: 10.1016/S1473-3099(15)00533-2. [DOI] [PubMed] [Google Scholar]
- 56.Kempf I, Fleury MA, Drider D, Bruneau M, Sanders P, Chauvin C, Madec J-Y, Jouy E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int J Antimicrob Ag. 2013;42(5):379–383. doi: 10.1016/j.ijantimicag.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 57.Olaitan AO, Dia NM, Gautret P, Benkouiten S, Belhouchat K, Drali T, Parola P, Brouqui P, Memish Z, Raoult D, et al. Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp enterica serotype Newport by pilgrims during Hajj. Int J Antimicrob Ag. 2015;45(6):600–604. doi: 10.1016/j.ijantimicag.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Frasson I, Bettanello S, De Canale E, Richter SN, Palu G. Serotype epidemiology and multidrug resistance patterns of Salmonella enterica infecting humans in Italy. Gut Pathog. 2016;8(1):26. doi: 10.1186/s13099-016-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies RH, Nicholas RA, McLaren IM, Corkish JD, Lanning DG, Wray C. Bacteriological and serological investigation of persistent Salmonella enteritidis infection in an integrated poultry organisation. Vet Microbiol. 1997;58(2–4):277–293. doi: 10.1016/S0378-1135(97)00157-0. [DOI] [PubMed] [Google Scholar]
- 60.Donado-Godoy P, Gardner I, Byrne BA, Leon M, Perez-Gutierrez E, Ovalle MV, Tafur MA, Miller W. Prevalence, risk factors, and antimicrobial resistance profiles of Salmonella from commercial broiler farms in two important poultry-producing regions of Colombia. J Food Prot. 2012;75(5):874–883. doi: 10.4315/0362-028X.JFP-11-458. [DOI] [PubMed] [Google Scholar]
- 61.Nayak R, Kenney PB, Keswani J, Ritz C. Isolation and characterisation of Salmonella in a Turkey production facility. Br Poult Sci. 2003;44(2):192–202. doi: 10.1080/0007166031000088370. [DOI] [PubMed] [Google Scholar]
- 62.Thakur S, Brake J, Keelara S, Zou M, Susick E. Farm and environmental distribution of Campylobacter and Salmonella in broiler flocks. Res Vet Sci. 2013;94(1):33–42. doi: 10.1016/j.rvsc.2012.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendrograms showing pattern analysis on the basis of Xba I-PFGE of the 16 Salmonella Senftenberg isolates obtained from broiler farms and their association with antimicrobial-resistance. The Dice coefficient was used to perform similarity analysis. aS, cloacal swabs; L, litter; F, feed. bR, resistance (dark pink); I, intermediate resistance (pink), S, sensitivity (light pink). Dotted lines indicate 90% similarity. (DOCX 271 kb)
Isolation rates and multidrug resistant (MDR) rates in different types of samples obtained during production cycles 1 and 2. Different lowercase (a/b) and capital letters (A/B) in the same row were used to indicate significant (P < 0.05) differences in isolation rates and MDR rates between production cycles 1 and 2, respectively. (DOCX 36 kb)
Data Availability Statement
The datasets analyzed in the present study are available from the corresponding author on reasonable request. Additional supporting files can be found in the supplementary material section.