Abstract
Objective(s):
Asthma, the most frequent chronic respiratory disease, results from a complex interaction between multiple genes and environmental factors. To date, more than 100 candidate genes and single nucleotide polymorphisms (SNPs) have been reported to be associated with asthma. One of the discovered genes related to asthma is ADAM33. However, the relationship between ADAM33 gene polymorphisms and asthma is controversial. The aim of this study was to investigate the association between four ADAM33 gene SNPs and susceptibility to asthma in patients from southwestern Iran.
Materials and Methods:
ADAM33 gene polymorphisms at positions T+1 (rs2280091), T1 (rs3918396), S1 (rs2280089), and F+1 (rs511898) were examined in 150 patients with asthma and 149 age- and sex-matched healthy controls with a PCR-RFLP method.
Results:
There were no differences between patients and controls in allelic or genotype frequencies of ADAM33 SNPs. We found no associations between allelic or genotype distribution of the SNPs and spirometry indices, concomitant involvement of other allergic diseases, or exposure to cigarette smoke. In contrast to H4 haplotype, which appeared to be protective against asthma, inheritance of H2 and H3 haplotypes increased the risk of asthma up to 2–3 folds.
Conclusion:
ADAM33 gene polymorphisms appear to play a partial role in asthma susceptibility, investigation of expression changes in this gene in response to environmental factors or the local formation of a soluble form of the molecule in the lung can be helpful to elucidate the impact of this molecule in the induction of asthma.
Key Words: ADAM33, Asthma, rs511898, rs3918396, rs2280089, rs2280091
Introduction
Asthma is the most frequent chronic respiratory disease, affecting more than 300 million people around the world (1). This heterogeneous disease is classified into allergic and nonallergic types, which share common symptoms. Generally, extrinsic irritants such as tobacco smoke, plant pollen, animal fur, dust mite feces, some kinds of food, exhaust fumes, or certain chemicals stimulate allergic asthma in genetically predisposed individuals, whereas nonallergic or intrinsic asthma is usually triggered by infections and physical or emotional stress (2). In contrast to the relatively stable prevalence of nonallergic asthma of around 3.4%–3.8%, the prevalence of allergic asthma increased from 5.0% in 1996 to 6.0% in 2006 and 7.3% in 2016 (1).
Asthma results from a complex interaction between multiple genes and environmental factors. So far more than 100 candidate genes and single nucleotide polymorphisms (SNPs) are reported to be associated with asthma (3). Familial aggregation of asthma with a higher concordance between monozygotic twins (0.74) than dizygotic twins (0.35), indicates the presence of a strong genetic influence in the induction of asthma (4).
In 2002, ADAM33 (disintegrin and metallopro-teinase domain-containing protein 33) was reported to be a genetic risk factor for susceptibility to asthma (5). The ADAM33 gene comprises 23 exons extending through a 14-kb segment on chromosome 20p13 and encodes a membrane-anchored metalloprotease which may exert its main effect by processing other molecules, e.g., growth factors and cytokines, linked to airway remodeling (6). In addition to the membrane-bound molecule, secreted and intracellular isoforms can be produced as further splice variants. Variants lacking the metalloproteinase domain have also been reported which may mediate other activities of the molecule. ADAM33 gene polymorphisms may affect the fate of ADAM33 transcripts through their effects on mRNA splicing, mRNA stability, and the selection of transcripts for export to the cytoplasm (7). ADAM33 is known to be involved in branching morphogenesis in the fetal lung, and polymorphic variations in this gene are associated with the pathophysiology of chronic obstructive pulmonary disease, airway remodeling, and hyperresponsiveness in asthma (8-10).
Because the strength of association between asthma and different SNPs in the ADAM33 gene was reported to vary in different populations, the aim of the present study was to investigate the association between four ADAM33 SNPs and susceptibility to asthma in a sample of patients from southwestern Iran.
Materials and Methods
Patients and controls
The participants were patients who were referred to allergy clinics affiliated with Shiraz University of Medical Sciences (Fars province, southwestern Iran) with a diagnosis of asthma according to Expert Panel Report 3 criteria (11). The study protocol was approved by the Ethics Committee of our university, and 150 patients with mild to moderate persistent asthma were included in the study after written informed consent was obtained from patients or their parents. Patients with any underlying disease except asthma were excluded from the study. Information was recorded about demographic characteristics, cigarette smoke exposure, family history of atopy and concomitant involvement of allergic rhinitis, atopic dermatitis, and urticaria.
By public announcement, a total of 149 age- and sex-matched (±1 year) unrelated healthy volunteers of the same ethnicity as the patients and with no personal or family history of asthma or other atopic diseases, were recruited as a control group and offered free screening for asthma. Blood samples (1 ml) for genetic analysis were obtained from patients and controls with EDTA as an anticoagulant.
Pulmonary function tests
Pulmonary function tests were performed by spirometry (Cosmed, Rome, Italy) on patients older than 6 years. Forced vital capacity (FVC), forced expiratory volume in 1 second (FEV1), ratio of forced expiratory volume in 1 second to forced vital capacity (FEV1/FVC) and peak expiratory flow (PEF) were measured, and spirometry indices <80% of the predicted values were considered abnormal (12).
Genotyping of ADAM33 SNPs
Genomic DNA was extracted from 200 µl of blood with a DNA extraction kit (GeNet Bio, Nonsan, South Korea) and four SNPs in the ADAM33 gene previously reported to be associated with asthma in some populations (5,13) were analyzed with a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method (Table 1).
Table 1.
ADAM33 gene SNP genotyping by PCR-RFLP: primers, restriction enzymes, and length of the digested product fragments
| ADAM33 SNP | Primer sequences |
Restriction
enzyme |
Recognition site | Fragment size (bp) |
|---|---|---|---|---|
| T+1 (rs2280089) (intron 20) |
F: AGGGTCTGGGAGAAATGGTG R: TCTTTGGAAGCTGAGCGATG |
MboII | …GAAGA(N)8↓… …CTTCT(N)7↑… |
GG: 424+132 bp AG: 424+132+123+301 bp AA: 301+132+123 bp |
| T1 (rs2280091) (exon 20) |
F: GTGAATATGGTCAGCAGGAG R: GTGACTTGGAGCAGATGG |
NcoI | …C↓CATGG… …GGTAC↑C… |
GG: 375 bp GA: 375+187+188 bp AA: 187+188 bp |
| S1 (rs3918396) (exon 19) |
F: GTGGCAGCATGGACAGT R: CAGGAGTAGGCTCAGGAAG |
BtsCI | …GGATGNN↓… …CCTAC↑NN… |
GG: 304 bp GA: 304+153+151 bp AA: 151+153 bp |
| F+1 (rs511898) (intron 6) |
F: AAATACGACTCGAGGC R: GGACTTCTCAACCCACGAG |
BsmBI | …CGTCTC(N)1↓… …GCAGAG(N)5↑… |
TT: 220 bp TC: 220+183+37 bp CC: 183+37 bp |
The polymorphic sequence-containing regions were amplified by PCR in a total volume of 25 μl containing 30 ng genomic DNA, 12.5 μl 2x master mix (Ampliqon, Odense, Denmark) and 0.5 μmol of each primer (Bioneer, Daejeon, South Korea). Cycling conditions were as follows: one cycle at 95 °C for 5 min, 30 cycles at 95 °C for 30 sec, 62 °C for 30 sec, 72 °C for 30 sec, and a final extension at 72 °C for 10 min. PCR products were digested with related restriction enzymes (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol, and digested products were then resolved on 2.5% agarose gel.
Statistical analysis
Allele, genotype, and haplotype frequencies of AMAM33 gene SNPs were calculated using Arlequin v. 3.1 software. Linkage disequilibrium (LD) among SNPs in healthy controls and Hardy-Weinberg compliance were analyzed using Haploview v. 4.2 software. Allele and genotype frequencies were compared between patients and controls – globally and after stratification based on age, sex, and both – using the chi-squared test, and odds ratios (OR) with a 95% confidence interval (CI) were calculated using Epi-info v. 7 software. Relationships between allele and genotype frequencies in each SNP and each spirometry parameter (based on a cut-off value of 80%), cigarette smoke exposure, family history of atopy, or concurrent involvement of other allergic diseases were also analyzed with the chi-squared test. Values of P<0.05 were considered statistically significant.
Results
The demographic and clinical characteristics along with the spirometry results in patients with asthma are shown in Table 2. Allele, genotype, and three-loci haplotype frequencies of the ADAM33 gene SNPs in patients and healthy controls are summarized in Table 3. There were no differences between patients and controls in allelic or genotype frequencies of the SNPs, even after data stratification based on age, sex, and both. We found no associations between allelic or genotype distribution of the SNPs and spirometry indices, family history of atopy, concomitant involvement of other allergic diseases, or exposure to cigarette smoke.
Table 2.
Demographic and clinical characteristics of patients with asthma
| Age (year) | <13 (n=44) | ≥13 (n=106) | ||||
|---|---|---|---|---|---|---|
| Sex | Female (n=22) | Male (n=22) | Female (n=68) | Male (n=38) | ||
| Mean age±SD (years) (Age range) |
8.95±2.44 (5 to 12) |
8.27±2.34 (5 to 12) |
44.17±16.52 (13 to 82) |
39.01±19.39 (13 to 82) |
||
| Family history | 9 | 12 | 29 | 23 | ||
| Cigarette smoke exposure | 19 | 14 | 38 | 11 | ||
| Concurrent allergic diseases | ||||||
| Allergic rhinitis | 12 | 12 | 33 | 29 | ||
| Eczema | 18 | 14 | 9 | 10 | ||
| Urticaria | 16 | 12 | 16 | 9 | ||
| Spirometry parameters | ||||||
| FEV1 <80% | 4 | 6 | 34 | 22 | ||
| FVC <80% | 4 | 9 | 35 | 20 | ||
| FEV1/FVC <80% | 0 | 0 | 3 | 4 | ||
| PEF <80% | 8 | 12 | 45 | 24 | ||
Table 3.
Allele, genotype, and haplotype frequencies of ADAM33 gene SNPs in Southwestern Iranian patients with asthma and healthy controls
| ADAM33 |
Patients
(n=150) |
Controls
(n=149) |
P -value | OR (CI95%) |
|---|---|---|---|---|
| Alleles | ||||
| T+1 | N (F%) | N (F%) | ||
| G | 221 (73.6%) | 234 (78.5%) | 0.19 | 0.7 (0.5 - 1.1) |
| A | 79 (26.4%) | 64 (21.5%) | ||
| T1 | ||||
| G | 82 (27.3%) | 90 (30.2%) | 0.49 | 0.8 (0.5 - 1.2) |
| A | 218 (72.7%) | 208 (69.8%) | ||
| S1 | ||||
| G | 248 (82.6%) | 255 (85.6%) | 0.39 | 0.8 (0.6 - 1.2) |
| A | 52 (17.4%) | 43 (14.4%) | ||
| F+1 | ||||
| T | 176 (58.6%) | 172 (57.7%) | 0.87 | 1.03 (0.75 - 1.4) |
| C | 124 (41.4%) | 126 (42.3%) | ||
| Genotypes | ||||
| T+1 | ||||
| GG | 77 (51.3%) | 92 (61.7%) | 0.1 | 0.6 (0.41 - 1.03) |
| GA | 67 (44.7%) | 50 (33.6%) | 1.5 (1 - 2.5) | |
| AA | 6 (4%) | 7 (4.7%) | 0.8 (0.2 - 2.5) | |
| T1 | ||||
| GG | 6 (4%) | 8 (5.4%) | 0.69 | 0.7 (0.24 - 2.17) |
| GA | 70 (46.7%) | 74 (49.7%) | 0.8 (0.56 - 1.39) | |
| AA | 74 (49.3%) | 67 (44.9%) | 1.1 (0.7 - 1.8) | |
| S1 | ||||
| GG | 99 (66%) | 106 (71.1%) | 0.41 | 0.7 (0.48 - 1.2) |
| GA | 50 (33.3%) | 43 (28.9%) | 1.2 (0.77 - 2.06) | |
| AA | 1 (0.6%) | --- | --- | |
| F+1 | ||||
| TT | 28 (18.6%) | 27 (18.1%) | 0.7 | 1 (0.57 - 1.86) |
| TC | 120 (80%) | 118 (79.2%) | 1 (0.58 - 1.84) | |
| CC | 2 (1.3%) | 4 (2.7%) | 0.4 (0.08 - 2.7) | |
| Haplotypes T+1/T1/S1 | ||||
| H1: G A G | 166 (55.33%) | 181 (60.79%) | 0.09 | 0.8008 (0.5784-1.1087) |
| H2: A G G | 77 (25.65%) | 44 (14.75%) | 0.00046 | 1.9933 (1.3205-3.0088) |
| H3: G A A | 50 (16.65%) | 21 (7.11%) | 0.00013 | 2.6381 (1.5411-4.5161) |
| H4: G G G | 5 (1.68%) | 24 (8.12%) | 0.0001 | 0.1935 (0.0728-0.5143) |
| H5: A A A | 2 (0.68%) | --- | 0.25 | --- |
| H6: A G A | --- | 14 (4.82%) | 0.000054 | --- |
| H7: G G A | --- | 8 (2.50%) | 0.0037 | --- |
| H8: A A G | --- | 6 (1.90%) | 0.015 | --- |
All SNPs met the criteria for the Hardy-Weinberg equilibrium except for rs511898 (Table 4). This SNP was omitted from subsequent linkage and haplotype analyses (14).
Table 4.
Deviation from Hardy-Weinberg equilibrium in the ADAM33 gene in patients from southwestern Iran with asthma and healthy controls
|
ADAm33
SNPs |
Position | Alleles |
HWE
|
|||||
|---|---|---|---|---|---|---|---|---|
|
Patients
|
Controls
|
|||||||
| Obs HET | Pred HET | P-value | Obs HET | Pred HET | P-value | |||
| T+1 | 3669480 | G:A | 0.447 | 0.388 | 0.1009 | 0.336 | 0.337 | 1.0000 |
| T1 | 3669587 | A:G | 0.467 | 0.397 | 0.0509 | 0.497 | 0.422 | 0.0474 |
| S1 | 3671118 | G:A | 0.333 | 0.287 | 0.0713 | 0.289 | 0.247 | 0.0574 |
| F+1 | 3674438 | T:C | 0.8 | 0.485 | <0.00001 | 0.792 | 0.488 | <0.00001 |
Obs HET: observed heterozygosity; Pred HET: predicted heterozygosity
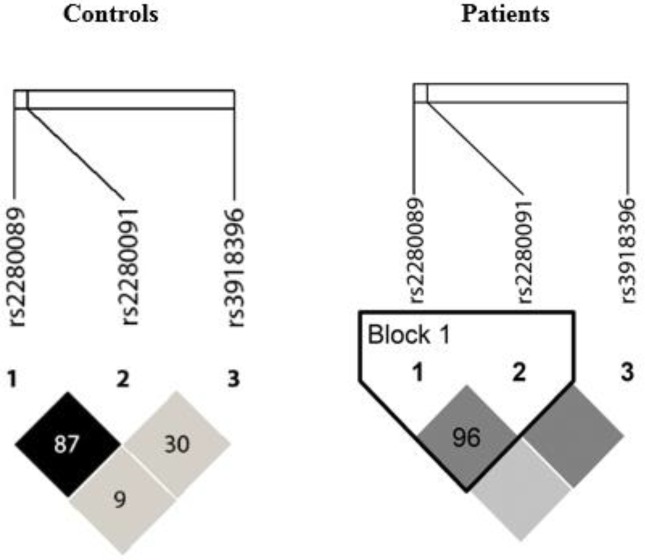
The results of linkage analysis for the three remaining SNPs showed a linkage between rs2280089 and rs2280091 (Figure 1). The results of haplotype analysis revealed that H5 (0.68%) was observed exclusively in patients, whereas H6 (4.8%), H7 (2.5%), and H8 (1.9%) were seen only in healthy controls. The frequency of H2 and H3 haplotypes were significantly higher in patients than controls (25.65% vs. 14.75%, P=0.00046 and 16.65% vs. 7.11%, P=0.00013, respectively), and inheritance of these haplotypes was associated with higher risk of asthma up to 2–3 folds. Haplotype H4 was more frequent among controls than patients (8.1% vs. 1.68%, P=0.0001) and inheritance of this haplotype was associated with decreased risk of asthma about one fifth in patients (Table 3).
Figure 1.
Linkage disequilibrium analysis of ADAM33 gene SNPs in patients with asthma and healthy controls from southwestern Iran
Discussion
Asthma is a chronic inflammatory disorder characterized by obstruction of the bronchial tubes and airway remodeling with increased smooth muscle mass, fibroblast activation, neovascularization, and epithelial alterations. ADAM33 is preferentially expressed in airway fibroblasts and smooth muscle cells in patients with asthma.
Although there are data showing a link between ADAM33 and asthma, the exact role of this gene in the pathophysiology of asthma is not entirely clear (5,6,15). The contribution of ADAM33 gene polymorphisms to the risk of asthma is controversial. Increased risk of asthma in persons with one or more ADAM33 SNP allele(s) has been reported in African American, US white, US Hispanic, Dutch white (16), Icelandic, UK (17), and Japanese populations (18).
We found no correlation between any of the four ADAM33 SNPs (T1, T+1, S1, and F+1) and asthma in a sample of patients from the population of southwestern Iran. The lack of association between ADAM33 polymorphisms and asthma was also reported in Australian (19), Chinese Li (20), Korean (21), Indian (22), and Turkish populations (23). In the only related report from Iran we are aware of, associations were found between the rs2280091 C allele and severe asthma, and between the rs2787094 G allele and moderate asthma in a population sample from northeastern Iran (24).
Our results suggest that ADAM33 gene polymorphisms play a restricted role in susceptibility to asthma. Although there is a possible linkage between undefined causal genes on the short arm of chromosome 20 and certain ADAM33 SNP alleles or haplotypes, increased levels of ADAM33 gene expression in response to environmental factors such as cigarette smoke or air pollutants might play a critical role in the induction of asthma. Moreover, the local formation of soluble ADAM33 in airways causes remodeling in the developing lungs, which through interaction with Th2-mediated inflammation makes patients more responsive to low concentrations of allergens (25). In this connection, there is evidence that in-utero and early-life tobacco smoke exposure influences the induction of TGF-β2, which in turn results in ADAM33 shedding and reduced lung function (26). However, in contrast to Reijmerink et al, who suggested a role for the interaction of cigarette smoke with certain ADAM33 polymorphisms in the induction of asthma (26), we found no association between the SNPs we studied and exposure to cigarette smoke in our patients with childhood or adulthood asthma.
In contrast to Jongepier et al., who found an association between the S2 minor allele and decreased FEV1 (27), and in agreement with El-Falaki et al (28), we found no association between any of the four SNPs analyzed here and spirometry parameters. Previously, one study reported an inverse correlation between ADAM33 protein levels in bronchoalveolar lavage fluids and FEV1 in patients with asthma (29). However, we are not aware of any reports of a functional correlation between any ADAM33 SNP alleles and ADAM33 protein levels in bronchial smooth muscle cells, bronchial lavage fluids, or serum in patients with asthma. Unlike Zhang et al., who reported associations between some ADAM33 polymorphisms and concomitant allergic rhinitis and asthma in the Chinese Han population (30), and Matsusue et al, who found a link between rs2853209 and atopic dermatitis in Japanese children (31), we found no association between ADAM33 SNPs and any spirometry indices. However, a potential limitation of our study is that we were unable to collect or process bronchoalveolar lavage fluid from our patients to analyze ADAM33 protein levels.
Like another study, we also found a linkage between rs2280089 and rs2280091 loci (32). As shown in Figure 1, this linkage was stronger in patients (r2 = 0.96) than controls (r2 = 0.87).
Although we found no link between asthma and any of the ADAM33 SNPs analyzed here, GGG haplotype was positively associated with the disease in our patients. This confirms the superiority of haplotype analysis over single-SNP analysis in studies that aim to elucidate the associations among different clinical, genetic, and environmental factors in complex genetic disorders such as asthma (33).
Conclusion
We found no link between asthma and T+1, T1, S1, or F+1 SNPs in the ADAM33 gene, however, AGG and GAA haplotypes of the first three SNPs were positively associated with the disease in our patients. ADAM33 gene SNPs appear to play a partial role in asthma susceptibility, and investigation of expression changes in this gene in response to environmental factors or the local formation of soluble form of the molecule in the lung can be helpful to elucidate the impact of this molecule in the induction of asthma.
Acknowledgment
The results presented in this paper were part of a student thesis for the MSc degree in Immunology (by Bent-Alhoda Hoseini-Pouya) and supported by a grant from Shiraz University of Medical Sciences, Shiraz, Iran (no: 7122). We thank K. Shashok (AuthorAID in the Eastern Mediterranean) for improving the use of English in the manuscript.
References
- 1.Backman H, Räisänen P, Hedman L, Stridsman C, Andersson M, Lindberg A, et al. Increased prevalence of allergic asthma from 1996 to 2006 and further to 2016 - results from three population surveys. Clin Exp Allergy. 2017;47:1426–1435. doi: 10.1111/cea.12963. [DOI] [PubMed] [Google Scholar]
- 2.Peters SP. Asthma phenotypes: nonallergic (intrinsic) asthma. J Allergy Clin Immunol Pract. 2014;2:650–652. doi: 10.1016/j.jaip.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Meyers DA. Genetics of Asthma and Allergy: What have we learned? J Allergy Clin Immunol. 2010;126:439–446. doi: 10.1016/j.jaci.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijanzadeh M, Mahesh PA, Ramachandra NB. An understanding of the genetic basis of asthma. Indian J Med Res. 2011;134:149–161. [PMC free article] [PubMed] [Google Scholar]
- 5.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 6.Yoshinaka T, Nishii K, Yamada K, Sawada H, Nishiwaki E, Smith K, et al. Identification and characterization of novel mouse and human ADAM33s with potential metalloprotease activity. Gene. 2002;282:227–236. doi: 10.1016/s0378-1119(01)00818-6. [DOI] [PubMed] [Google Scholar]
- 7.Powell RM, Wicks J, Holloway JW, Holgate ST, Davies DE. The splicing and fate of ADAM33 transcripts in primary human airways fibroblasts. Am J Respir Cell Mol Biol. 2004;31:13–21. doi: 10.1165/rcmb.2003-0330OC. [DOI] [PubMed] [Google Scholar]
- 8.Reijmerink NE, Kerkhof M, Koppelman GH, Gerritsen J, de Jongste JC, Smit HA, et al. Smoke exposure interacts with ADAM33 polymorphisms in the development of lung function and hyperresponsiveness. Allergy. 2009;64:898–904. doi: 10.1111/j.1398-9995.2009.01939.x. [DOI] [PubMed] [Google Scholar]
- 9.Figarska SM, Vonk JM, van Diemen CC, Postma DS, Boezen HM. ADAM33 gene polymorphisms and mortality. A prospective cohort study. PLoS One. 2013;8:e67768. doi: 10.1371/journal.pone.0067768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holgate ST, Davies DE, Rorke S, Cakebread J, Murphy G, Powell RM, Holloway JW. ADAM 33 and its association with airway remodeling and hyperresponsiveness in asthma. Clin Rev Allergy Immunol. 2004;27:23–34. doi: 10.1385/CRIAI:27:1:023. [DOI] [PubMed] [Google Scholar]
- 11.National Asthma Education and Prevention Program. Expert Panel Report 3 (EPR-3): guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120:S94–S138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 12.Pulmonary function testing. Clin Privil White Pap. 2012;38:1–15. [PubMed] [Google Scholar]
- 13.Liang S, Wei X, Gong C, Wei J, Chen Z, Deng J. A disintegrin and metalloprotease 33 (ADAM33) gene polymorphisms and the risk of asthma: a meta-analysis. Hum Immunol. 2013;74:648–657. doi: 10.1016/j.humimm.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Nair AK, Sugunan D, Kumar H, Anilkumar G. Case-control analysis of SNPs in GLUT4, RBP4 and STRA6: association of SNPs in STRA6 with type 2 diabetes in a South Indian population. PLoS One. 2010;5:e11444. doi: 10.1371/journal.pone.0011444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma N, Tripathi P, Awasthi S. Role of ADAM33 gene and associated single nucleotide polymorphisms in asthma. Allergy Rhinol (Providence) 2011;2:e63–e70. doi: 10.2500/ar.2011.2.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, et al. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112:717–722. doi: 10.1016/s0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- 17.Blakey J, Halapi E, Bjornsdottir US, Wheatley A, Kristinsson S, Upmanyu R, et al. Contribution of ADAM33 polymorphisms to the population risk of asthma. Thorax. 2005;60:274–276. doi: 10.1136/thx.2004.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota T, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, et al. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin Exp Allergy. 2006;36:884–891. doi: 10.1111/j.1365-2222.2006.02522.x. [DOI] [PubMed] [Google Scholar]
- 19.Kedda MA, Duffy DL, Bradley B, O’Hehir RE, Thompson PJ. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet. 2006;14:1027–1036. doi: 10.1038/sj.ejhg.5201662. [DOI] [PubMed] [Google Scholar]
- 20.Shen B, Lin R, Wang CC, Rei J, Sun Y, Yang YL, Lin YY. ADAM33 gene polymorphisms identified to be associated with asthma in a Chinese Li population. Biomed Rep. 2017;6:323–328. doi: 10.3892/br.2017.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, Park HS, Park SW, Jang AS, Uh ST, Rhim T, et al. ADAM33 polymorphism: association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy. 2004;34:860–865. doi: 10.1111/j.1365-2222.2004.01977.x. [DOI] [PubMed] [Google Scholar]
- 22.Bijanzadeh M, Ramachandra NB, Mahesh PA, Mysore RS, Kumar P, Manjunath BS, Jayaraj BS. Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188:415–422. doi: 10.1007/s00408-010-9247-2. [DOI] [PubMed] [Google Scholar]
- 23.Bora E, Arikan-Ayyildiz Z, Firinci F, Çankaya T, Giray-Bozkaya Ö, Uzuner N, Ülgenalp A. ADAM33 Gene polymorphisms are not associated with asthma in Turkish children. Pediatr Allergy Immunol Pulmonol. 2012;25:97–100. [Google Scholar]
- 24.Karimi MR, Faridhosseini R, Abbaszadegan MR, Azad FJ, Shirkani A, Riyahi A, Montazar M, Gholamin M. Association of ADAM33 gene polymorphisms with allergic asthma. Iran J Basic Med Sci . 2014;17:716–721. [PMC free article] [PubMed] [Google Scholar]
- 25.Davies ER, Kelly JF, Howarth PH, Wilson DI, Holgate ST, Davies DE, et al. Soluble ADAM33 initiates airway remodeling to promote susceptibilityfor allergic asthma in earlylife. JCI Insight. 2016;1:e87632. doi: 10.1172/jci.insight.87632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reijmerink NE, Kerkhof M, Koppelman GH, Gerritsen J, de Jongste JC, Smit HA, et al. Smoke exposure interacts with ADAM33 polymorphisms in the development of lung function and hyperresponsiveness. Allergy. 2009;64:898–904. doi: 10.1111/j.1398-9995.2009.01939.x. [DOI] [PubMed] [Google Scholar]
- 27.Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, et al. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34:757–760. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- 28.El-Falaki MM, Wilson MM, Ezzat GM, Mokhtar DA, El Baz MS, Hamed DH. A disintegrin and metalloproteinase 33 (ADAM33) gene polymorphism association with asthma in Egyptian children. Egypt J Med Human Genet . 2013;14:55–62. [Google Scholar]
- 29.Lee JY, Park SW, Chang HK, Kim HY, Rhim T, Lee JH, et al. A disintegrin and metalloproteinase 33 protein in patients with asthma: Relevance to air flow limitation. Am J Respir Crit Care Med. 2006;173:729–735. doi: 10.1164/rccm.200409-1175OC. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Su D, Zhang X, Sui H, Jin L, Lü F, Zhang J. Association of ADAM33 gene polymorphisms with adult concomitant allergic rhinitis and asthma in Chinese Han population. Mol Biol Rep. 2009;36:1505–1509. doi: 10.1007/s11033-008-9343-z. [DOI] [PubMed] [Google Scholar]
- 31.Matsusue A, Kiyohara C, Tanaka K, Sasaki S, Miyake Y. ADAM33 genetic polymorphisms and risk of atopic dermatitis among Japanese children. Clin Biochem. 2009;42:477–483. doi: 10.1016/j.clinbiochem.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Raby BA, Silverman EK, Kwiatkowski DJ, Lange C, Lazarus R, Weiss ST. ADAM33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol. 2004;113:1071–1078. doi: 10.1016/j.jaci.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 33.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]