Abstract
Background & objectives:
Clinical outcome after hepatitis B virus (HBV) exposure varies extremely from spontaneous clearance to chronic hepatitis B and often progresses to liver cirrhosis (LC) and hepatocellular carcinoma (HCC). Host genetic factor plays an important role in the regulation of immune response. This study was aimed to investigate whether HLA class II DQA1 and DQB1 gene polymorphism were associated with chronic hepatitis B infection and in the development of HBV-related LC and HCC.
Methods:
DQA1 and DQB1 allele polymorphism were studied in 187 patients with HBV-related liver diseases (which included 73 chronic hepatitis B, 84 LC and 30 HCC patients) and 109 controls who had spontaneously recovered from HBV infection using polymerase chain reaction amplification with sequence-specific primers.
Results:
Our data suggested that DQA1*0101/2/4 [odds ratio (OR)=2.78; Pc=0.003], DQA1*0103 (OR=2.64; Pc=0.0007) and DQB1*0302/3 (OR=2.15; Pc=0.01) were associated with the protection from chronic HBV infection, whereas DQB1*0402 (OR=0.25; Pc=0.001) showed susceptible effect on chronic HBV infection. DQB1*0601 (OR=3.73; Pc=0.006) conferred protective effect from developing LC; similarly, DQB1*0302/3 (OR=5.53; Pc=0.05) and DQB1*0402 (OR=0.00; Pc=0.001) conferred protective effect from developing HCC. However, DQA1*0601 and DQB1*0503 showed susceptible effect on chronic HBV infection; these associations were no longer significant after Bonferroni correction.
Interpretation & conclusions:
Our results revealed HLA-DQA1*0101/2/4 - DQA1*0103 - DQB1*0302/3 and DQB1*0601 as protective and DQB1*0402 as risk alleles. The study suggests that various subtypes of HLA-DQA1 and DQB1 are associated with both HBV clearance and development of chronic HBV infections.
Keywords: Chronic hepatitis B, hepatitis B virus, hepatocellular carcinoma, human leucocyte antigen, liver cirrhosis, polymorphism
Hepatitis B virus (HBV) is one of the most common causes of liver diseases including cirrhosis and hepatocellular carcinoma (HCC)1. During the lifetime, despite the fact that only 2-10 per cent of HBV-infected individuals develop chronic complications of liver disease, the clinical outcomes differ with 15-40 per cent higher risk of developing liver cirrhosis (LC) and HCC2. Primary HBV infection in the host may be either symptomatic or asymptomatic, where young children are mostly asymptomatic and adults self-limited, with viral clearance and development of lasting immunity to reinfection3. Some primary HBV infections in adults (<5%) do not resolve and develop into persistent infections3. Among the chronic hepatitis B, 20 per cent develop LC (a condition where regenerative nodules and fibrosis coexist with severe liver injury) and less than five per cent develop HCC with long-term disease progression. However, the factors determining the chronicity of primary HBV infection and its disease progression to LC and HCC are not clearly understood.
It has been shown that outcome of the HBV infection is not because of the variation in the virulence of viral strains4, it may rather be because of host-immune response5. Furthermore, a study conducted in HBV infected twins in China showed a strong genetic association with the outcome of HBV infection and indicated that host genetic factors might play a role in influencing the clinical phenotype6. First-degree relatives of HBV-related HCC patients were supposed to be at increased risk of HCC7. Thus, there was a probability that immunogenetic factors might influence disease severity and different clinical outcomes.
Several studies have shown association of HLA class II genes with the outcome of HBV infections8,9,10,11,12. However, mostly, the results were inconsistent due to disparities in the study design, sample size and ethnicity. Many researchers have investigated the association of HLA genes with HBV-related HCC13,14,15,16,17. There is a paucity of Indian HLA studies in these patients. Therefore, the current study was undertaken to investigate the HLA allele polymorphism associated with HBV infection and disease progression to LC and HCC. The study was designed to characterize HLA-DQA1 and DQB1 gene polymorphism in chronic HBV infection and their possible association with the development of HBV-related LC and HCC in Indian patients.
Material & Methods
Patients were selected from the outpatient department of Medicine, Lok Nayak Jai Prakash Narayan Hospital, New Delhi, India, from February 2010 to February 2013. The sample size was calculated by using the online sample size calculator, Raosoft (http://www.raosoft.com/samplesize.html). A total of 296 HBV cases were genotyped and analyzed in this study. The patients were categorized into two groups, 187 HBV-related liver disease (Group I) with persistent hepatitis B surface antigen (HBsAg) positivity for more than six months including 73 chronic hepatitis B [male/female = 53/20; mean age = 29.0±13.3 yr; hepatitis B e antigen (HBeAg) positive 39], 84 LC (male/female = 69/15; mean age = 40.8±14.6 yr; HBeAg positive 57) and 30 HCC (male/female = 27/3; mean age = 55.2±11.7 yr; HBeAg positive 11) and 109 individuals who have spontaneously recovered (male/female = 82/27; mean age = 29.8±11.7 yr) from HBV infection serve as controls (Group II) (Table I). Informed written consent was taken from all the patients before enrolling in the study.
Table I.
Baseline clinical, demographical and laboratory characteristics of hepatitis B virus-infected patients
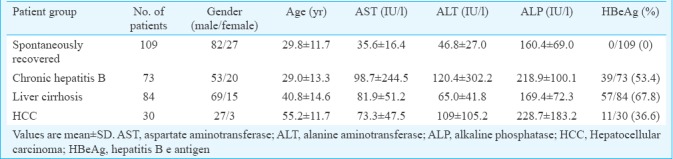
The following demographic and clinical parameters were recorded at the time of peripheral blood collection: gender, age, serum aspartate aminotransferase (AST) levels, alanine aminotransferase (ALT) levels, alkaline phosphatase levels and serological HBV markers. Individuals who were found to be positive for HBsAg for more than six months, with persistent or intermittent elevation in ALT/AST levels, HBV DNA viral load >105 copies/ml and liver biopsy showing chronic hepatitis (necroinflammatory score ≥4) were diagnosed as chronic hepatitis B as per the American Association for the Study of Liver Diseases (AASLD) guidelines18, while the spontaneously recovered group was identified as individuals positive for anti-HBs and anti-HBc antibody and negative for HBsAg and HBeAg. LC was diagnosed on the basis of showing the presence of oesophagal varices by endoscopic examination, and clinical and biochemical/radiological evidence showing small and nodular liver in advanced cirrhosis along with increased echogenicity and irregular appearing areas. HCC was diagnosed as per the European Association for the Study of the Liver (EASL) diagnostic criteria19 based on either fine needle aspiration cytology or by arterialization on colour Doppler, magnetic resonance imaging and alpha-foetoprotein >400 ng/ml. The study was approved by the Institutional Ethical Committee of Maulana Azad Medical College, New Delhi.
Serological testing: Blood samples (5 ml) were collected from all the participants and serum separated and stored at 20°C. Enzyme-linked immunosorbent assay was used to detect serum HBsAg, anti-HBc, anti-HBs and HBeAg according to the manufacturer's instruction (Radim Diagnostics, Italy).
DNA extraction: Genomic DNA was extracted from peripheral blood cells using QIAamp Blood Kit (Qiagen, Germantown, USA). All DNA samples were diluted to 100 ng/μl and stored at 4°C in Tris-EDTA buffer before use.
HLA-DQA1 and DQB1 allele polymorphism: HLA-DQA1 and DQB1 polymorphisms were detected by the sequence-specific primer (SSP)-PCR designed by Olerup et al20 and synthesized by Sigma-Aldrich, USA. In each PCR, an internal positive control was included that amplified human growth hormone gene. The 5’-primer (5’TGC CAA GTG GAG CAC CCA A3’) and 3’-primer (5’GCA TCT TGC TCT GTG CAG AT3’) gave rise to 796 base pair (bp) fragment. A total amount of 25 μl PCR reaction mixture was prepared which contained 1 μl of genomic DNA (100 ng/μl), 2.5 μl of 10× reaction buffer (1% Triton X-100, 500 mm KCl, 100 mm Tris-HCl, pH 9.0), 2 μl of MgCl2(25 mmol/l), 0.25 μl of dNTP (10 mmol/l), 1 μl of each SSP (5 μmol/l), 0.2 μl of the internal positive control each (5 μmol/l), 0.1 μl of Taq polymerase (5 U/μl) and 16.75 μl of molecular grade water. PCR amplification was carried out in Biometra T-Gradient Thermal Cycler (Goettingen Germany). The PCR cycling conditions for HLA-DQA1 and DQB1 alleles were as follows: predenaturation at 94°C for 4 min, denaturation at 94°C for 60 sec, annealing at 65°C for 60 sec, extension at 72°C for 60 sec, repetition for 35 cycles and final extension at 72°C for five minutes. PCR products were electrophoresed in 2 per cent agarose gels stained with ethidium bromide. The gels were examined under ultraviolet transillumination (Vilber Lourmat Gel documentation system) and documented. PCR results were interpreted based on the presence or absence of specific amplified product with specific bp size and with the presence of internal amplification control.
Statistical analysis: HLA-DQA1 and DQB1 alleles were calculated by direct count. The phenotypic frequencies of the alleles were compared between HBV carriers and spontaneously recovered groups using Chi-square test or Fisher's exact test when required. A large number of alleles were tested, so there may be a chance of significant association when using multiple comparisons. Those P values were corrected by Bonferroni correction (Pc=P×the number of alleles observed)21. Odds ratios (OR) with 95 per cent confidence intervals were calculated.
Results
Comparison of frequency of HLA-DQA1 and DQB1 allele between groups I and II: HLA-DQA1*0101/2/4 showed a significant difference between spontaneously recovered and persistent HBV infection. The frequency of individuals carrying the DQA1*0101/2/4 allele was 66.0 per cent among spontaneously recovered, whereas it was only 41.1 per cent among persistent infections (χ2=17.05, P=0.00003, Pc=0.003, OR=2.78) suggesting that this allele was protective against HBV infection. Similarly, DQA1*0103 was significantly increased in spontaneously recovered individuals compared to those with persistent infections (59.6 vs. 35.8%, χ2=15.79, P=0.00007, Pc=0.0007, OR=2.64) indicating protective effect of this allele against HBV infection. DQA1*0104 and DQA1*0601 showed significant difference between spontaneously recovered and persistent infection (71.5 vs. 57.7%, χ2=5.62, P=0.017, Pc=0.18, OR=1.84 and 18.3 vs. 9.6%, χ2=4.68, P=0.030, Pc=0.33, OR=2.11, respectively), showing some protective effects against HBV infection. However, after Bonferroni correction, this association was no longer found to be significant (Table II).
Table II.
Frequency of HLA-DQA1 alleles in hepatitis B virus infections and control group
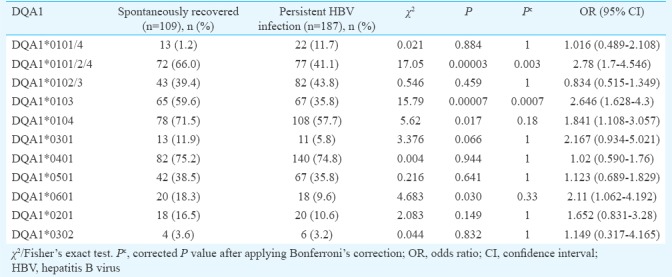
Phenotypic frequency of HLA-DQB1 alleles is shown in Table III. In spontaneously recovered individuals, the frequencies of DQB1*0302/3 were significantly increased compared with the persistent infection (56.8 vs 37.9%, χ2=9.95 P=0.001, Pc=0.013, OR=2.15). Conversely, DQB1*0402 was significantly increased in persistent HBV infection compared with spontaneously recovered (27.8 vs 9.1%, χ2=15.1, P=0.0001, Pc=0.001, OR=0.25). This difference was significant after Bonferroni correction also, which suggested a susceptible effect on HBV infection. DQB1*0501, DQB1*0602 and DQB1*0604 showed significant difference between spontaneously recovered and persistent infection (3.6 vs. 0.5%, χ2=4.07, P=0.04, Pc=0.55, OR=7.08; 8.2 vs. 1.6%, χ2=7.83, P=0.005, Pc=0.06, OR=5.52 and 7.3 vs. 2.1%, χ2=4.78, P=0.02, Pc=0.36, OR=3.62, respectively), showing some protective effects against HBV infection. However, after Bonferroni correction, this association was no longer found to be significant.
Table III.
Frequency of HLA-DQB1 alleles in hepatitis B virus infections and control group
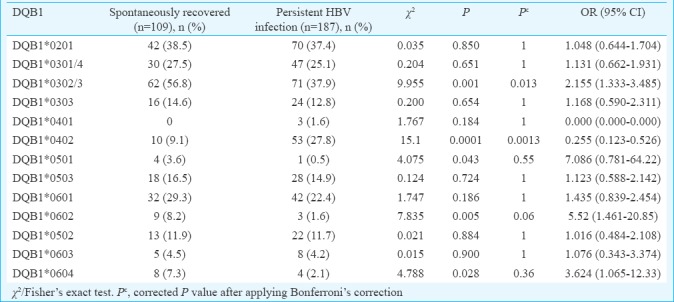
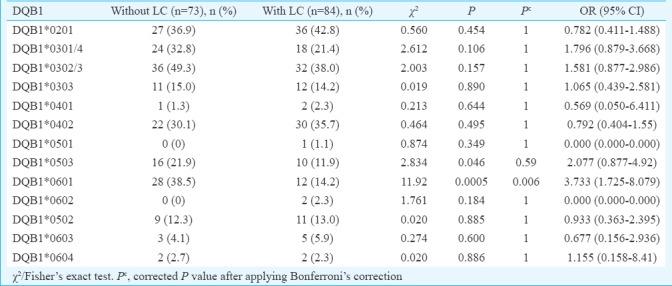
Comparison of phenotypic frequency of HLA-DQA1 and DQB1 alleles in persistent HBV infection with and without liver cirrhosis: The phenotype frequency of DQA1*0601 was significantly increased in persistent infection without LC (13.6%) compared to those with LC (3.5%) suggesting a protective effect of this allele against the development of LC. However, this difference was not significant after Bonferroni correction (χ2=5.27, P=0.02, Pc=0.23, OR=4.28). No other HLA-DQA1 alleles showed association (Table IV). DQB1*0601 was significantly increased in persistent infection without LC (38.5%) compared to those with LC (14.2%), which suggested protective effect of this allele against the development of LC. This difference was significant after Bonferroni correction also (χ2=11.92, P=0.0005, Pc=0.006, OR=3.73). Similarly, DQB1*0503 showed significant difference between persistent infection without LC and with LC (21.9 vs. 11.9%, χ2=2.83, P=0.04, Pc=0.59, OR=2.07) showing protective effect of this allele against the development of LC. However, this difference was not significant after Bonferroni correction (Table V).
Table IV.
Frequency of HLA-DQA1 alleles in hepatitis B virus infection with or without liver cirrhosis
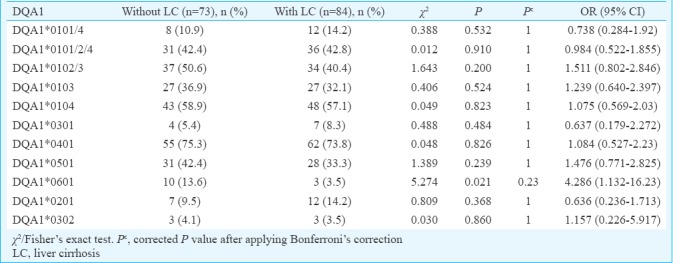
Table V.
Frequency of HLA-DQB1 alleles in hepatitis B virus infection with or without liver cirrhosis
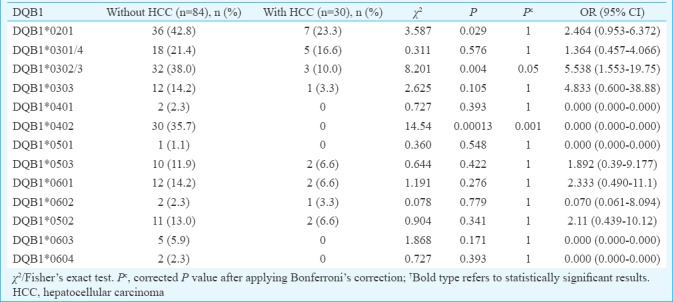
Comparison of phenotypic frequency of HLA-DQA1 and DQB1 alleles in persistent HBV infection with and without HCC: DQA1*0601 was significantly increased in HBV infection with HCC (16.6%) compared to those without HCC (3.5%), which suggested susceptible effect of this allele in the development of HCC (χ2=5.80, P=0.015, Pc=0.16, OR=0.18). However, this difference was not significant after Bonferroni correction (Table VI). Conversely, DQB1*0302/3 and DQB1*0402 were significantly increased in patients with HBV infection without HCC compared with those with HCC (38.0 vs. 10.0%, χ2=8.20, P=0.004, Pc=0.05, OR=5.53 and 37.5 vs. 0.0%, χ2=14.54, P=0.0001, Pc=0.001, respectively) suggesting a protective effect of these alleles for the development of HCC (Table VII).
Table VI.
Frequency of HLA-DQA1 alleles in hepatitis B virus infection with or without hepatocellular carcinoma (HCC)
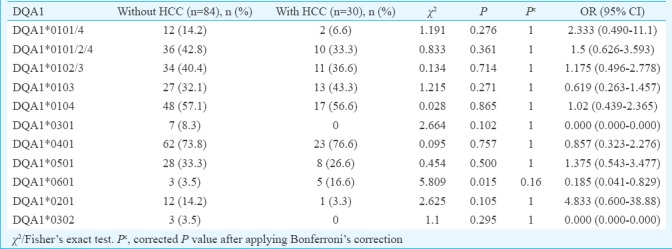
Table VII.
Frequency of HLA-DQB1 alleles in hepatitis B virus infection with or without hepatocellular carcinoma
Discussion
Our results indicate that certain HLA-DQA1 and DQB1 alleles were associated with persistent HBV infection and in the development of HBV-related LC. We found that DQA1*0101/2/4, DQA1*0103 and DQB1*0302/3 showed a protective effect against the development of chronic HBV infection. These finding were consistent with reported findings in Japanese population22 and in Korean population12 while DQB1*0402 was associated with chronic HBV infection. Studies from south India23 and western India24 reported a strong association between HLA class II alleles and chronicity of HBV infection. The mechanism underlying these associations is not clearly understood. HLA class II molecules present antigen in the presence of cytokine to CD4+ lymphocytes. Among HLA class II antigens, DQ molecules are unique and many of the variable amino acid residues are located on the α-helical part of the antigen-binding site25. Hence, it may be hypothesized that DQA1*0101/2/4 molecule binds and presents antigens more effectively, resulting in vigorous immune response towards HBV infection. Conversely, the association of DQB1*0402 with chronic HBV infection may be because of its inability to bind and present antigens effectively. However, further investigation needs to be done to confirm these hypotheses.
It was found that DQA1*0104, DQA1*0601, DQB1*0501, DQB1*0602 and DQB1*0604 were significantly increased in controls compared to those with persistent infection showing some protective effects from chronic HBV infection. However, these associations were not significant after Bonferroni correction. Because several HLA alleles were tested in each individual and the same data were used for comparing frequency, it was possible that one of the alleles would deviate significantly. Hence, to overcome this error, the P value was corrected by Bonferroni inequality method21. In various other studies, DQA1*0102, DQA1*030126,27 and DQB1*0302, DQB1*0609, DQB1*0503, DQB1*030312,28 alleles have been reported to be showing protective effect from developing chronic HBV infection.
Some investigators reported that HLA class II alleles were associated with development of end-stage liver disease caused by different aetiology and had a complex relationship between them29,30,31,32. In this study, we compared the HLA class II DQA1 and DQB1 allele frequency between patients with persistent HBV infection with and without LC. It was found that the phenotypic frequencies of DQA1*0601 and DQB1*0503 were significantly increased in HBV carriers without LC when compared with LC suggesting a protective role against the development of LC. DQB1*0601 was significantly decreased in persistent HBV infection with LC compared to those without LC, suggesting a strong protective role against the development of LC.
Some previous studies have investigated HLA class II allele association with HCC from China11,15, Egypt33 and Hong Kong34 showing conflicting results. In this study, we compared the HLA class II DQA1 and DQB1 allele frequency between patients with persistent HBV infection with and without HCC and found that the phenotypic frequency of DQA1*0601 was significantly increased in persistent infection with HCC compared to without HCC suggesting a susceptible effect for the development of HCC. However, this significance was lost after Bonferroni correction. Conversely, the phenotypic frequency of DQB1*0302/3 and DQB1*0402 was significantly increased in persistent infection without HCC when compared with HCC suggesting a protective role against the development of HCC. This difference was significant even after Bonferroni correction.
In HLA association studies, once candidate genetic polymorphisms are identified in a ‘derivation cohort’, these findings are re-tested and confirmed in a second separate ‘validation cohort’. The lack of a validation cohort was a limitation of our study.
In conclusion, three HLA-DQA1 alleles and four DQB1 alleles were found to be associated with development of chronic HBV infection, LC and HCC; DQA1*0101/2/4, DQA1*0103 and DQB1*0302/3 were associated with protection from developing chronic HBV infection, whereas DQB1*0402 was associated with susceptibility to chronic HBV infection. DQB1*0601 conferred protective effect from the development of LC, whereas DQA1*0601 showed susceptible effect for the development of HCC in patients with HBV infection. Conversely, DQB1*0302/3 and DQB1*0402 conferred protective effect from the development of HCC.
Footnotes
Financial support & sponsorship: This study was funded by the Indian Council of Medical Research, Department of Health Research, Ministry of Health & Family Welfare, Government of India, New Delhi.
Conflicts of Interest: None.
References
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Purcell RH. The discovery of the hepatitis viruses. Gastroenterology. 1993;104:955–63. doi: 10.1016/0016-5085(93)90261-a. [DOI] [PubMed] [Google Scholar]
- 3.Wright TL, Lau JY. Clinical aspects of hepatitis B virus infection. Lancet. 1993;342:1340–4. doi: 10.1016/0140-6736(93)92250-w. [DOI] [PubMed] [Google Scholar]
- 4.Cacciola I, Cerenzia G, Pollicino T, Squadrito G, Castellaneta S, Zanetti AR, et al. Genomic heterogeneity of hepatitis B virus (HBV) and outcome of perinatal HBV infection. J Hepatol. 2002;36:426–32. doi: 10.1016/s0168-8278(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 5.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 6.Xu BY, Wang YM, Deng GH, Huang YP, Zhong LH, Liu GD, et al. The primary comparative analysis between the host genetic factors and their relationships with clinical phenotype of HBV infected twins. Zhonghua Yi Xue Za Zhi. 2004;84:189–93. [PubMed] [Google Scholar]
- 7.Yu MW, Chang HC, Liaw YF, Lin SM, Lee SD, Liu CJ, et al. Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. J Natl Cancer Inst. 2000;92:1159–64. doi: 10.1093/jnci/92.14.1159. [DOI] [PubMed] [Google Scholar]
- 8.Thio CL, Carrington M, Marti D, O’Brien SJ, Vlahov D, Nelson KE, et al. Class II HLA alleles and hepatitis B virus persistence in African Americans. J Infect Dis. 1999;179:1004–6. doi: 10.1086/314684. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, Kaslow RA. HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology. 2004;39:978–88. doi: 10.1002/hep.20142. [DOI] [PubMed] [Google Scholar]
- 10.Wu YF, Wang LY, Lee TD, Lin HH, Hu CT, Cheng ML, et al. HLA phenotypes and outcomes of hepatitis B virus infection in Taiwan. J Med Virol. 2004;72:17–25. doi: 10.1002/jmv.10557. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Cheng B. Association of polymorphisms of human leucocyte antigen-DQA1 and DQB1 alleles with chronic hepatitis B virus infection, liver cirrhosis and hepatocellular carcinoma in Chinese. Int J Immunogenet. 2007;34:373–8. doi: 10.1111/j.1744-313X.2007.00702.x. [DOI] [PubMed] [Google Scholar]
- 12.Cho SW, Cheong JY, Ju YS, Oh DH, Suh YJ, Lee KW. Human leukocyte antigen class II association with spontaneous recovery from hepatitis B virus infection in Koreans: Analysis at the haplotype level. J Korean Med Sci. 2008;23:838–44. doi: 10.3346/jkms.2008.23.5.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zervas J, Karvountzis G, Theodoropoulos G. HLA antigens and hepatocellular carcinoma. Gastroenterology. 1980;79:601. [PubMed] [Google Scholar]
- 14.Chan SH, Simons MJ, Oon CJ. HLA antigen in Chinese patients with hepatocellular carcinomas. J Natl Cancer Inst. 1980;65:21–3. [PubMed] [Google Scholar]
- 15.Donaldson PT, Ho S, Williams R, Johnson PJ. HLA class II alleles in Chinese patients with hepatocellular carcinoma. Liver. 2001;21:143–8. doi: 10.1034/j.1600-0676.2001.021002143.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Cai M, Li G, Wei D, Tan Y. Expression of HLA-I antigens in primary hepatocellular carcinoma. Hua Xi Yi Ke Da Xue Xue Bao. 2001;32:495–6. 504. [PubMed] [Google Scholar]
- 17.Pellegris G, Ravagnani F, Notti P, Fissi S, Lombardo C. B and C hepatitis viruses, HLA-DQ1 and -DR3 alleles and autoimmunity in patients with hepatocellular carcinoma. J Hepatol. 2002;36:521–6. doi: 10.1016/s0168-8278(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 18.Lok ASF, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507–39. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 19.Bruix J, Sherman M, Llovet JM, Beaugrand M, Lencioni R, Burroughs AK, et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. J Hepatol. 2001;35:421–30. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 20.Olerup O, Aldener A, Fogdell A. HLA-DQB1 and -DQA1 typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Tissue Antigens. 1993;41:119–34. doi: 10.1111/j.1399-0039.1993.tb01991.x. [DOI] [PubMed] [Google Scholar]
- 21.Etymologia: Bonferroni correction. Emerg Infect Dis. 2015;21:289. doi: 10.3201/eid2102.ET2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884–92. doi: 10.1093/hmg/ddr301. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher GJ, Samuel P, Christdas J, Gnanamony M, Ismail AM, Anantharam R, et al. Association of HLA and TNF polymorphisms with the outcome of HBV infection in the South Indian population. Genes Immun. 2011;12:552–8. doi: 10.1038/gene.2011.32. [DOI] [PubMed] [Google Scholar]
- 24.Amarapurpar DN, Patel ND, Kankonkar SR. HLA class II genotyping in chronic hepatitis B infection. J Assoc Physicians India. 2003;51:779–81. [PubMed] [Google Scholar]
- 25.Marsh SG, Bodmer JG. HLA-DR and-DQ epitopes and monoclonal antibody specificity. Immunol Today. 1989;10:305–12. doi: 10.1016/0167-5699(89)90086-8. [DOI] [PubMed] [Google Scholar]
- 26.Lu LP, Liu Y, Li XW, Sun GC, Zhu XL, Wu YZ, et al. Association of polymorphisms of human leucocyte antigen -DRB1 and -DQA1 allele with outcomes of hepatitis B virus infection in Han population of north China. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2006;28:134–42. [PubMed] [Google Scholar]
- 27.Jiang YG, Wang YM. Study on the polymorphism of human leucocyte antigen-DRB1, -DQA1 and -DQB1 alleles in patients with hepatitis B. Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25:337–40. [PubMed] [Google Scholar]
- 28.Xi-Lin Z, Te D, Jun-Hong L, Liang-Ping L, Xin-Hui G, Ji-Rong G, et al. Analysis of HLA-DQB1 gene polymorphisms in asymptomatic HBV carriers and chronic hepatitis B patients in the Chinese Han population. Int J Immunogenet. 2006;33:249–54. doi: 10.1111/j.1744-313X.2006.00607.x. [DOI] [PubMed] [Google Scholar]
- 29.Tillmann HL, Chen DF, Trautwein C, Kliem V, Grundey A, Berning-Haag A, et al. Low frequency of HLA-DRB1*11 in hepatitis C virus induced end stage liver disease. Gut. 2001;48:714–8. doi: 10.1136/gut.48.5.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donaldson PT, Baragiotta A, Heneghan MA, Floreani A, Venturi C, Underhill JA, et al. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: A large-scale study. Hepatology. 2006;44:667–74. doi: 10.1002/hep.21316. [DOI] [PubMed] [Google Scholar]
- 31.Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: An old story now reviving. Hepatology. 2011;54:714–23. doi: 10.1002/hep.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin B, Wang J, Chen J, Liang Y, Yang Z, Zhong R. Association of human leukocyte antigen class II with susceptibility to primary biliary cirrhosis: A systematic review and meta-analysis. PLoS One. 2013;8:e79580. doi: 10.1371/journal.pone.0079580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Chennawi FA, Auf FA, Metwally SS, Mosaad YM, El-Wahab MA, Tawhid ZE. HLA-class II alleles in Egyptian patients with hepatocellular carcinoma. Immunol Invest. 2008;37:661–74. doi: 10.1080/08820130802111605. [DOI] [PubMed] [Google Scholar]
- 34.Li PK, Leung NW, Poon AS, Wong KC, Chan TH, Lai KN. Molecular genetics of major histocompatibility complex class II genes in hepatocellular carcinoma. Dig Dis Sci. 1995;40:1542–6. doi: 10.1007/BF02285206. [DOI] [PubMed] [Google Scholar]


