Abstract
Pancreatic cancer is currently one of the deadliest of the solid malignancies, whose incidence and death rates are increasing consistently during the past 30 years. Ribonucleotide reductase (RR) is a rate-limiting enzyme that catalyzes the formation of deoxyribonucleotides from ribonucleotides, which are essential for DNA synthesis and replication. In this study, 23 small interfering RNAs (siRNAs) against RRM2, the second subunit of RR, were designed and screened, and one of them (termed siRRM2), with high potency and good RNase-resistant capability, was selected. Transfection of siRRM2 into PANC-1, a pancreatic cell line, dramatically repressed the formation of cell colonies by inducing remarkable cell-cycle arrest at S-phase. When combining with doxorubicin (DOX), siRRM2 improved the efficacy 4 times more than applying DOX alone, suggesting a synergistic effect of siRRM2 and DOX. Moreover, the combined application of siRRM2-loaded lipid nanoparticle and DOX significantly suppressed the tumor growth on the PANC-1 xenografted murine model. The inhibition efficiency revealed by tumor weight at the endpoint of the treatment reached more than 40%. Hence, siRRM2 effectively suppressed pancreatic tumor growth alone or synergistically with DOX. This study provides a feasible target gene, a drug-viable siRNA, and a promising therapeutic potential for the treatment of pancreatic cancer.
Keywords: pancreatic cancer, RRM2, siRNA, doxorubicin, small interfering RNA, RNA interference, LNP, lipid nanoparticle, combined therapy, synergistic effect
Graphical Abstract
Introduction
Pancreatic cancer is a disease in which malignant (cancer) cells form in the tissues of the pancreas. It is currently one of the deadliest of the solid malignancies, whose incidence and death rates are increasing consistently during the past 30 years.1, 2 Worldwide, the incidence ranges from 1 to 10 cases per 100,000 people, and it is generally higher in developed countries and among men.3, 4 It was predicted that pancreatic cancer will become the second leading cause of cancer-related death by 2030 in the United States, ranking second only to lung cancer.1, 5 Pancreatic cancer typically does not cause symptoms until it has grown, so it is most frequently diagnosed in advanced stages rather than early in the course of the disease.4 As a result, the cancer often has metastasized to other organs with very poor prognosis. There are various treatments for pancreatic cancer, including surgery (resection), chemotherapy, and chemoradiotherapy.4 However, present therapeutic strategies have not substantially improved the survival of patients over the past several decades, revealed by its resistance to the therapy, that more than 80% of patients relapse after resection, and that the 5-year survival rate has remained at only ∼5%.6 Therefore, new treatments are urgently needed for patients with pancreatic cancer.
RNAi is a fundamental pathway in eukaryotic cells by which small interfering RNA (siRNA) is able to mediate targeted mRNA transcript cleavage, repress gene expression, and compromise gene function within living cells.7 Given the ability to knock down, in essence, any gene of interest, RNAi via siRNAs has generated a great deal of interest in both basic research and clinical application. siRNA-based therapeutics7, 8, 9, 10, 11 have shown promise for treating genetic diseases (e.g., transthyretin amyloidosis, hemophilia, and porphyria), metabolic diseases (e.g., hypercholesterolemia), virus infection diseases (hepatitis B virus [HBV], Ebola, HIV, and RSV [respiratory syncytial virus]), ophthalmic diseases (non-arteritic anterior ischemic optic neuropathy [NAION], age-related macular degeneration [AMD], and diabetic macular edema [DME]), and various solid tumors (e.g., hepatocellular carcinoma, melanoma, and pancreatic cancer). siRNA therapeutics for oncology that have entered clinical trials include ALN-KSP, CALAA-01, TKM-PLK1, Atu027, DCR-MYC, SiG12D-LODER, and siRNA-EPHA2-DOPC.12 In addition, siRNA has been used to circumvent multiple drug resistance (MDR), to enhance the chemosensitivity of tumor cells to chemotherapeutics.13, 14, 15, 16 siRNA nanoplexes also have been administered sequentially with conventional chemotherapeutics in order to block multiple signaling networks simultaneously.17, 18
Ribonucleotide reductase (RR) is a rate-limiting enzyme that catalyzes the formation of deoxyribonucleotides from ribonucleotides, which is essential for DNA synthesis and replication.19 Human RR consists of two subunits, RRM1 and RRM2, and the expression of both proteins is required for enzymatic activity. In mammalian cells, RRM2 is expressed only in the late G1 or early S phase of the cell cycle, whereas the levels of RRM1 remain relatively consistent throughout the cell cycle.20, 21 It was demonstrated that elevated RRM2 activity played a pivotal role in cellular response to DNA damage, tumor progression and invasion, angiogenesis, and the increase in drug-resistant properties.22, 23, 24, 25 Overexpression of RRM2 is observed in many kinds of human cancers. It was identified as a diagnostic marker and an established anti-cancer therapeutic target.26, 27 Moreover, it was reported that CALAA-01, a tumor-targeted nanodrug containing anti-RRM2 siRNA, mediated effective gene silencing in human beings and alleviated the symptom of melanoma.28 It was also observed that siRNA targeting RRM2 could be used for treating head and neck cancer29 (including oral squamous cell carcinoma30), ovarian cancer,31 gastric adenocarcinoma,32 hepatocellular carcinoma,33 colorectal cancers,34 etc. Hence, siRNA against RRM2 displayed versatile anti-tumor activity in various solid tumors.
In this study, we explored the anti-tumor effect of siRNA against RRM2 on PANC-1, a human pancreatic carcinoma and epithelial-like cell line. 23 siRNAs targeting RRM2 were designed and screened. One of them (siRNA-04M) was finally selected, termed siRRM2, and used in the following experiments. The influences of siRRM2 on cell morphology, colony formation, and cell-cycle arrest were evaluated. By combining with doxorubicin (DOX), a well-defined anti-tumor chemotherapeutic, the siRNA’s performances on cell proliferation (in vitro) and tumor growth (in vivo) were further investigated.
Results
siRNA Activity Screening and Stability Evaluation
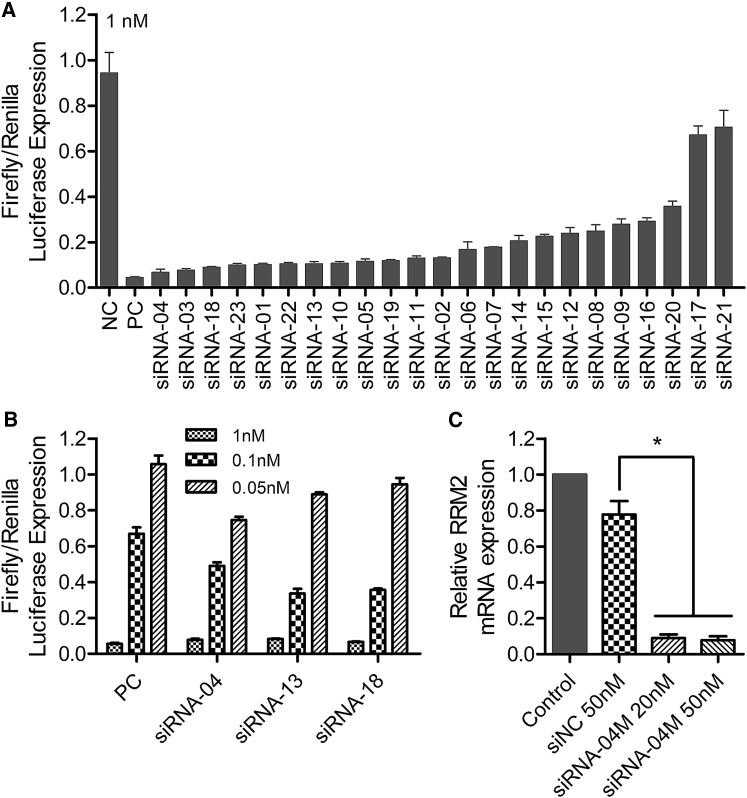
23 siRNAs targeting RRM2 (numbered from siRNA-01 to siRNA-23) were designed and selected with a high-throughput screening system called siQuant.35 For the first round of screening, 23 siRNAs were respectively transfected into HEK293A cells with lipofectamine 2000 at 1 nM. As a result, 14 of them displayed more than 80% silencing efficiency, and 8 of them showed approximately 90% knockdown efficiency (Figure 1A). Then three siRNAs (siRNA-04, siRNA-13, and siRNA-18) were selected to perform the second round of screening at three transfection concentrations of 1, 0.1, and 0.05 nM. It was observed that siRNA-04 achieved higher gene-silencing efficiency at the lowest transfection concentration (0.05 nM) (Figure 1B).
Figure 1.
siRNA Screening and Activity Validation
(A) The first round selection of 23 siRNAs at 1 nM with siQuant in HEK293A. (B) The second round selection of 3 siRNAs at three transfection concentrations of 1, 0.1, and 0.05 nM. (C) RRM2 mRNA expression of PANC-1 after being treated with siRNA-04M at 20 and 50 nM. NC and PC represent negative control (a scramble siRNA) and positive control (an siRNA showing good gene-silencing efficiency), respectively. Data were shown as mean ± SD. *p < 0.05, n = 3.
To stabilize the siRNA, precise chemical modifications with methoxy group and/or fluorine at 2′ of riboses of certain nucleotides were introduced to the siRNAs (as shown in our patent application file36). RNase-resistant assay was performed to evaluate their stability in serum (Figure S1). Data showed that both full-length and truncated versions of siRNA-04 without chemical modification were observed within 24 hr. Unfortunately, we failed to observe the full-length siRNA-04 without modification when incubating for 48 and 72 hr. In contrast, siRNA-04M with stabilization modification chemistry was resistant to RNase attack in serum, as siRNA still remained in full length even after 72-hr incubation. Hence, siRNA-04 itself is stable to some extent, and chemical modification further enhances its stability significantly.
Furthermore, to validate the siRNA’s silencing activity against the endogenous targeting mRNA, instead of the sequence on the luciferase reporter system, and to evaluate the influence of chemical modification on the siRNA’s potency, chemically modified siRNA-04 (siRNA-04M) was transfected into PANC-1 cell, a human pancreatic carcinoma and epithelial-like cell line, and mRNA expression was analyzed with real-time qPCR. Data demonstrated that siRNA-04M effectively inhibited the expression of RRM2 mRNA at the concentrations of 20 and 50 nM (Figure 1C).
On-Target Activity and Off-Target Effect of Anti-RRM2 siRNAs
siRNAs potentially may trigger off-target effects via several different mechanisms,37 such as the following: (1) the passenger strand of the siRNA mediates gene silencing in a complete-match manner (siRNA’s working pattern); (2) either the passenger or guide strand of the siRNA mediates gene silencing via seed region matching (a microRNA [miRNA]-like pathway); and (3) gene disturbance resulting from an immune response stimulated by the siRNA molecule. Evaluation of on-target and off-target effects is of great importance for siRNA therapeutics development,38 since off-target effects may trigger serious toxicity in vivo.39
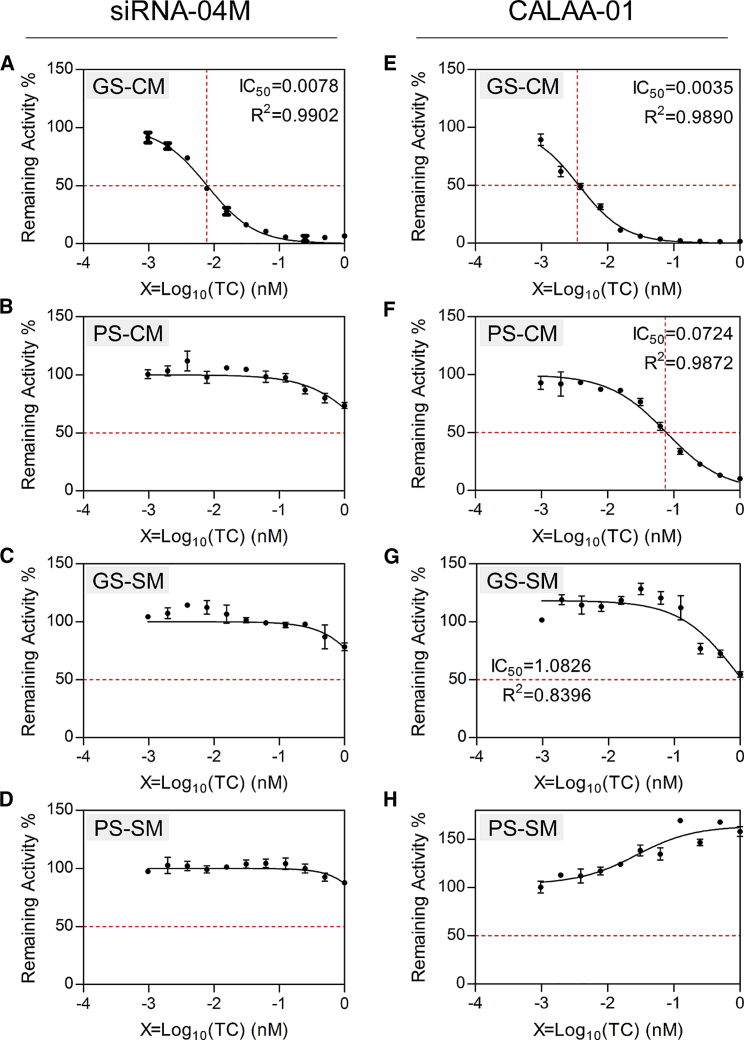
psiCHECK, a more sensitive activity evaluation system than siQuant, was applied to determine the tiniest change in gene silencing. Meanwhile, CALAA-01 is an siRNA therapeutic that has been clinically studied in phase I for the treatment of solid tumor.28 The siRNA against RRM2 used in this clinical study was included as a control in this assay. GS-CM or PS-CM indicates the guide strand (GS) or passenger strand (PS) of the siRNA match with its targeting mRNA in a complete-match (CM) manner. The GS is the desired gene-silencing modulator, representing siRNA’s on-target activity. GS-SM or PS-SM mean the guide or PS of the siRNA works via seed region matching, an miRNA-like pathway. Hence, gene silencing mediated by PS-CM, GS-SM, or PS-SM all represent siRNA’s off-target effects. Data showed that the on-target IC50 of siRNA-04M in the HEK293A cell was 0.0078 nM, comparable with the IC50 of CALAA-01 (0.0035 nM) (Figure 2). No silencing activity was observed for all other three forms for siRNA-04M. However, significant off-target gene knockdown was observed for CALAA-01, as the IC50 values of PS-CM and GS-SM were 0.0724 and 1.0826 nM, respectively (Figure 2). Therefore, siRNA-04M is superior to CALAA-01 in terms of their on-target and off-target effects, supporting the following experiments with siRNA-04M both in vitro and in vivo.
Figure 2.
On-Target Activity and Off-Target Effect of siRNA-04M and siRNA Contained in CALAA-01
siRNA that works by its guide strand (GS) and completely matching (CM) with its targeting mRNA represents on-target activity (A and E). If it works by its passenger strand (PS) completely matching (CM) with its targeting mRNA (B and F), or either its GS or its PS matching with its targeting mRNA in the seed region (SM, seed match) (C, D, G, and H), an off-target effect may be generated. IC50 was calculated with GraphPad Prism 5 software and is shown in the upper-right corner. Data were shown as mean ± SD. Data were duplicated three times.
Toxicity Evaluation of siRNA
Cytotoxicity of the siRNA was further evaluated in PANC-1 cells with a 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay. Free uptake of the siRNA was applied at the concentrations of 0.4, 2, 10, 50, 250, 500, and 1,000 nM. Cell viability was well maintained for all treatment concentrations, suggesting good biocompatibility of siRNA-04M (Figure S2).
In addition, in vivo toxicity, including cytokine inducement, of siRNA-04M was thoroughly investigated. Lipopolysaccharide (LPS) was included as a positive control. siRNA and LPS were all dosed at 5 mg/kg, via intravenous and intraperitoneal injection, respectively. Data revealed that LPS triggered significant cytokine release in vivo. The concentration of TNF-α (tumor necrosis factor alpha), IFN-γ (interferon gamma), IL-6 (interleukin-6), KC (keratinocyte-derived cytokine, or CXCL1 [chemokine (C-X-C motif) ligand 1]), MCP-1 (monocyte chemoattractant protein-1, or CCL2 [chemokine (C-C motif) ligand 2]), GM-CSF (granulocyte-macrophage colony-stimulating factor, or CSF2 [colony-stimulating factor 2]), and IL-12p70 were all dramatically increased in circulation. The ratios of spleen to body weight were also elevated significantly at 24 and 48 hr post-treatment. However, siRNA-04M did not induce any cytokine release, influence liver and kidney function, and had no impact on organ coefficients (Figures S3–S5). Furthermore, the siRNA was dosed at a higher dose of 10 mg/kg via intravenous (i.v.) injection in another test. Concentrations of cytokines (IL-6 and TNF-α) and several biochemistry parameters (ALT [alanine transaminase], AST [aspartate aminotransferase], TP [total protein], and LDH [lactate dehydrogenase]) were recorded at 24 hr post-treatment (Figure S6). It was also proven that the siRNA was well tolerated by animals, although a higher dose of siRNA has been applied.
In consideration, siRRM-04M with high potency, good stability, no off-target effect, and no toxicity was finally selected as the lead compound in this study. siRNA-04M is termed siRRM2 and used in the following experiments.
Influence of RRM2 Knockdown on Colony Formation
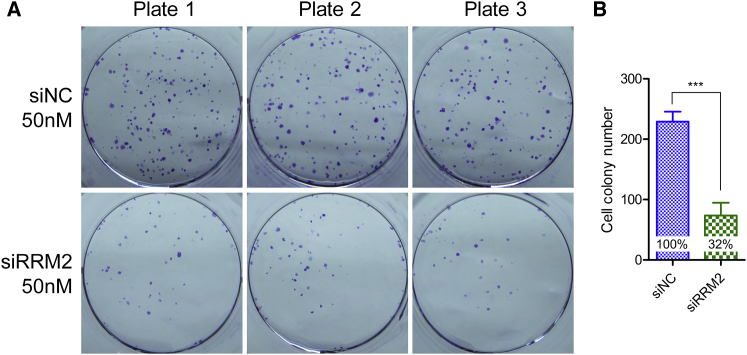
Colony formation assay was performed to evaluate the efficacy of RRM2 knockdown. siNC (a scramble siRNA without any targeting gene in mouse, rat, monkey, and human beings) was used as a control. Data revealed that the cells treated with siRRM2 showed a remarkable reduction of colonies compared with the cells treated with siNC (Figure 3A), suggesting RRM2 knockdown significantly influenced cell proliferation. The quantitative analysis data proved that the colonies decreased approximately 70% after treatment with siRRM2 (Figure 3B).
Figure 3.
Cell Colonies of PANC-1 Cells Treated with siNC or siRRM2 at 50 nM
(A) The digital images of cell colonies. (B) Quantification analysis of the colonies. Data were shown as mean ± SD. ***p < 0.001, n = 3. The inserted percentages represent the relative colony numbers for these two treatments. The colony number of the cells treated with siNC was set as 100%, by normalizing to which, the relative colony number of cells treated with siRRM2 was calculated.
Cell-Cycle Arrest Induced by Knockdown of RRM2
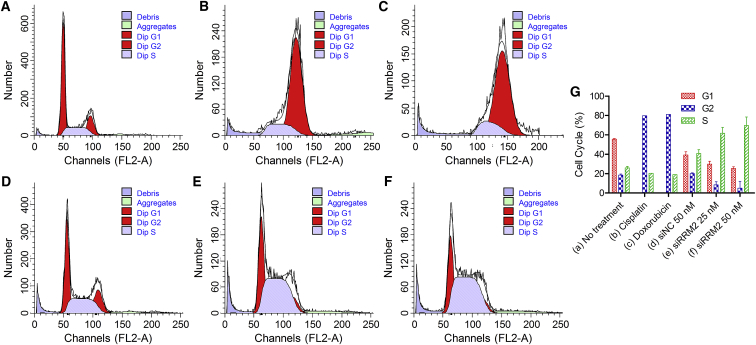
To explore the mechanism of siRRM2’s influence on cell proliferation, cell-cycle arrest assay was performed. Here, two classical antineoplastic drugs, cisplatin (Pt) and DOX, were used as positive controls at the doses of 25 and 0.2 μg/mL, respectively. The transfection concentrations of siRRM2 were 20 and 50 nM. Here, siNC (50 nM) was also included as negative control. Data manifested that both Pt and DOX induced significant G2/M checkpoint arrest (Figures 4B and 4C). Pt is an inorganic platinum complex, which induces cytotoxicity by inhibiting DNA synthesis by conforming DNA adducts. These DNA adducts can activate several signal transduction pathways, including Erk, p53, p73, and MAPK, which culminates in the activation of apoptosis.40, 41 As a result, it will induce G2 arrest.41 Doxorubicin, an antibiotic anthracycline, is commonly considered to exert its anti-tumor activity at two fundamental levels, altering DNA and producing free radicals to trigger apoptosis of cancer cells through DNA damage. Doxorubicin-induced G2/M checkpoint arrest is attributed to elevated cyclin G2 (CycG2) expression and phospho-modification of proteins in the ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) signaling pathways.42, 43, 44 In addition, compared with the controls of no treatment and siNC 50 nM, cells treated with siRRM2 (50 and 25 nM) displayed a remarkable and dose-dependent increase in S-phase populations; and the G2/M-phase population decreased significantly (Figures 4E and 4F). This was consistent with the S-phase cell-cycle arrest matched with the mechanism of action, and it was in line with the reported data.31, 45 Synthesis of RRM2 protein is regulated in a cell-cycle-dependent fashion. It begins to rise in late G1 and reach the highest level during S-phase when DNA replication occurs. Therefore, when RRM2 expression is repressed by siRNA, the protein level will decrease in G1/S phase, resulting in the block of conversion of ribonucleoside 5′-diphosphates into their corresponding 2′-deoxyribonucleotides and the inhibition of DNA synthesis and replication.
Figure 4.
Cell-Cycle Arrest Induced by Cisplatin, Doxorubicin, and siRRM2
(A–F) The cell cycle distribution of untreated cells (A) and cells treated with cisplatin (B), doxorubicin (C), siNC at 50 nM (D), siRRM2 at 25 nM (E), and siRRM2 at 50 nM (F). (G) Percentages of cell populations in G1, G2, and S phases of the cells shown in (A)–(F). Data were shown as mean ± SD. n = 1 for cells treated with cisplatin and doxorubicin; n = 2 for untreated cells and the cells treated with siNC and siRRM2.
Impact of siRRM2 Transfection on Cell Morphology
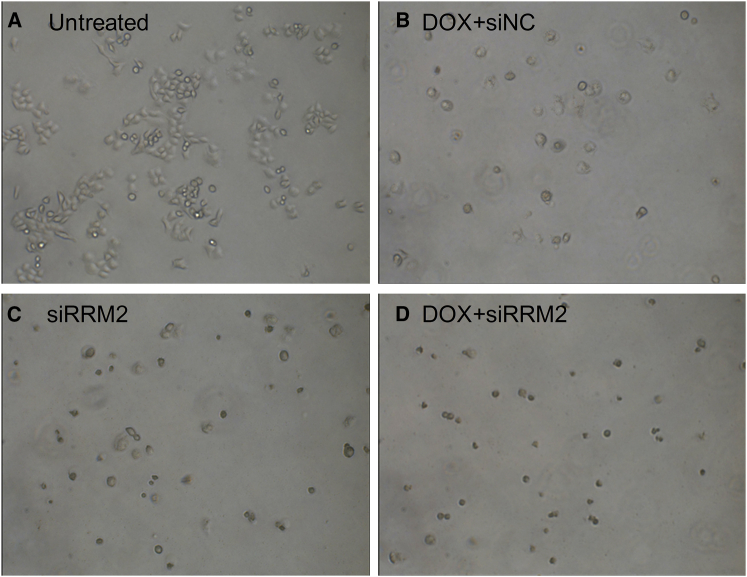
siRRM2 and DOX were further used to explore their influence on cell morphology. Data displayed that all three kinds of treatment induced remarkable cell shrinkage and death (Figures 5B–5D). Compared with untreated cells, cell numbers for the cells treated with DOX and/or siRRM2 were significantly reduced. Moreover, cells treated with DOX (0.2 μg/mL) and siNC (50 nM) showed a little bit better cell morphology than cells treated with siRRM2 alone (50 nM) and cells treated with siRRM2 combined with DOX. Combination treatment of siRRM2 and DOX caused almost all cells’ death, suggesting a promising synergistic effect of siRRM2 and DOX on anti-proliferation activity.
Figure 5.
Cell Morphology of Cells with Various Treatments
(A–D) Cells without treatment (A), cells treated with DOX plus siNC (B), cells treated with siRRM2 alone (C), and cells treated with DOX plus siRRM2 (D).
Cell Viability for the Cells Treated with siRRM2 Alone or Combined with DOX
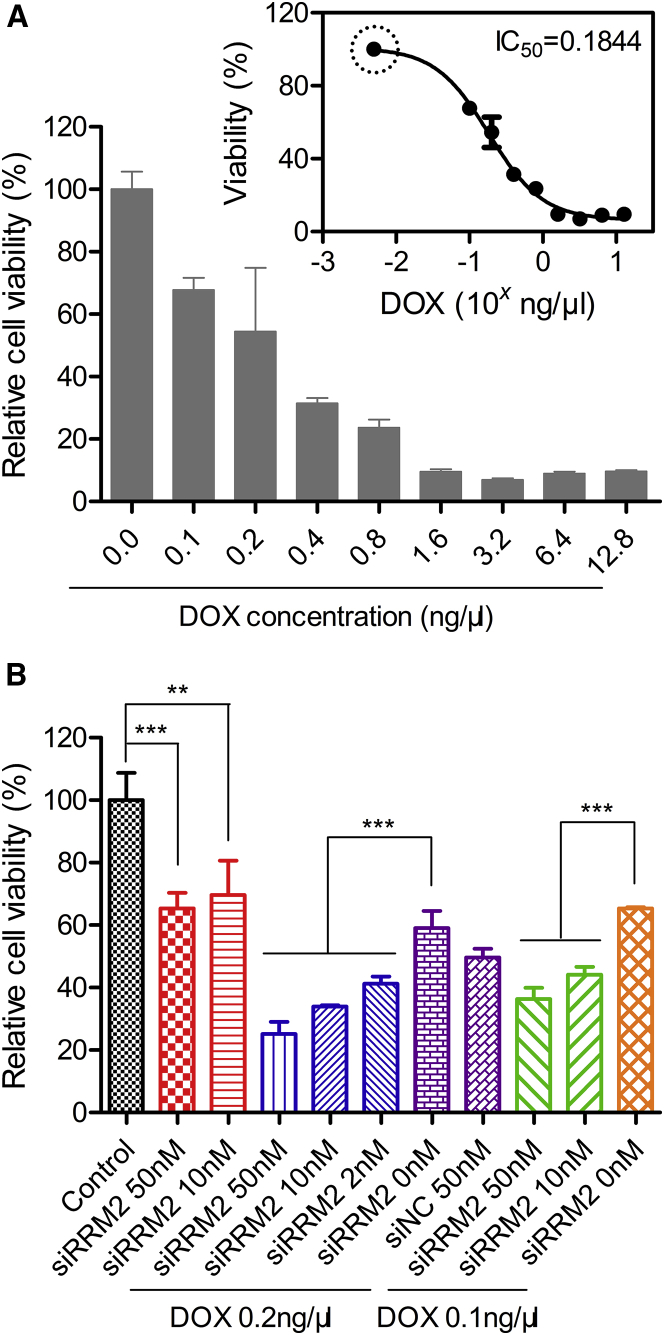
MTT assay was performed to evaluate the capability of siRRM2 in inhibiting tumor cell proliferation. First, DOX was applied to PANC-1 at 0.0, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4, and 12.8 ng/μL. Cell growth was repressed dose dependently compared with the cells without treatment. Cells treated with 0.1 and 3.2 ng/μL DOX exhibited ∼32% and ∼93% inhibition of cell viability, respectively. More than 90% inhibition efficiency was achieved when DOX was applied at >1.6 ng/μL (Figure 6A). It was worthy to note that viabilities of the cells treated at doses of 6.4 and 12.8 ng/μL were comparable with those of the cells treated at doses of 1.6 and 3.2 ng/μL. That is because DOX itself can be excited at ∼545 nm, although the highest excitation wavelength for DOX is around 480 nm. The more DOX that is added into the cells, the stronger the signal interference that is observed.
Figure 6.
Cell Viability of PANC-1 Cells Evaluated with MTT Assay
(A) Cell viability of PANC-1 cells treated with DOX alone at various concentrations. Inserted graph shows the fitting curve of the cell viability, by which the IC50 was calculated. The data marked with a circle means that the data point is generated by reasonable assumption. (B) Cell viability of PANC-1 treated with siRRM2 alone or combined with DOX. Data were shown as mean ± SD. **p < 0.01, ***p < 0.001, n = 6 (A) or 5 (B).
According to the result calculated by a fitting function of cell viability, the IC50 of DOX was ∼0.18 ng/μL (Figure 5A, inserted graph). The fitting curve was generated with GraphPad Prism software according to the formula of Y = Bottom + (Top − Bottom)/(1 + 10ˆ((X − LogIC50))). Moreover, cells treated with 0.1 ng/μL DOX (the lowest concentration we applied) remained at ∼70% cell viability. Since it did not reach the upper limit of cell viability, fitting function could not be generated in a reasonable way. Untreated cells were equal to 0.0 ng/μL DOX being added. However, a concentration of 0.0 ng/μL cannot be transferred to a Log value for plotting on the fitting function. Alternatively, to obtain an accurate IC50, we assumed that the cells treated with 0.005 ng/μL (an extremely low concentration) DOX displayed ∼100% cell viability (as marked with circle in the inserted graph). As a result, a well-fitting curve with an R2 (coefficient of determination) of ∼0.95 was generated, and the IC50 was successfully produced.
Second, siRRM2 was applied to cells alone or synergistically with DOX (Figure 6B). When siRRM2 was added alone to PANC-1 cells at 50 and 10 nM (transfection concentration), 35% and 30% growth inhibition were achieved, respectively. When siRRM2 (50, 10, and 2 nM) was co-added into the cells with 0.2 ng/μL DOX, they caused 75%, 66%, and 59% losses of cell viability, respectively. In addition, if the DOX concentration was reduced to 0.1 ng/μL, siRRM2 transfected at 50 and 10 nM still resulted in 64% and 56% losses of cell viability, respectively. Moreover, if we fixed the siRNA transfection concentration at 50 nM, applying 0.2 and 0.1 ng/μL DOX enhanced the inhibition efficiency from 35% (without DOX treatment) to 75% and 64%, respectively. When the concentration of siRRM2 was reduced to 10 nM, applying 0.2 and 0.1 ng/μL DOX elevated the inhibition efficiency from 30% (without DOX treatment) to 66% and 56%, respectively. Therefore, the combined use of the siRNA and DOX could achieve approximately two times higher efficacy than use of the siRNA alone. On the other hand, when siRRM2 was applied at 50 nM, 0.2 or 0.1 ng/μL DOX could induce 75% or 64% of cells to lose their viability, while 0.8 or 0.4 ng/μL DOX was needed if we wanted to achieve the same efficacy by DOX alone. This demonstrated that combination application of siRRM2 and DOX improved the efficacy by ∼4 times more than using DOX alone. Overall, cell viability data revealed a strong synergistic effect of siRRM2 and DOX on inhibiting the proliferation of PANC-1 cells (Figure 6B).
In Vivo Tumor Growth Inhibition
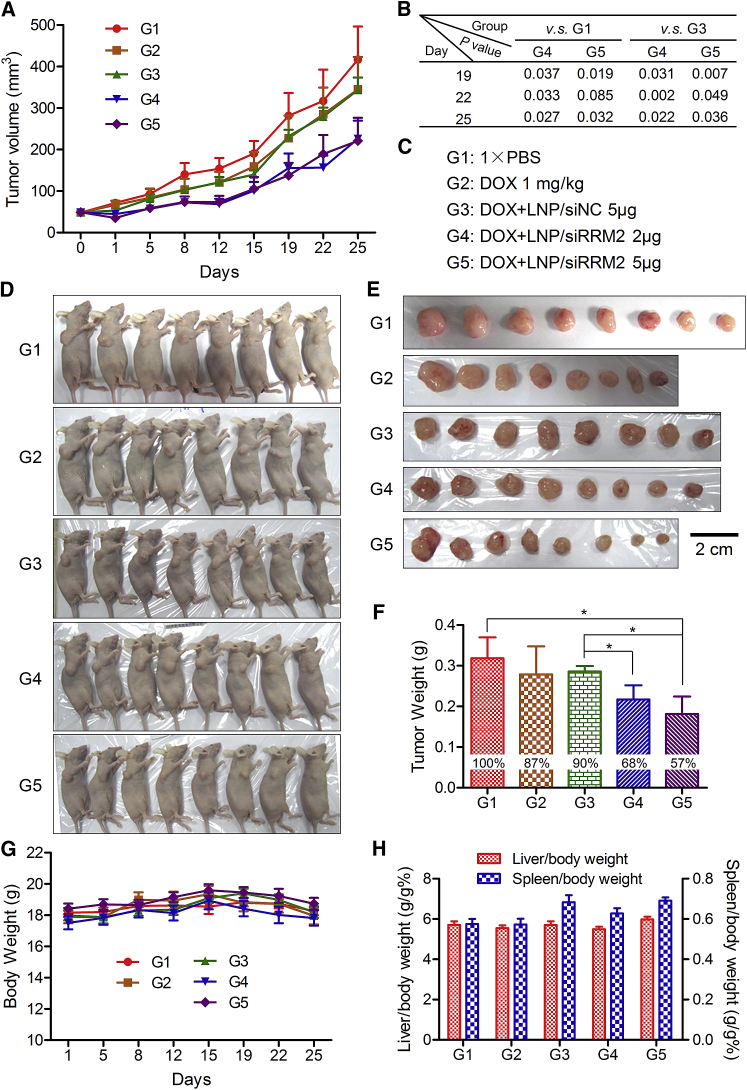
To explore the potential of combination treatment of siRRM2 and DOX in pancreatic cancer therapy, tumor growth suppression was evaluated with PANC-1 tumor-bearing BALB/c nude mice. Mice were randomly divided into five groups when tumor volumes reached ∼50 mm3. Then the following formulations were administered twice weekly into the mice: group 1, normal saline; group 2, DOX (1.0 mg/kg) alone; group 3, DOX (1.0 mg/kg) combined with lipid nanoparticle (LNP)/siNC (5 μg/tumor); group 4, DOX (1.0 mg/kg) combined with LNP/siRRM2 (2 μg/tumor); and group 5, DOX (1.0 mg/kg) combined with LNP/siRRM2 (5 μg/tumor). DOX and LNP/siRNA complexes were administered via intraperitoneal (i.p.) and peritumoral injections, respectively. LNP used in this assay is a novel lipid-based delivery system that has exhibited excellent siRNA delivery efficacy in vivo (data not shown).
Data revealed that DOX alone could slightly suppress tumor growth and the combination of siRRM2 and DOX remarkably enhanced the inhibition efficiency of tumor growth (Figure 7A). For tumor volumes at day 19, p values of group 5 versus group 1, and group 5 versus group 3 were 0.019 and 0.007, respectively (Figure 7B). The tumor volumes of groups 4 and 5 at day 25 were significantly smaller than groups 1 and 3, as all four p values were less than 0.05 (Figures 7A and 7B).
Figure 7.
Tumor Growth Inhibitions by DOX Alone or Combined with siRRM2 in PANC-1 Xenograft Tumor Murine Model
(A) Tumor growth curves for five group mice with various treatments. (B) Statistical analysis results for the tumor volumes at days 19, 22, and 25. (C) Treatment information for these five groups of mice. (D and E) Digital pictures of the whole bodies of the mice (D) and the isolated tumor tissues (E) at the end time point (day 25). Scale bar, 2 cm. (F) Average tumor weights for five groups of mice at the end time point. The inserted percentages in the histograms represent the relative tumor weight by normalizing to the average tumor weight of group 1 that was treated with 1× PBS. (G) Body weights of the mice during the whole treatment course. (H) Organ coefficients of the liver and the spleen at the end time point. Data were shown as mean ± SEM. *p < 0.05, n = 8.
Digital pictures of whole bodies and isolated tumors also provided similar information (Figures 7D and 7E). More importantly, tumor weights recorded at the end time point demonstrated that tumor suppression efficiencies of groups 2–5, compared to group 1, reached ∼13%, ∼10%, ∼32%, and ∼43%, respectively (Figure 7F). The differences between group 5 and group 1, group 5 and group 3, as well as group 4 and group 3 were all significant. These data revealed that (1) DOX alone could inhibit pancreatic tumor growth to an extent; (2) the combination treatment of DOX and siRRM2 dramatically improved the suppression efficiency compared to applying DOX alone; (3) the synergistic effect was attributed to siRRM2, because DOX combined with siNC exhibited a comparable tumor inhibition as with DOX alone, and because the dose-dependent effect for the siRRM2 was clearly observed in this assay. In addition, body weight, organ coefficients (the ratio of liver and body weight and the ratio of spleen and body weight) proved that all treatments did not cause obvious adverse effects during the whole treatment course (Figures 7G and 7H). Clinical observation was also performed during the treatment course, and no abnormal behavior was observed.
Discussion
Pancreatic cancer is a cancer type with a high degree of malignancy. It will become the second leading cause of cancer-related death by 2030 in the United States. DOX is an anthracycline antibiotic and a first-line anti-neoplastic drug for the treatment of a wide variety of cancers. RRM2, a pivotal gene involved in carcinogenesis, is a new molecular marker for the diagnosis and clinical outcomes of cancer, and a potential therapeutic target as well. LNP is a powerful and well-studied siRNA delivery system. LNP used in this study is a novel lipid-based nucleic acid delivery system, and previously it has been demonstrated to be a potent siRNA transporting tool with an ED50 of ∼0.05 mg/kg when used for liver-targeting delivery. On the other hand, the combination of several different anti-tumor reagents has become an effective and feasible treatment method for pancreatic cancer.46, 47, 48, 49 In this study, DOX and LNP-loaded siRRM2 were combinedly given to tumor-bearing mice. Tumor growth was significantly inhibited with this combination strategy. A strong synergistic effect has been observed and achieved, as applying the siRNA reduced the required dose of DOX while achieving the same efficacy as when applying DOX alone, and, in turn, DOX also enhanced the anti-tumor effect of the siRNA (Figures 6 and 7).
In conclusion, in this study we designed 23 siRNAs against RRM2, a pivotal gene involved in DNA synthesis and replication, and one of them was selected as the lead sequence. It was observed that both siRRM2 alone and DOX alone could induce significant cell growth inhibition, as suggested by the data of cell colony formation, cell-cycle arrest, cell morphology, and cell viability. Combined application of siRRM2 and DOX dramatically enhanced the anti-tumor efficacy by ∼4 times more than applying DOX alone. Moreover, a tumor growth inhibition assay performed in a PANC-1 xenograft tumor model demonstrated that the combination of siRRM2 and DOX significantly suppressed the tumor growth in vivo. The inhibition efficiency revealed by tumor weight at the endpoint reached >40%. Therefore, this work offers a good siRNA molecule against RRM2. This siRNA significantly inhibited the growth of pancreatic cancer cells in vitro and in vivo, regardless of applying it alone or in combination with DOX. The regimen explored in this work constitutes a safe and feasible salvage therapy in patients with pancreatic cancer, exhibiting good potential for clinical translation in the future.
Materials and Methods
Materials
siNC (catalog: SR-NC001, without any targeting gene in mouse, rat, and human) and siRRM2 (catalog: SR3204M, targeting ribonucleotide reductase M2 [RRM2]), were synthesized by Suzhou Ribo Life Science (Kunshan, China). To enhance their stability, several bases of their sequences were modified with methoxy group or fluorine at the 2′ site hydroxyl groups. Lipofectamine 2000, DMEM, Opti-MEM, fetal bovine serum (FBS), penicillin, streptomycin, trypsin, and TRIzol were purchased from Life Technologies (Molecular Probes, Eugene, OR, USA). Cell culture plates and serological pipettes were from Corning (NY, USA). Reverse transcription kit and real-time PCR kit (UltraSYBR Mixture) were purchased from Promega (Fitchburg, WI, USA) and Beijing ComWin Biotech (Beijing, China), respectively. Golden View used for staining nucleic acid in gel was purchased from Beijing BioDee BioTech (Beijing, China). Loading buffer was provided by Takara Bio (Japan). Agrose was from Oxoid (UK, part of Thermo Fisher Scientific). Crystal Violet Staining Solution was purchased from Beyotime Biotechnology (Haimen, Jiangsu, China). RNAlater, doxorubicin, Pt, and propidine iodide (PI) were purchased from Sigma-Aldrich (St. Louis, MO, USA). PI is a popular red fluorescent nuclear and chromosome counterstain. RNase A was provided by TransGen Biotech (Beijing, China). Pentobarbital sodium was provided by Peking University Laboratory Animal Center.
Cell Culture
PANC-1, a pancreatic carcinoma cell line, was obtained from the Cell Resource Center of Peking Union Medical College (Beijing, China). HEK293A, a human embryonic kidney cell line, was kept in our lab. Both cells were cultured with DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Activity Screening of siRNAs with siQuant or psiCHECK Assay
siRNAs targeting RRM2 were designed by scientists from Suzhou Ribo Life Science and screened with the siQuant system, a previously reported and well-defined siRNA validation method.35 Briefly, human embryonic kidney cells (HEK293A) were grown in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. The cells were seeded into 24-well plates at a density of ∼5 × 104 cells/well 1 day before transfection. Reporter plasmid (firefly luciferase expression plasmid, 100 ng/well) carrying siRNA target was transfected into the cells at approximately 60% confluence using lipofectamine 2000 (1 μL/well), together with pRL-TK control vector (renilla luciferase expression plasmid, 50 ng/well), also with siRNA against RRM2 or scramble siRNA (negative control [NC]) or positive control siRNA (positive control [PC]). The transfection concentration of siRNA against RRM2 was 1, 0.1, or 0.05 nM. 24 hr after transfection, the activities of both luciferases were determined by a fluorometer (Synergy HT, BioTek Instruments, Winooski, VT, USA). The firefly luciferase signal was normalized to the renilla luciferase signal for each individual well, and the silencing efficacy of each siRNA was calculated by comparison with the sample treated with NC siRNA. All experiments were performed in triplicates and repeated at least twice.
For the on-target and off-target evaluation of siRNAs, psiCHECK dual-luciferase reporter system (Promega, Madison, WI, USA),50 a more sensitive reporter system than siQuant, was applied. Both firefly and renilla luciferase expression sequences were constructed in one plasmid. The target of siRNA was inserted into the psiCHECK-2 plasmid at the 3′ UTR of renilla luciferase. siRNA was transfected into HEK293A at a series of transfection concentrations of 1.0000, 0.5000, 0.2500, 0.1250, 0.0625, 0.0313, 0.0156, 0.0078, 0.0039, 0.0020, and 0.0010 nM. The activity of both luciferases was determined 24 hr after transfection by a Synergy HT fluorometer. Cells were lysed with Passive Lysis Buffer (Promega). Firefly and renilla luciferase activities were evaluated with the Dual-Luciferase Reporter Assay system (Promega). Renilla luciferase activity was normalized to the firefly luciferase activity, in contrast to the siQuant system. Then siRNA activity was calculated by comparison with the NC sample. Then the fitting curve was generated; the IC50 was calculated with GraphPad Prism 5.0 software.
Validation of the siRNA’s Activity in PANC-1 with RT-PCR
To validate the silencing efficiency of siRNAs targeting RRM2, qRT-PCR was performed in PANC-1. Cells were plated in 12-well plates (1 × 105 per well) 1 day before transfection and incubated in DMEM to approximately 60% confluence. Then siRNAs were transfected into the cells with lipofectamine 2000 at the concentrations of 20 and 50 nM. 24 hr later, total RNA of the cells was harvested with Trizol according to the manufacturer’s protocol. Then, cDNA was prepared by two-step reverse transcription. First, 1 μg total RNA was incubated for 10 min at 70°C, followed by immediately transferring onto ice. Then 2 μL 10× reaction buffer; 4 μL MgCl2 (25 mM); 2 μL dNTP (10 mM); 0.5 μL RNase inhibitor; and 1 μL (15 U) AMV reverse transcription enzyme, 1 μL oligo dT, and the desired volume of ddH2O were added into the precooled tube (20 μL in total), followed by a reaction at 42°C for 30 min and 95°C for 5 min, respectively. Then the cDNA solution was diluted 5 times by adding 80 μL ddH2O. A real-time PCR reaction system (10 μL 2× UltraSYBR Mixture, 0.5 μL forward primer [10 μM], 0.5 μL reverse primer [10 μM], 5 μL cDNA solution, and 4 μL ddH2O) was prepared and first hot-started for 10 min at 95°C before 40 cycles of 30 s at 95°C, 30 s at 59°C, and 30 s at 72°C. After the melting procedure was completed, the samples were stored at 4°C. The expression level of RRM2 was analyzed by the Ct (cycle threshold) values using standard protocol.
Colony Formation Assay
Colony formation assay was employed to evaluate the influence of siRRM2 on the proliferation of PANC-1. PANC-1 cells were plated in 6-well plates (300 cells/well in 2 mL DMEM) 1 day before transfection. siRRM2 and irrelevant siRNA (siNC) were added into each well at the transfection concentration of 50 nM. Triplicates were used in this assay. After incubation at 37°C for 4 hr, 4 mL fresh DMEM containing 10% FBS was added. Then the medium was replaced once every 3 days with 4 mL fresh DMEM containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. 10 days after transfection, the colonies were washed twice with 1× PBS, followed by fixing and staining for 30 min with a solution containing 0.1% (w/v) crystal violet and 20% (v/v) ethanol. Then the colonies were washed again to remove excess crystal violet with 1× PBS. Finally, after the dishes were dry, digital images of the colonies were acquired using a camera, and colony numbers were counted.
FACS Assay for Cell-Cycle Arrest
Fluorescence-activated cell sorting (FACS) was used to examine the cell cycle of the cells treated with various formulations. PANC-1 cells were plated in 6-well plates (2 × 105 per well) 1 day before transfection, and they were cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin to 60%–70% confluence. After replacing DMEM with Opti-MEM, a reduced serum medium, siRRM2 (25 or 50 nM) and irrelevant siRNA (siNC, 50 nM) were transfected into each well at the desired concentrations with lipofectamine 2000. DOX and Pt were included as positive controls. The transfection concentrations of doxorubicin and Pt were 0.2 and 25 μg/mL, respectively. After incubation at 37°C for 4 hr, 2 mL fresh DMEM containing 10% FBS was added, followed by further incubation for 68 hr. Then the cells were washed once with ice-cold 1× PBS and re-suspended in 0.5 mL 1× PBS. Then 3.0 mL 70% cold EtOH was added into the cells, followed by fixing the cells at −20°C for 3 hr. Subsequently, the cells were centrifuged at 2,000 rpm/min for 5 min and washed once with 5 mL 1× PBS containing 1% FBS (50 μL). Then the cells were centrifuged at 2,000 rpm/min for 5 min and re-suspended in 200 μL PBS and 6 μL RNase (20 mg/mL), and 20 μL of a solution containing 500 μg/mL PI and 10% Triton X-100 was added. The mixture was gently blended, kept in a dark place, and incubated at 37°C for 30 min. Finally, the cells were centrifuged at 2,000 rpm/min for 5 min, and the precipitates were re-suspended in 600 μL PBS before proceeding to FACS assay. The cell cycle profiles were analyzed using a BD Biosciences FACScan (Becton Dickinson, San Jose, CA, USA) with MODFIT software. At least 10,000 cells in each sample were analyzed to obtain a measurable signal. All measurements were performed using the same instrument setting.
Influence of siRRM2 on Cellular Morphology
To investigate the influence of siRNA targeting RRM2 (siRRM2) on the cellular morphology of PANC-1, cells were planted in 6-well plates (2 × 105 per well) 1 day before transfection. siRRM2 was transfected into the cell with lipofectamine 2000 at the concentration of 50 nM. Doxorubicin (0.2 μg/mL) and Pt (25 μg/mL) were used as positive controls. In addition, cells treated with doxorubicin (0.2 μg/mL) combined with siRRM2 (50 nM) or siNC (50 nM) were included as an additional group. 72 hr later, the cells were observed using an inverted fluorescence microscope (Olympus X71, Olympus, Tokyo, Japan), and digital images were acquired and analyzed with the software package provided by Olympus.
MTT Assay
An MTT assay was employed to evaluate the synergistic effect of siRRM2 and DOX in PACN-1 cells. The cells were seeded at 4,000 cells/100 μL DMEM/well on a 96-well plate, and they were allowed to adhere overnight at 37°C. First, we explored the anti-proliferation efficacy of treatment with DOX alone. The concentrations of DOX were 0.0, 0.1, 0.2, 0.4, 0.8, 1.6, and 3.2 ng/μL, respectively. Second, we evaluated the synergistic effect of combination treatment with siRRM2 and DOX. Here, the concentration of DOX was fixed at 0.2 ng/μL for panel 1; siRRM2 was co-transfected at 50, 10, and 2 nM, respectively. DOX alone (0.2 ng/μL) and DOX (0.2 ng/μL) plus siNC (50 nM) were included as controls. For panel 2, DOX was fixed at 0.1 ng/μL; siRRM2 was co-added into the cells at 50 and 10 nM, respectively. Meanwhile, cells treated with siRRM2 alone (50 and 10 nM) were also explored. 4 hr after transfection, 200 μL fresh complete DMEM was added. 68 hr after transfection, 200 μL medium was removed, and 10 μL MTT (5 mg/mL) was added into each well, followed by another incubation of 4 hr under the same incubator conditions. Then, the entire medium was removed and 150 μL DMSO was added and incubated for 10 min at 37°C to dissolve formazan. Finally, the absorbance was read at 545 nm, and the absolute absorbance (ODnet545) was generated as OD545 − OD545woc. OD545woc represents the average value of OD545 collected from six wells with the same solutions to the experimental plates but without any cells.
To compare the relative viability, all the data were presented as the mean percentage ± SD of six or five replicates compared with the value of mock-treated cells. The cell viability was calculated as follows: cell viability (%) = (OD545(sample)/OD545(control)) × 100, where OD545(sample) is the absorbance at 545 nm of the transfected cells and OD545(control) is the absorbance at 545 nm of the mock control (nontransfected cells). IC50 (the half maximal inhibitory concentration) of DOX was calculated according to a fitting curve of cell viability. The fitting curve was generated with GraphPad Prism software according to the function of Y = Bottom + (Top − Bottom)/(1 + 10ˆ((X − LogIC50))), whose R2 (coefficient of determination) was ∼0.95.
In addition, MTT was also employed to evaluate the cytotoxicity of naked siRNA. Here, siRNA was applied to the cell at the concentrations of 1,000, 500, 250, 50, 10, 2, and 0.4 nM, respectively, without transfection reagent. Cell viability was determined at 24 hr post-treatment according to the above-mentioned protocol.
Tumor Growth Suppression In Vivo
Animals were purchased from Vital River Laboratory Animal Technology and maintained in Peking University Laboratory Animal Center (an AAALAC-accredited and specific-pathogen-free [SPF] experimental animal facility). All procedures involving experimental animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Peking University.
Male BALB/c nude mice, 6–8 weeks old and weighing 18–22 g, were used to evaluate the anti-tumor effect of siRNA-loaded LNPs combined with DOX. First, 5 × 106 PANC-1 cells (in 100 μL) were injected subcutaneously into the right axillary fossa of the mice. The tumor volume was monitored by measuring the perpendicular diameter of the tumor using calipers, and it was calculated according to the following formula: tumor volume (mm3) = 0.5 × length × width.2 When the volumes of the tumors had increased to ∼50 mm3, mice were randomly divided into five groups (eight mice per group), followed by administration of the following formulations, respectively: (1) normal saline, (2) DOX (1.0 mg/kg), (3) DOX (1.0 mg/kg) combined with LNP/siNC (5 μg/tumor), (4) DOX (1.0 mg/kg) combined with LNP/siRRM2 (2 μg/tumor), and (5) DOX (1.0 mg/kg) combined with LNP/siRRM2 (5 μg/tumor). DOX and LNP/siRNA complexes were administered twice weekly via i.p. and peritumoral injections, respectively. 25 days later, mice were sacrificed by cervical dislocation, the tumors were isolated, and the digital images were acquired. The weights of tumor, liver, and spleen were also recorded.
The LNP/siRNA complex was prepared by Suzhou Ribo Life Science (Kunshan, China). The proposed LNP is a novel lipid-based breakthrough reagent for in vivo RNAi delivery, with greatly improved performance and up to 85% knockdown achieved using microgram levels of siRNA (data not shown; Y.H., unpublished data). It is composed of a proprietary ionizable amino lipid, PEGylated lipid, and cholesterol. The average particle size of LNP/siRNA, as determined by dynamic light scattering (ZEN3500, Malvern Instruments, UK), was approximately 80 nm. Encapsulation efficiency was calculated by determining unencapsulated siRNA content by measuring the fluorescence upon the addition of RiboGreen (Molecular Probes, Eugene, OR) to the LNP slurry (Fi) and comparing this value to the total siRNA content obtained upon lysis of the LNPs by 1% Triton X-100 (Ft), where percentage encapsulation = (Ft − Fi)/Ft × 100. As a result, the encapsulation efficiency of LNP/siRNA was higher than 90%.
Statistical Analysis
The data were expressed as the mean ± SD or mean ± SEM, as indicated in the figure legends. The statistical variance was calculated by a t test; p < 0.05 was considered statistically significant.
Author Contributions
Y.H., X.J., and S.G. designed and led the research. Y.H., X.J., S.Z., X.W., Y.-H.W., J.-L.J., L.G., B.H., N.L., H.B., J.Z., and T.Y. performed the research. L.G., S.Z., H.B., J.Z., and Q.C. were involved in the animal study. X.-H.X. was involved in discussion of the data and provided insightful suggestions. Y.H., X.J., S.G., and H.-Y.Z. analyzed the data. Y.H. summarized the data, prepared the figures, and wrote the paper. All authors discussed results and commented on the paper.
Conflicts of Interest
X.J., J.-L.J., L.G., B.H., N.L., J.Z., H.B., S.G., and H.-Y.Z. are employees of Suzhou Ribo Life Science Co. Ltd.
Acknowledgments
This work was supported by the Beijing Institute of Technology Research Fund Program for Young Scholars and the Fundamental Research Funds for the Central Universities, the Hunan Provincial Natural Science Foundation of China (2018JJ1019), the Hu-Xiang Young Talent Program, the National Drug Program of China (2015ZX09102-023-002 and 2014ZX09304313-001), and the Jiangsu Provincial Natural Science Foundation of China (BK2011381). It was also financially supported by Suzhou Ribo Life Science Co. Ltd.
Footnotes
Supplemental Information includes Supplemental Materials and Methods and six figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.08.003.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Garrido-Laguna I., Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 3.Ryan D.P., Hong T.S., Bardeesy N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 4.Wolfgang C.L., Herman J.M., Laheru D.A., Klein A.P., Erdek M.A., Fishman E.K., Hruban R.H. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 6.Hidalgo M. Pancreatic cancer. N. Engl. J. Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 7.Bobbin M.L., Rossi J.J. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y. Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics. Mol. Ther. Nucleic Acids. 2017;6:116–132. doi: 10.1016/j.omtn.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-Targeted Therapeutics. Cell Metab. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y.Y., Liang Z.C. Asialoglycoprotein Receptor and Its Application in Liver-targeted Drug Delivery. Prog. Biochem. Biophys. 2015;42:501–510. [Google Scholar]
- 11.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barata P., Sood A.K., Hong D.S. RNA-targeted therapeutics in cancer clinical trials: Current status and future directions. Cancer Treat. Rev. 2016;50:35–47. doi: 10.1016/j.ctrv.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Yhee J.Y., Song S., Lee S.J., Park S.G., Kim K.S., Kim M.G., Son S., Koo H., Kwon I.C., Jeong J.H. Cancer-targeted MDR-1 siRNA delivery using self-cross-linked glycol chitosan nanoparticles to overcome drug resistance. J. Control. Release. 2015;198:1–9. doi: 10.1016/j.jconrel.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Lin G., Zhu W., Yang L., Wu J., Lin B., Xu Y., Cheng Z., Xia C., Gong Q., Song B., Ai H. Delivery of siRNA by MRI-visible nanovehicles to overcome drug resistance in MCF-7/ADR human breast cancer cells. Biomaterials. 2014;35:9495–9507. doi: 10.1016/j.biomaterials.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 15.Shen J., Wang Q., Hu Q., Li Y., Tang G., Chu P.K. Restoration of chemosensitivity by multifunctional micelles mediated by P-gp siRNA to reverse MDR. Biomaterials. 2014;35:8621–8634. doi: 10.1016/j.biomaterials.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Deng Z.J., Morton S.W., Ben-Akiva E., Dreaden E.C., Shopsowitz K.E., Hammond P.T. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano. 2013;7:9571–9584. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura K., Abu Lila A.S., Matsunaga M., Doi Y., Ishida T., Kiwada H. A double-modulation strategy in cancer treatment with a chemotherapeutic agent and siRNA. Mol. Ther. 2011;19:2040–2047. doi: 10.1038/mt.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis S.M., Taylor C.A., Tang T., Liu Z., Zheng Q., Dondero R., Thompson J.E. SNS01-T modulation of eIF5A inhibits B-cell cancer progression and synergizes with bortezomib and lenalidomide. Mol. Ther. 2014;22:1643–1652. doi: 10.1038/mt.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elledge S.J., Zhou Z., Allen J.B. Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem. Sci. 1992;17:119–123. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- 20.Engström Y., Eriksson S., Jildevik I., Skog S., Thelander L., Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J. Biol. Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 21.Nordlund P., Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y.W., Jones T.L., Martin S.E., Caplen N.J., Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J. Biol. Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuta E., Okuda H., Kobayashi A., Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim. Biophys. Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duxbury M.S., Ito H., Zinner M.J., Ashley S.W., Whang E.E. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- 25.Zhang K., Hu S., Wu J., Chen L., Lu J., Wang X., Liu X., Zhou B., Yen Y. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol. Cancer. 2009;8:11. doi: 10.1186/1476-4598-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa T., Maeda D., Kume H., Homma Y., Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57:885–892. doi: 10.1111/j.1365-2559.2010.03725.x. [DOI] [PubMed] [Google Scholar]
- 27.Cerqueira N.M., Pereira S., Fernandes P.A., Ramos M.J. Overview of ribonucleotide reductase inhibitors: an appealing target in anti-tumour therapy. Curr. Med. Chem. 2005;12:1283–1294. doi: 10.2174/0929867054020981. [DOI] [PubMed] [Google Scholar]
- 28.Davis M.E., Zuckerman J.E., Choi C.H., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman M.A., Amin A.R., Wang X., Zuckerman J.E., Choi C.H., Zhou B., Wang D., Nannapaneni S., Koenig L., Chen Z. Systemic delivery of siRNA nanoparticles targeting RRM2 suppresses head and neck tumor growth. J. Control. Release. 2012;159:384–392. doi: 10.1016/j.jconrel.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto K., Nakashiro K., Tanaka H., Tokuzen N., Hamakawa H. Ribonucleotide reductase M2 is a promising molecular target for the treatment of oral squamous cell carcinoma. Int. J. Oncol. 2015;46:1971–1977. doi: 10.3892/ijo.2015.2912. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M., Wang J., Yao R., Wang L. Small interfering RNA (siRNA)-mediated silencing of the M2 subunit of ribonucleotide reductase: a novel therapeutic strategy in ovarian cancer. Int. J. Gynecol. Cancer. 2013;23:659–666. doi: 10.1097/IGC.0b013e318287e2b3. [DOI] [PubMed] [Google Scholar]
- 32.Kang W., Tong J.H., Chan A.W., Zhao J., Wang S., Dong Y., Sin F.M., Yeung S., Cheng A.S., Yu J., To K. Targeting ribonucleotide reductase M2 subunit by small interfering RNA exerts anti-oncogenic effects in gastric adenocarcinoma. Oncol. Rep. 2014;31:2579–2586. doi: 10.3892/or.2014.3148. [DOI] [PubMed] [Google Scholar]
- 33.Gao J., Chen H., Yu Y., Song J., Song H., Su X., Li W., Tong X., Qian W., Wang H. Inhibition of hepatocellular carcinoma growth using immunoliposomes for co-delivery of adriamycin and ribonucleotide reductase M2 siRNA. Biomaterials. 2013;34:10084–10098. doi: 10.1016/j.biomaterials.2013.08.088. [DOI] [PubMed] [Google Scholar]
- 34.Liu X., Zhang H., Lai L., Wang X., Loera S., Xue L., He H., Zhang K., Hu S., Huang Y. Ribonucleotide reductase small subunit M2 serves as a prognostic biomarker and predicts poor survival of colorectal cancers. Clin. Sci. (Lond.) 2013;124:567–578. doi: 10.1042/CS20120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Q., Thonberg H., Zhang H.Y., Wahlestedt C., Liang Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem. Biophys. Res. Commun. 2004;325:243–249. doi: 10.1016/j.bbrc.2004.09.222. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, H., Gao, S., and Huang, Y. (September 2017). Small interfering RNA, pharmaceutical composition, and application thereof. World Intellectual Property Organization (WIPO) patent WO/2017/036398.
- 37.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 38.Song X., Wang X., Ma Y., Liang Z., Yang Z., Cao H. Site-Specific Modification Using the 2′-Methoxyethyl Group Improves the Specificity and Activity of siRNAs. Mol. Ther. Nucleic Acids. 2017;9:242–250. doi: 10.1016/j.omtn.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018;9:723. doi: 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddik Z.H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 41.Sorenson C.M., Eastman A. Mechanism of cis-diamminedichloroplatinum(II)-induced cytotoxicity: role of G2 arrest and DNA double-strand breaks. Cancer Res. 1988;48:4484–4488. [PubMed] [Google Scholar]
- 42.Zimmermann M., Arachchige-Don A.S., Donaldson M.S., Dallapiazza R.F., Cowan C.E., Horne M.C. Elevated cyclin G2 expression intersects with DNA damage checkpoint signaling and is required for a potent G2/M checkpoint arrest response to doxorubicin. J. Biol. Chem. 2012;287:22838–22853. doi: 10.1074/jbc.M112.376855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Y.P., Liao J.Z., Lu Y.Q., Tian D.A., Ye F., Zhao P.X., Xiang G.Y., Tang W.X., He X.X. MiR-375 and Doxorubicin Co-delivered by Liposomes for Combination Therapy of Hepatocellular Carcinoma. Mol. Ther. Nucleic Acids. 2017;7:181–189. doi: 10.1016/j.omtn.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porciani D., Tedeschi L., Marchetti L., Citti L., Piazza V., Beltram F., Signore G. Aptamer-Mediated Codelivery of Doxorubicin and NF-κB Decoy Enhances Chemosensitivity of Pancreatic Tumor Cells. Mol. Ther. Nucleic Acids. 2015;4:e235. doi: 10.1038/mtna.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuckerman J.E., Hsueh T., Koya R.C., Davis M.E., Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J. Invest. Dermatol. 2011;131:453–460. doi: 10.1038/jid.2010.310. [DOI] [PubMed] [Google Scholar]
- 46.Lee S., Oh S.Y., Kim B.G., Kwon H.C., Kim S.H., Rho M.H., Kim Y.H., Rho M.S., Jeong J.S., Kim H.J. Second-line treatment with a combination of continuous 5-fluorouracil, doxorubicin, and mitomycin-C (conti-FAM) in gemcitabine-pretreated pancreatic and biliary tract cancer. Am. J. Clin. Oncol. 2009;32:348–352. doi: 10.1097/COC.0b013e31818c08ff. [DOI] [PubMed] [Google Scholar]
- 47.Yin F., Yang C., Wang Q., Zeng S., Hu R., Lin G., Tian J., Hu S., Lan R.F., Yoon H.S. A Light-Driven Therapy of Pancreatic Adenocarcinoma Using Gold Nanorods-Based Nanocarriers for Co-Delivery of Doxorubicin and siRNA. Theranostics. 2015;5:818–833. doi: 10.7150/thno.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin F., Hu K., Chen Y., Yu M., Wang D., Wang Q., Yong K.T., Lu F., Liang Y., Li Z. SiRNA Delivery with PEGylated Graphene Oxide Nanosheets for Combined Photothermal and Genetherapy for Pancreatic Cancer. Theranostics. 2017;7:1133–1148. doi: 10.7150/thno.17841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin L., Fan Y., Gao F., Jin L., Li D., Sun W., Li F., Qin P., Shi Q., Shi X., Du L. UTMD-Promoted Co-Delivery of Gemcitabine and miR-21 Inhibitor by Dendrimer-Entrapped Gold Nanoparticles for Pancreatic Cancer Therapy. Theranostics. 2018;8:1923–1939. doi: 10.7150/thno.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han X., Yang F., Cao H., Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054–3064. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.