Abstract
Modern medicine is in the midst of a revolution driven by “big data,” rapidly advancing computing power, and broader integration of technology into healthcare. Highly detailed and individualized profiles of both health and disease states are now possible, including biomarkers, genomic profiles, cognitive and behavioral phenotypes, high-frequency assessments, and medical imaging. Although these data are incredibly complex, they can potentially be used to understand multi-determinant causal relationships, elucidate modifiable factors, and ultimately customize treatments based on individual parameters. Especially for neurodegenerative diseases, where an effective therapeutic agent has yet to be discovered, there remains a critical need for an interdisciplinary perspective on data and information management due to the number of unanswered questions. Biomedical informatics is a multidisciplinary field that falls at the intersection of information technology, computer and data science, engineering, and healthcare that will be instrumental for uncovering novel insights into neurodegenerative disease research, including both causal relationships and therapeutic targets and maximizing the utility of both clinical and research data. The present study aims to provide a brief overview of biomedical informatics and how clinical data applications such as clinical decision support tools can be developed to derive new knowledge from the wealth of available data to advance clinical care and scientific research of neurodegenerative diseases in the era of precision medicine.
Keywords: Informatics, Neurodegenerative disease, Biomarker, Clinical decision support, Precision medicine, Evidence-based practice, Alzheimer's disease, Learning healthcare, Machine learning
1. Biomedical informatics applications for precision management of neurodegenerative disease
The practice of medicine is in the midst of a modern-day revolution, driven by “big data,” rapidly advancing computing power, and broader integration of technology into health-care service provision. It is anticipated that from 2014 to 2024, health-care information technology job growth is expected to substantially outpace job growth among other industries [1], due in part to the deluge of health data generated by new technologies. These data can create highly detailed and individualized profiles of both health and disease states, which can potentially be used to understand multi-determinant causal relationships, elucidate modifiable factors, and ultimately customize treatments based on individual parameters. In the case of neurodegenerative diseases (NDDs), where many questions are still unanswered and effective therapies have remained elusive, there is a critical need for an interdisciplinary perspective on data and information management to derive novel insights, generate new knowledge, improve care, and facilitate treatment discovery. Biomedical informatics (BMIs) is a multidisciplinary field that falls at the intersection of information technology, computer and data science, engineering, and healthcare; this interdisciplinary intersection will play an integral role in the future of medicine. The present study aims to provide a brief overview of BMIs and how novel clinical data applications can be developed to derive new knowledge from existing data to advance clinical care and scientific research of NDD in the era of precision medicine (PM). The generation and organization of big data and its application in healthcare settings depend on a scientific infrastructure that anticipates both the needs of these types of data, as well as, how they may be used. The National Institutes of Health and the National Institute of General Medical Sciences support awards such as the Center for Biomedical Research Excellence grants to support big data infrastructure and advance data science. The Center for Neurodegeneration and Translational Neuroscience is a Center for Biomedical Research Excellence–supported neuroscience enterprise with a Data Management and Statistics Core that serves as a platform for investigating how to apply big data to PM.
1.1. What is PM?
PM has recently been defined as “an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person.” [2]. A primary aim of PM is to link individuals with the best possible treatment for an individual's disease in the hope of improving clinical outcomes, and ultimately, patient health. Effective implementation of PM into clinical practice requires integration of translational research from a diverse array of data sources to ensure that the PM approach is firmly rooted in empirical evidence. Although individualized approaches to clinical care have been present for decades, (e.g., matching blood transfusions or solid organ transplants based on blood type), the wealth of data available in modern medicine, with all its technological advances, moves the potential for truly precise interventions far beyond what has historically been possible. NDDs present significant opportunity for development of PM interventions [3], not only because of the wealth of genetic information now available [4], [5] but also because of the concurrent growth in biomarker discovery [6] and the ability to characterize the cognitive and behavioral phenotype in rich detail. Moreover, the historical approaches (e.g., one-size-fits-all treatment) have almost universally failed to uncover an effective therapeutic agent [7], which may in part be due to the incredible diversity in disease manifestations that can result from the same underlying pathology.
1.2. Biomarkers of neurodegeneration for PM
Definitive diagnosis of NDD requires positive identification of the pathologic changes occurring in the brain, which for most NDD begin decades before the onset of observable symptoms. As a result, there is considerable interest in the discovery and validation of reliable biomarkers that could be used to improve diagnostic accuracy, especially early in the disease process before the full clinical syndrome is manifest [8]. At present, body fluid analysis and brain imaging are the two principal sources for biomarker data.
1.3. Fluid biomarkers
Cerebrospinal fluid (CSF) has been a prominent target for discovery of potential biomarkers, given the possibility that it provides molecular insights into pathologic processes within the brain. For example, amyloid β-42 and tau were two of the earliest validated biomarkers in Alzheimer's disease (AD) [9], which has been refined to separate tau into total tau and phosphorylated tau [10], [11]. These CSF markers have demonstrated good sensitivity to AD pathology and are widely used in both clinical practice and research. However, limited specificity [12], [13] has mitigated their utility as stand-alone diagnostics, especially at preclinical stages [14]. Coupled with the invasive nature of CSF studies, recent efforts have focused on identification of potential biomarkers in peripheral fluids (e.g., saliva, blood).
Though there is considerable appeal in a validated blood test for AD pathology, most efforts to date have not been successful [15]. Recent developments, however, have shown significant promise, with high rates of overall classification accuracy [16]. If replicated and independently validated, the simplicity of a blood test would have significant clinical utility. Once integrated with the clinical history, a blood test with high sensitivity would make an excellent screening tool that could be used to quickly and efficiently rule out the presence of pathology or prompt for additional diagnostic testing.
1.4. Imaging biomarkers
Brain imaging is also a widely used biomarker, including both structural and functional imaging. For example, magnetic resonance imaging can be used to measure both regional (e.g., medial temporal structures in AD; frontal atrophy in frontotemporal dementia) and whole-brain atrophy, both of which can be used to inform differential diagnosis [17]. Several molecular imaging techniques have also been validated for detecting AD pathology (specifically β amyloid), including Pittsburgh compound B [18], and fluorine-18 labeled radiotracers such as florbetapir [19], [20], florbetaben, and flutemetamol [21]. Cerebral glucose metabolism has also been widely used (e.g., fluorodeoxyglucose positron emission tomography), both for identification of early AD-related changes [22] and differentiating them from other NDDs (e.g., frontotemporal dementia) [23], [24]. Tau imaging is increasingly used to identify the state of tau aggregation in the course of AD [25], which may be particularly beneficial very early in the disease process [26]. The noninvasive, or minimally invasive, nature of imaging studies makes them among the first biomarkers to be reviewed in the clinical setting. When integrated with additional diagnostic studies such as neuropsychological evaluation or fluid biomarkers, imaging studies can be particularly informative, both for ruling conditions in, and ruling conditions out.
1.5. Genomic data
Genetic information is commonly used to assist clinicians and researchers in guiding differential diagnosis and is also the central component of PM (e.g., pharmacogenomics). Genetic information is especially useful in identifying at-risk individuals long before symptoms emerge. For clinical trials and studies focusing on prevention of disease, this is especially important because it increases the likelihood of including those at highest risk of disease. Although there are specific genes associated with causing early onset AD (e.g., genes encoding amyloid precursor protein and presenilin 1 and 2) and increasing the risk of late onset AD (e.g., the e4 allele of the apolipoprotein E) [27], the development of AD most likely has a polygenic determination [28]. Similarly, several genetic markers have also been associated with idiopathic Parkinson's disease [29], dementia with Lewy bodies [30], [31], and frontotemporal dementia [32]. A common theme across diseases is that there is considerable heterogeneity in the possible genetic determinants, most are polygenic disorders, and there is growing evidence of genetic overlap between diseases [28], [31], [33].
1.6. Cognitive and behavioral markers
Detailed characterization of the clinical presentation, including both the cognitive and behavioral phenotypes are core components of a comprehensive diagnostic work up, whether in clinic or in research participation. At present, active assessments (i.e., an active and intentional approach to measuring cognitive functioning and quantification of behavior) such as cognitive screenings performed in clinic or detailed neuropsychological evaluation are often used both to inform differential diagnosis and to establish baselines of functioning that can be used to monitor disease progression over time. The major advantage of active assessments is the level of control retained by clinicians and researchers over what measures are used, when they are administered, and under what circumstances. Moreover, they can be used to construct a very rich snapshot of functioning that cuts across several domains of ability, and there is an extensive literature base for these measures.
The challenges with active assessments, however, are that not only are they labor intensive, slow, and inefficient [34], [35], but most are insensitive to change in the earliest phases of the disease [8], when intervention may be most effective. They are also grossly lacking in temporal resolution, limiting the conclusions that can be drawn at the time of assessment, forcing clinicians and researchers to either estimate previous levels of functioning or make inferences about the pattern of change between assessments (e.g., that decline follows a uniformly linear pattern). The ecological validity of most of these tools is also quite poor, requiring additional inferences about how observed patterns of performance translate to real-world functioning.
One solution is to make better use of technology in the measurement of human behavior via portable and wearable devices (e.g., phones, smart watches, and sensors), which would address many of the limitations of active assessments. Leveraging portable device platforms would foster the development of remote assessments that could reduce the demand on patients and research participants to come to clinic and provide higher frequency data. Portable platforms would also increase the opportunities to collect data and maintain closer contact with patients, which could detect significant changes sooner and allow intervention earlier if needed. In addition, portable technology would also allow for the development and validation of passive data collection (PDC) methods that could be used to automatically “capture” data from everyday behaviors (e.g., embedded background applications, sensors, and so on). These data would generate an incredibly rich characterization of everyday cognition and real-world behavior, and because they are tied directly to an individual through a specific device, the data would be nearly as unique as a genetic sequence.
When integrated with active assessments, these continuous or high-frequency data capture methods that would dramatically enrich the overall cognitive and behavioral profile, and many of the shortcomings associated with traditional active assessments would be mitigated. Although PDC methods would generate a substantial amount of “noise” along with the behavioral data, much of this could be automatically filtered out with concurrent development of purpose-built informatics applications. Moreover, they could be used much in the same ways as traditional biomarkers to detect the presence of underlying NDD. With proper validation and sufficient classification accuracy, continuous data capture also has incredibly high clinical utility and efficiency because they are by definition, computer applications for quantifying everyday behaviors. They do not place significant demands on the local healthcare system and require minimal clinician input once deployed.
1.7. What is BMIs?
BMIs has been defined by the American Medical Informatics Association as “the interdisciplinary field that studies and pursues the effective uses of biomedical data, information, and knowledge for scientific inquiry, problem solving, and decision-making, motivated by efforts to improve human health” [36]. A core emphasis of BMI is on development of technology-driven resources for the storage, access, use, and dissemination of health-care data to derive new information and generate new knowledge. There is currently considerable interest and ongoing work in comprehension, integration, and usability of diverse data sources in BMI, which aligns closely with NDD and PM. Several distinct, but closely related application areas have been defined by the American Medical Informatics Association. Two of which are particularly important in the study of NDD—translational bioinformatics and clinical research informatics.
Translational bioinformatics is defined as “the development of storage, analytic, and interpretive methods to optimize the transformation of increasingly voluminous biomedical and genomic data into proactive, predictive, preventive, and participatory health.” Given the increasing precision with which biomedical data are now being collected, the sheer amount of data that is generated and its complexity, extracting insights that can be used to inform clinical practice is a challenge requiring substantial computing power. However, BMI applications can be developed to detect and translate the deeply embedded patterns within these data into predictive treatments tailored to the individual, ultimately leading to improvement in patient outcomes.
Related to translational bioinformatics and somewhat of a counterpart, clinical research informatics is defined as “the use of informatics in the discovery and management of new knowledge relating to health and disease.” Although highly relevant for clinical trials, clinical research informatics also emphasizes the continued utilization of amassed data and secondary use and re-use of data, such as electronic health records and those data aggregated over the course of routine clinical care, for ongoing research purposes (e.g., patient registries, collaborative knowledge-bases). Even though these clinical data are even more complex relative to the highly controlled and well-characterized data amassed in a research setting, they serve as the foundation of the learning health-care system [37]. So, while translational informatics focuses on moving research evidence from bench-to-bedside to support evidence-based practice, clinical research informatics aims to generate complementary practice-based evidence.
1.8. Advancing translational neuroscience research with BMIs
Over the past several decades, it has become increasingly apparent that NDD cannot be reduced to a single determinant and attention has shifted to integration of multidisciplinary data (e.g., biomarker data, imaging data, genetic data, and cognitive and behavioral phenotypes). The major challenge, however, is that these “multi-omic” assessments generate a set of incredibly complex “big” data. When studied at the level of the individual patient, these data can be manually integrated relatively easily, but elucidating patterns within even modest sample sizes quickly exceed what is feasible with manually guided approaches. Efficiently and effectively separating the signal from the noise in such data sets requires dedicated computing power with purpose-built BMIs applications to analyze the data and derive novel insights, which is especially critical for PM.
1.9. Mapping functional networks
NDD is not typified by focal lesions, but rather disruption of complex neuroanatomical networks [38], [39], evident even in early disease stages [40]. Computational network analysis is a mathematical modeling approach capable of integrating diverse data sets and mapping distributed networks by identifying shared elements (i.e., nodes) and establishing the relationships between them (i.e., edges). It has been effectively utilized to map both structural and functional connections in the brain (e.g., [41]) and uncover common genetic pathways in AD [42]. With sufficient longitudinal data, the evolution of disease over time can also be modeled [43]. The ability to model and predict disease course is an essential component of PM.
Computational network analysis has also been effectively used to study diseases that may be best characterized as continua instead of distinct categorical states and for syndromes with heterogeneous clinical presentations [44], [45]; both of these are characteristic of NDD. The ability of network analyses to integrate highly diverse and complex data sets is a tremendous strength. As data of increasing granularity become widely available, it may soon be possible to uncover precise causal relationships driving NDD, which may lead to the discovery of an effective therapeutic agent.
1.10. Clinical decision support tools
Clinical decisions support (CDS) tools are applications built to deliver filtered information to providers at the point of care to enhance health and health care [46]. In essence, a CDS tool is an application that filters and translates empirical evidence into an immediately usable format to guide clinicians in their clinical decision-making. By integrating patient-specific parameters (e.g., genetics, biomarker profiles), CDS applications can be used to drive PM interventions. CDS applications are not a replacement for clinical judgment, but rather an additional source of information to be integrated with clinical judgment in an effort to formulate personalized treatment recommendations; CDS applications are a means by which evidence-based practice can be realized.
In AD and related NDD, determining specific treatments is not yet an issue because there is not an effective therapeutic agent to slow or reverse the underlying pathologic process; however, there is a considerable need to develop tools capable of empirically deriving individualized risk profiles. Formulating specific diagnoses based on a set of easily observed/assessed clinical characteristics (e.g., biomarkers, behavioral phenotypes) and subsequently developing predictive algorithms of the patient's disease course will be particularly important. In clinical trials, where recruitment is focusing on very early phases of the disease process, it is critical to study those likely to manifest the disease of interest. Being able to accurately identify those with the highest risk based on individual parameters will increase recruitment accuracy, and therefore, the odds of discovering an effective therapeutic agent.
Machine learning (ML) is a data-driven approach to generating such predictive algorithms using a set of training data that are then validated in independent data sets [47], [48]. In NDD, ML could be effectively used to develop statistical models to establish individual risk profiles and make specific diagnoses based on a set of individual clinical characteristics. In supervised learning approaches, researchers identify a specific endpoint or outcome of interest from a set of data and allow the program to find and evaluate the underlying patterns in the remaining data that predict those endpoints. For example, to develop an algorithm for determining in vivo amyloid status (i.e., the outcome) based on set of easily observed clinical characteristics (i.e., the predictors), an initial set of training data would be required that includes amyloid status (e.g., florbetapir scan) and potential clinical characteristics of interest (e.g., volumetric neuroimaging, cognitive data). The patterns of performance among the predictors that optimize accurate classification of the output would then be used to generate an algorithm that could be applied to a novel data set containing the same predictors. When provided with performance feedback (i.e., was the classification correct or incorrect?), the algorithms are iteratively refined to reduce error rates, effectively “learning” from experience. While similar classification models can be built using classical statistical methods (e.g., logistic regression), hypothesis-driven approaches require researchers to select predictors of interest based on an a priori assumption; in complex data sets, this can be particularly challenging and may not generate the most efficient models.
Even in supervised learning, there is still a need to identify and label a specific output. In highly complex data sets, however, there are likely valuable insights embedded within the data that may be missed using a supervised approach. In unsupervised learning approaches, researchers allow the program to explore the underlying structure of the entire data set without any predetermined endpoint labels; unsupervised learning is especially useful for mining extremely large and complex data sets, dimension reduction, and uncovering patterns of data not readily apparent to researchers through classical hypothesis testing [48], [49]. Both supervised and unsupervised learning methods are being used to extract valuable insights and develop predictive algorithms from both data and metadata. When appropriately implemented, classification and predictions derived from hierarchical, or deep, learning methods have demonstrated impressive accuracy comparable with or surpassing human-level accuracy [50], [51], [52] and are approaching Bayes error rates.
Historically focused on imaging data, systematically capturing cognitive and behavioral data alongside additional biological data sources (e.g., structural and functional neuroimaging, genetic data, CSF) and subjecting these data to ML as well may lead to identification of precision biomarkers associated with specific cognitive and behavioral phenotypes. The resulting models could be used to generate an empirically derived probability of underlying pathology based solely on the clinical presentation, which has significant utility for both clinical trial recruitment and clinical management. For example, of data of this type might be used to predict evolution of cognitively normal individuals into amyloid-positive states, and thus eligibility for clinical trial participation and eventually for preventative treatment. If integrated into an electronic medical record along with natural language processing application to extract information from clinical notes (e.g., health record phenotyping), classification models could be deployed autonomously, and probabilities updated in real time as new notes are generated. Already validated for use in screening for common conditions (e.g., type 2 diabetes) [53] and with fully automated approaches rivaling manual methods in accuracy [54], extending these applications to NDD to generate CDS tools to guide treatment selection, especially if compounds or therapeutics are developed that have proven efficacy for specific subsets of the disease population, would facilitate the practice of PM.
1.11. Transition from state of wellness to disease
One of the major challenges facing NDD is the blurred boundary between a state of health and one of disease, which in part is due to the difficulty detecting the subtle changes associated with the transition, as well as clear characterization of the earliest disease states. Using a complex network approach, however, it may be possible to identify points of vulnerability (i.e., tipping points) associated with a development of a clinical syndrome following a period of stability (e.g., Hofmann et al, 2016) [55]. In NDD especially, where individual difference factors (e.g., cognitive reserve) can influence the relationship between manifest symptoms and disease burden, being able to identify factors associated with, or even preceding, these pathologic transitions may increase opportunities for intervention. Such approaches have already been used with neuroimaging data [56], [57]. High-throughput data streams and user-generated data sources (e.g., wearables) providing behavioral data in real-world settings could then be integrated with clinical data sources (e.g. electronic medical records). ML-derived algorithms could then be utilized to automatically monitor individual data streams and flag patterns of behavioral that suggest an emerging transition from healthy to disease states well before the clinical syndrome emerges.
1.12. Modern database infrastructure
In general, development of CDS tools, whether it be for prediction of disease or for identification of a specific therapeutic agent, has required prohibitively large sample sizes to achieve sufficient stability and reliability. This is especially the case for identifying the very subtle signals in the earliest phases of disease. In the absence of distributed collaborative networks contributing to a shared data repository, it has been difficult to aggregate samples of this size. Furthermore, when sufficiently powered studies were completed, the data were collected over a predetermined period of time, using prespecified tools, in a restricted sample, to address an a priori hypothesis, with highly limited access beyond the original study investigators. On study completion, these data were then typically siloed and ended up sitting idle.
Moving these data sets, however, to cloud-based repositories promotes real-time, open-access data sharing and collaboration, a movement which has been endorsed by federal funding agencies and journal editors [58], [59], [60]. Creating a repository to house data collected during the course of routine clinical care alongside data from ongoing molecular and genetic research and clinical trials that are collaboratively aggregated by multiple users would foster the development of novel measurement tools, establishment of clinical normative data, and facilitation of knowledge discovery via multidisciplinary hypothesis-driven, as well as, data-driven research (e.g., deep learning, data mining, and ML). Providing open access to data will also help address issues of transparency in research, reproducibility of results, and independent replication, especially when paired with simultaneous sharing of software and code [61], [62], [63].
Although the costs associated with cloud-based storage were at one point prohibitive for all but the most well-funded studies or larger enterprise organizations, publicly available services such as Amazon's Web Services have brought leading edge technology within reach for most organizations. Ultimately, the utility of research and clinical data are inherently tied to the accessibility of the data, both within studies and across studies and utilization of modern data management can dramatically increase accessibility. Making these resources open-access and built using flexible platforms furthers accessibility and promotes longevity, allowing perpetual use and re-use of the data and integration of new data, maximizing return on financial investments.
2. Conclusion
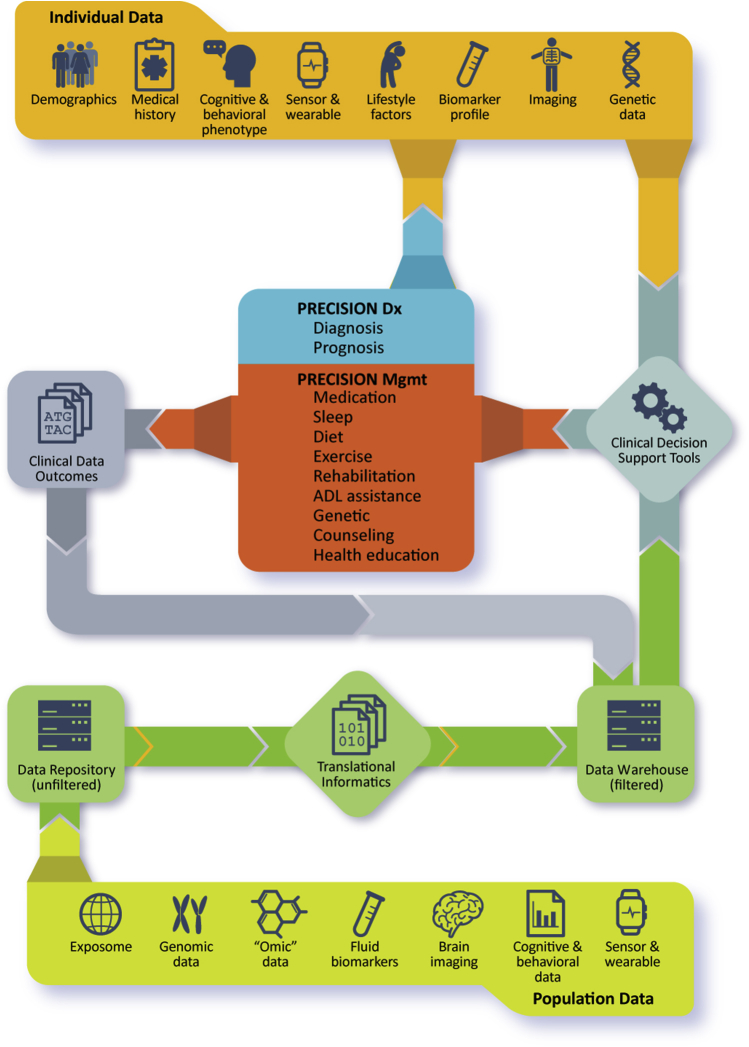
As the era of personalized medicine continues to evolve and rich individual data sets become increasingly accessible, development of cloud-based repositories that collaboratively aggregate data will rapidly advance basic, translational, and clinical science. Creating dedicated BMIs applications with advanced analytic capability designed identify subtle patterns within those data will bring targeted interventions for NDD based on individually determined risk profiles within reach. Fig. 1 presents a high-level overview, tying these concepts together to show how data from population studies can be aggregated and used to develop clinical decision support tools, which can then be deployed in clinical settings, using individual patient data to guide clinical decision-making. The clinical outcomes resulting from these decisions are then returned to the data set, which are used to refine and improve treatment models and decision support tools, completing the learning health environment. Ultimately, developing comprehensive and integrated data environments will lead to improved patient outcomes and facilitate knowledge discovery, including disease mechanisms, causal factors, and novel therapeutics.
Research in Context.
-
1.
Systematic review: Modern medicine is in the midst of a revolution driven by “big data,” rapidly advancing computing power, and broader integration of technology into healthcare. Highly detailed and individualized profiles of both health and disease states are now possible, including biomarkers, genomic profiles, cognitive and behavioral phenotypes, high-frequency assessments, and medical imaging.
-
2.
Interpretation: These individual profiles can be used to understand multi-determinant causal relationships, elucidate modifiable factors, and ultimately customize treatments based on individual parameters, which is especially important for neurodegenerative diseases, where an effective therapeutic agent has yet to be discovered.
-
3.
Future directions: By collaboratively aggregating these data in structured repositories, biomedical informatics can be used to develop clinical applications and decision support tools to derive new knowledge from the wealth of available data to advance clinical care and scientific research of neurodegenerative disease in the era of precision medicine.
Fig. 1.
Precision diagnosis and management of neurodegenerative disease relies on integration of population-level data and individual data derived from multiple sources. Effective utilization will require informatics applications to aggregate and filter data, which can drive development of clinical decision support tools for deployment in clinical settings. Developing tools to systematically capture clinical outcomes at the point of service delivery will facilitate refinement of the clinical decision support tools, creating a learning healthcare system and closing the loop in evidence-based care.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (Grant: P20GM109025) and Keep Memory Alive, Las Vegas, NV. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Keep Memory Alive.
References
- 1.U.S. Bureau of Labor Statistics . 2016. Medical Records and Health Information Technicians Job Outlook.https://www.bls.gov/ooh/healthcare/medical-records-and-health-information-technicians.htm#tab-6 Available at: [Google Scholar]
- 2.Genetics Home Reference . 2017. What is Precision Medicine? Help Me Understand Genet Precis Med.https://ghr.nlm.nih.gov/primer/precisionmedicine/definition Available at: [Google Scholar]
- 3.Tan L., Jiang T., Tan L., Yu J.-T. Toward precision medicine in neurological diseases. Ann Transl Med. 2016;4:104. doi: 10.21037/atm.2016.03.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy J., Orr H. The genetics of neurodegenerative diseases. J Neurochem. 2006;97:1690–1699. doi: 10.1111/j.1471-4159.2006.03979.x. [DOI] [PubMed] [Google Scholar]
- 5.Tsuji S. Genetics of neurodegenerative diseases: Insights from high-throughput resequencing. Hum Mol Genet. 2010;19:R65–R70. doi: 10.1093/hmg/ddq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeromin A., Bowser R. Biomarkers in neurodegenerative diseases. Adv Neurobiol. 2017;15:491–528. doi: 10.1007/978-3-319-57193-5_20. [DOI] [PubMed] [Google Scholar]
- 7.Cummings J.L., Morstorf T., Zhong K. Alzheimer's disease drug-development pipeline: Few candidates, frequent failures. Alzheimers Res Ther. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lista S., Molinuevo J.L., Cavedo E., Rami L., Amouyel P., Teipel S.J. Evolving evidence for the value of neuroimaging methods and biological markers in subjects categorized with subjective cognitive decline. J Alzheimers Dis. 2015;48:S171–S191. doi: 10.3233/JAD-150202. [DOI] [PubMed] [Google Scholar]
- 9.Galasko D., Chang L., Motter R., Clark C.M., Kaye J., Knopman D. High cerebrospinal fluid tau and low amyloid beta42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch Neurol. 1998;55:937–945. doi: 10.1001/archneur.55.7.937. [DOI] [PubMed] [Google Scholar]
- 10.Blennow K., Hampel H. CSF markers for incipient Alzheimer's disease. Lancet Neurol. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- 11.Mattsson N., Zetterberg H. Alzheimer's disease and CSF biomarkers: Key challenges for broad clinical applications. Biomark Med. 2009;3:735–737. doi: 10.2217/bmm.09.65. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie C., Smailagic N., Noel-Storr A.H., Ukoumunne O., Ladds E.C., Martin S. In: Ritchie C., editor. Vol. 3. John Wiley & Sons, Ltd; Chichester, UK: 2017. p. CD010803. (CSF tau and the CSF tau/ABeta Ratio for the Diagnosis of Alzheimer's Disease Dementia and Other Dementias in People with Mild Cognitive Impairment (MCI)). Cochrane Database Syst Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blennow K. CSF biomarkers for mild cognitive impairment. J Intern Med. 2004;256:224–234. doi: 10.1111/j.1365-2796.2004.01368.x. [DOI] [PubMed] [Google Scholar]
- 14.Koyama A., Okereke O.I., Yang T., Blacker D., Selkoe D.J., Grodstein F. Plasma amyloid-β as a predictor of dementia and cognitive decline. Arch Neurol. 2012;69:824–831. doi: 10.1001/archneurol.2011.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toledo J.B., Shaw L.M., Trojanowski J.Q. Plasma amyloid beta measurements - a desired but elusive Alzheimer's disease biomarker. Alzheimers Res Ther. 2013;5:8. doi: 10.1186/alzrt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ovod V., Ramsey K.N., Mawuenyega K.G., Bollinger J.G., Hicks T., Schneider T. Amyloid β concentrations and stable isotope labeling kinetics of human plasma specific to central nervous system amyloidosis. Alzheimers Dement. 2017;13:841–849. doi: 10.1016/j.jalz.2017.06.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisoni G.B., Fox N.C., Jack C.R., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klunk W.E., Engler H., Nordberg A., Wang Y., Blomqvist G., Holt D.P. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 19.Villemagne V.L., Ong K., Mulligan R.S., Holl G., Pejoska S., Jones G. Amyloid imaging with 18F-Florbetaben in Alzheimer disease and other dementias. J Nucl Med. 2011;52:1210–1217. doi: 10.2967/jnumed.111.089730. [DOI] [PubMed] [Google Scholar]
- 20.Wong D.F., Rosenberg P.B., Zhou Y., Kumar A., Raymont V., Ravert H.T. In Vivo imaging of amyloid deposition in Alzheimer disease using the Radioligand 18F-AV-45 (Flobetapir F 18) J Nucl Med. 2010;51:913–920. doi: 10.2967/jnumed.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinne J.O., Wong D.F., Wolk D.A., Leinonen V., Arnold S.E., Buckley C. [18F]Flutemetamol PET imaging and cortical biopsy histopathology for fibrillar amyloid β detection in living subjects with normal pressure hydrocephalus: pooled analysis of four studies. Acta Neuropathol. 2012;124:833–845. doi: 10.1007/s00401-012-1051-z. [DOI] [PubMed] [Google Scholar]
- 22.Bloudek L.M., Spackman D.E., Blankenburg M., Sullivan S.D. Review and meta-analysis of biomarkers and diagnostic imaging in Alzheimer's disease. J Alzheimers Dis. 2011;26:627–645. doi: 10.3233/JAD-2011-110458. [DOI] [PubMed] [Google Scholar]
- 23.Marcus C., Mena E., Subramaniam R.M. Brain PET in the diagnosis of Alzheimer's disease. Clin Nucl Med. 2014;39 doi: 10.1097/RLU.0000000000000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster N.L., Heidebrink J.L., Clark C.M., Jagust W.J., Arnold S.E., Barbas N.R. FDG-PET improves accuracy in distinguishing frontotemporal dementia and Alzheimer's disease. Brain. 2007;130:2616–2635. doi: 10.1093/brain/awm177. [DOI] [PubMed] [Google Scholar]
- 25.Johnson K.A., Schultz A., Betensky R.A., Becker J.A., Sepulcre J., Rentz D. Tau positron emission tomographic imaging in aging and early Alzheimer disease. Ann Neurol. 2016;79:110–119. doi: 10.1002/ana.24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckley R.F., Hanseeuw B., Schultz A.P., Vannini P., Aghjayan S.L., Properzi M.J. Region-specific association of subjective cognitive decline with tauopathy independent of global β-amyloid burden. JAMA Neurol. 2017;74:1455. doi: 10.1001/jamaneurol.2017.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso Vilatela M.E., López-López M., Yescas-Gómez P. Genetics of Alzheimer's disease. Arch Med Res. 2012;43:622–631. doi: 10.1016/j.arcmed.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Desikan R.S., Fan C.C., Wang Y., Schork A.J., Cabral H.J., Cupples L.A. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017;14:e1002258. doi: 10.1371/journal.pmed.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein C., Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerreiro R., Ross O.A., Kun-Rodrigues C., Hernandez D.G., Orme T., Eicher J.D. Investigating the genetic architecture of dementia with Lewy bodies: a two-stage genome-wide association study. Lancet Neurol. 2018;17:64–74. doi: 10.1016/S1474-4422(17)30400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerreiro R., Escott-Price V., Darwent L., Parkkinen L., Ansorge O., Hernandez D.G. Genome-wide analysis of genetic correlation in dementia with Lewy bodies, Parkinson's and Alzheimer's diseases. Neurobiol Aging. 2016;38:214. doi: 10.1016/j.neurobiolaging.2015.10.028. e7–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blauwendraat C., Wilke C., Simón-Sánchez J., Jansen I.E., Reifschneider A., Capell A. The wide genetic landscape of clinical frontotemporal dementia: systematic combined sequencing of 121 consecutive subjects. Genet Med. 2018;20:240–249. doi: 10.1038/gim.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari R., Wang Y., Vandrovcova J., Guelfi S., Witeolar A., Karch C.M. Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer's and Parkinson's diseases. J Neurol Neurosurg Psychiatr. 2017;88:152–164. doi: 10.1136/jnnp-2016-314411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins F.S., Riley W.T. NIHs transformative opportunities for the behavioral and social sciences. Sci Transl Med. 2016;8:366ed14. doi: 10.1126/scitranslmed.aai9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J.B., Barr W.B. The technology crisis in neuropsychology. Arch Clin Neuropsychol. 2017;32:541–554. doi: 10.1093/arclin/acx050. [DOI] [PubMed] [Google Scholar]
- 36.Kulikowski C.A., Shortliffe E.H., Currie L.M., Elkin P.L., Hunter L.E., Johnson T.R. AMIA Board white paper: definition of biomedical informatics and specification of core competencies for graduate education in the discipline. J Am Med Inform Assoc. 2012;19:931–938. doi: 10.1136/amiajnl-2012-001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medicine I of M (US) R on E-B. Olsen L., Aisner D., McGinnis J.M. National Academies Press; US: 2007. The Learning Healthcare System. [PubMed] [Google Scholar]
- 38.Dennis E.L., Thompson P.M. Functional brain connectivity using fMRI in aging and Alzheimer's disease. Neuropsychol Rev. 2014;24:49–62. doi: 10.1007/s11065-014-9249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daianu M., Jahanshad N., Nir T.M., Toga A.W., Jack C.R., Weiner M.W. Breakdown of brain connectivity between normal aging and Alzheimer's disease: A structural k -core network analysis. Brain Connect. 2013;3:407–422. doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minati L., Chan D., Mastropasqua C., Serra L., Spanò B., Marra C. Widespread alterations in functional brain network architecture in amnestic mild cognitive impairment. J Alzheimers Dis. 2014;40:213–220. doi: 10.3233/JAD-131766. [DOI] [PubMed] [Google Scholar]
- 41.Rubinov M., Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Talwar P., Silla Y., Grover S., Gupta M., Agarwal R., Kushwaha S. Genomic convergence and network analysis approach to identify candidate genes in Alzheimer's disease. BMC Genomics. 2014;15:199. doi: 10.1186/1471-2164-15-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oxtoby N.P., Garbarino S., Firth N.C., Warren J.D., Schott J.M., Alexander D.C. Data-driven sequence of changes to anatomical brain connectivity in sporadic Alzheimer's disease. Front Neurol. 2017;8:580. doi: 10.3389/fneur.2017.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borsboom D., Rhemtulla M., Cramer A.O.J., van der Maas H.L.J., Scheffer M., Dolan C.V. Kinds versus continua: a review of psychometric approaches to uncover the structure of psychiatric constructs. Psychol Med. 2016;46:1–13. doi: 10.1017/S0033291715001944. [DOI] [PubMed] [Google Scholar]
- 45.Borsboom D., Cramer A.O. Network analysis: an integrative approach to the structure of psychopathology. Annu Rev Clin Psychol. 2013;9:91–121. doi: 10.1146/annurev-clinpsy-050212-185608. [DOI] [PubMed] [Google Scholar]
- 46.HealthIT.gov . 2013. What is Clinical Decision Support (CDS)? Policymaking, Regul Strateg.https://www.healthit.gov/policy-researchers-implementers/clinical-decision-support-cds Available at: [Google Scholar]
- 47.Hastie T, Tibshirani R, Friedman JH(Jerome H. The elements of statistical learning: data mining, inference, and prediction. 2009. doi:10.1007/b94608.
- 48.Jordan M.I., Mitchell T.M. Machine learning: Trends, perspectives, and prospects. Science. 2015;349:255–260. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- 49.Altman R. Artificial intelligence (AI) systems for interpreting complex medical datasets. Clin Pharmacol Ther. 2017;101:585–586. doi: 10.1002/cpt.650. [DOI] [PubMed] [Google Scholar]
- 50.Plis S.M., Hjelm D.R., Slakhutdinov R., Allen E.A., Bockholt H.J., Long J.D. Deep learning for neuroimaging: A validation study. Front Neurosci. 2014;8:229. doi: 10.3389/fnins.2014.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vieira S., Pinaya W.H.L., Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neurosci Biobehav Rev. 2017;74:58–75. doi: 10.1016/j.neubiorev.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Akkus Z., Galimzianova A., Hoogi A., Rubin D.L., Erickson B.J. Deep learning for brain MRI segmentation: State of the art and future directions. J Digit Imaging. 2017;30:449–459. doi: 10.1007/s10278-017-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson A.E., Kerr W.T., Thames A., Li T., Xiao J., Cohen M.S. Electronic health record phenotyping improves detection and screening of type 2 diabetes in the general United States population: A cross-sectional, unselected, retrospective study. J Biomed Inform. 2016;60:162–168. doi: 10.1016/j.jbi.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu S., Ma Y., Gronsbell J., Cai T., Ananthakrishnan A.N., Gainer V.S. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc. 2018;25:54–60. doi: 10.1093/jamia/ocx111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofmann S.G., Curtiss J., Mcnally R.J. A complex network perspective on Clinical Science. Perspect Psychol Sci. 2016;11:597–605. doi: 10.1177/1745691616639283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beheshti I., Demirel H., Matsuda H., Alzheimer's Disease Neuroimaging Initiative Classification of Alzheimer’s disease and prediction of mild cognitive impairment-to-Alzheimer's conversion from structural magnetic resource imaging using feature ranking and a genetic algorithm. Comput Biol Med. 2017;83:109–119. doi: 10.1016/j.compbiomed.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 57.Dallora A.L., Eivazzadeh S., Mendes E., Berglund J., Anderberg P. Machine learning and microsimulation techniques on the prognosis of dementia: A systematic literature review. PLoS One. 2017;12:e0179804. doi: 10.1371/journal.pone.0179804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taichman D.B., Sahni P., Pinborg A., Peiperl L., Laine C., James A. Data sharing statements for clinical trials. JAMA. 2017;317:2491. doi: 10.1001/jama.2017.6514. [DOI] [PubMed] [Google Scholar]
- 59.Taichman D.B., Backus J., Baethge C., Bauchner H., de Leeuw P.W., Drazen J.M. Sharing Clinical Trial Data — A proposal from the International Committee of Medical Journal Editors. N Engl J Med. 2016;374:384–386. doi: 10.1056/NEJMe1515172. [DOI] [PubMed] [Google Scholar]
- 60.Announcement: Where are the data? Nature. 2016;537:138. doi: 10.1038/537138a. [DOI] [PubMed] [Google Scholar]
- 61.Johnson A.E., Stone D.J., Celi L.A., Pollard T.J. The MIMIC Code Repository: Enabling reproducibility in critical care research. J Am Med Inform Assoc. 2018;25:32–39. doi: 10.1093/jamia/ocx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doel T., Shakir D.I., Pratt R., Aertsen M., Moggridge J., Bellon E. GIFT-Cloud: A data sharing and collaboration platform for medical imaging research. Comput Methods Programs Biomed. 2017;139:181–190. doi: 10.1016/j.cmpb.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gewin V. Data sharing: An open mind on open data. Nature. 2016;529:117–119. doi: 10.1038/nj7584-117a. [DOI] [PubMed] [Google Scholar]