Summary
We demonstrate that GRHL2 plays pivotal roles in the maintenance of tumorigenicity, cancer stemness and epithelial phenotype of oral cancer cells, in which miR-200 family genes partially mediate the effect of GRHL2 on oral squamous cell carcinomas formation.
Abstract
Grainyhead-like 2 (GRHL2) is one of the three mammalian homologues of Drosophila Grainyhead involved in epithelial morphogenesis. We recently showed that GRHL2 also controls normal epithelial cell proliferation and differentiation. In this study, we investigated the role of GRHL2 in oral carcinogenesis and the underlying mechanism. GRHL2 expression was elevated in cells and tissues of oral squamous cell carcinomas (OSCCs) compared with normal counterparts. Knockdown of GRHL2 resulted in the loss of in vivo tumorigenicity, cancer stemness and epithelial phenotype of oral cancer cells. GRHL2 loss also inhibited oral cancer cell proliferation and colony formation. GRHL2 regulated the expression of miR-200 family and Octamer-binding transcription factor 4 (Oct-4) genes through direct promoter DNA binding. Overexpression of miR-200 genes in the oral cancer cells depleted of GRHL2 partially restored the epithelial phenotype, proliferative rate and cancer stemness, indicating that miR-200 genes in part mediate the functional effects of GRHL2. Taken together, this study demonstrates a novel connection between GRHL2 and miR-200, and supports protumorigenic effect of GRHL2 on OSCCs.
Introduction
Grainyhead-like 2 (GRHL2) is one of the three known mammalian homologues of Drosophila Grainyhead (GRH), along with GRHL1 and GRHL3, which are involved in epithelial morphogenesis and barrier formation (1–4). In addition, we have demonstrated that GRHL2 plays a unique role in control of cellular proliferation and differentiation through transcriptional regulation of its target genes, including hTERT, PCNA and epidermal differentiation complex genes (5,6). GRHL2 enhances the replicative potential of normal human keratinocytes in part by delaying the loss of telomerase activity during the replication period (5). Furthermore, GRHL2 overexpression interferes with normal keratinocyte differentiation by suppressing the expression of the epidermal differentiation complex genes, e.g. involucrin, filaggrin and loricrin genes (6). Given these biological functions, it was presumed that aberrant overexpression of GRHL2 would lead to epithelial hyperplasia with inadequate terminal differentiation. In fact, expression of GRHL2 is enhanced in epithelial hyperproliferative lesions, such as psoriasis and atopic dermatitis, which also present with abnormal keratinocyte differentiation (6).
Our earlier study identified GRHL2 as a transcription factor regulating the hTERT gene expression in oral squamous cell carcinoma (OSCC) cells, indicating its possible connection with the development of human oral cancer (7). Cancer-related role of GRHL2 has been reported by several studies. GRHL2 overexpression in NIH3T3 cells leads to tumorigenic conversion of cells (8); GRHL2 gene amplification is linked with recurrence of tumors in hepatocellular carcinomas (9); GRHL2 inhibits death-receptor triggered apoptotic signal in cancer cells (10); GRHL2 expression is associated with poor relapse-free survival and increased metastatic potential in breast cancers (11). On the contrary, there are also reports suggesting tumor suppressive effects of GRHL2. Its expression level declines in gastric cancers; GRHL2 inhibits epithelial–mesenchymal transition (EMT), which is generally viewed as a mechanism to promote tumor metastasis and malignancy (12). GRHL2 expression is suppressed in tumors exhibiting mesenchymal phenotype, and GRHL2 overexpression leads to mesenchymal–epithelial transition (MET) through inhibition of ZEB1 (13,14). Thus, involvement of GRHL2 in cancer is controversial in part due to variable levels of GRHL2 expression associated with varying cancers and due to its inhibitory role in EMT, which is linked with malignant cancer.
Head and neck squamous cell carcinoma, including OSCC, is one of the leading causes of cancer-related death worldwide. Local progression and lymph node involvement are the major causes of OSCC-related mortality and the incidence of distal organ metastasis is relatively rare compared with other cancers (15). Cells from OSCC demonstrate epithelial phenotype. However, the mechanism responsible for the local progression of OSCC is unclear. GRHL2 is found to be an epithelial-specific transcriptional factor and can induce keratinocyte proliferation. In this study, we further explore the novel effect of GRHL2 on the induction of the epithelial plasticity and stemness characteristics, which results in restricting the dissemination but promoting the local progression of this cancer.
Materials and methods
Cells and cell culture
Primary normal human oral keratinocytes (NHOK) were established from oral epithelium of normal buccal mucosa or gingiva obtained from nine healthy patients. Normal human oral fibroblasts (NHOF) were established from the explant cultures of the gingival connective tissues. Normal human epidermal keratinocytes (NHEK) were isolated from neonatal foreskin tissues. Detailed methods for primary cell culture establishment and maintenance can be found elsewhere (16). NHOK cultures were maintained at subconfluence levels to prevent terminal differentiation, as reported in our previous study (17). Dental pulp stem cells were obtained from healthy dental pulp of extracted teeth and cultured in α-MEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 20mM l-glutamine (Invitrogen). HOK-16B cells were cultured in Keratinocyte Growth Medium (KGM) (Lonza, Allendale, NJ) (18). The SCC4, SCC9 and SCC15 cancer cell lines were purchased from the ATCC (Manassas, VA) and the UM-SCC cell lines were kindly provided by Dr. T. Carey, University of Michigan, Ann Arbor. HOK-16B-BaP-T, SCC4, SCC9 and SCC15 cancer cell lines were cultured in DMEM/Ham’s F-12 (Invitrogen) supplemented with 10% FBS and 0.4 µg/ml hydrocortisone, whereas the UM-SCC cell lines were cultured in DMEM supplemented with 10% FBS.
To overexpress GRHL2 in cells, we utilized a retroviral vector (LXSN-GRHL2) including human GRHL2 cDNA sequence or its control vector (LXSN) according to methods described elsewhere (5,19). Endogenous GRHL2 in SCC4 and SCC15 cells was knocked down using the lentiviral vector (LV-ShGRHL2) expressing short hairpin RNA (shRNA) against the GRHL2 target sequence (7), or independent GRHL2 shRNA lentiviral virus (denoted LV2-ShGRHL2) from a commercial source (sc-77606-V, Santa Cruz Biotech). Stable knockdown of endogenous GRHL2 were achieved through single-cell culture selection assay or puromycin selection (1 µg/ml) for 2 weeks. To overexpress the miR-200 genes, genomic fragments encoding cluster 1 (miR-200b, -200a and -429) and cluster 2 (miR-200c and -141) were cloned into pMSCV-puro using the primers from Korpal et al. (20). p19 embryonic carcinoma cell line cell was purchased from ATCC (Manassas, VA) and grown in α-MEM supplemented with 7.5% bovine serum and 2.5% FBS. Retinoic acid-induced differentiation was performed according to the method described by Gill et al. (21). Mouse embryonic stem cells were cultured on gelatinized tissue culture dishes in mouse embryonic stem cells growth medium containing 1000 units/ml LIF. For embryoid body formation, cells were collected by trypsinization, washed and resuspended in mouse embryonic stem cells growth medium without LIF (22).
All cell lines were tested for mycoplasma, using the MycoAlert Detection Kit (Cambrex, Rockland, ME). After establishing the culture, the cells were regularly monitored for their authenticity by phenotypic assessment, e.g. morphology, growth kinetics and intercellular growth pattern, as well as molecular markers, including those of cellular differentiation and proliferation, senescence and immortalization.
Tumor spheroid assay
SCC4/ShGRHL2 and SCC4/EGFP cells were suspended in defined serum-free medium composed of DMEM⁄ F12 (Invitrogen), N2 supplement (Invitrogen), 10ng/ml human recombinant basic fibroblast growth factor (bFGF) and 10ng/ml epidermal growth factor (EGF) (R&D Systems, Minneapolis, MN) at 1000 cells/ml and seeded in six-well low attachment plate (Costar, NY, NY). Culture medium was replenished once every 3 days. Suspended tumor spheroids could be visible within the first week after seeding and were dissociated by mechanical trituration and Accutase® solution (Sigma, St Louis, MO), followed by replating cells with the same number in fresh spheroid medium. The spheroids were collected and re-suspended to generate secondary spheroids. All experiments were conducted using triplicate samples for statistical validation.
qRT-PCR
Total RNA was isolated from the cultured cells using the RNeasy Mini kit (Qiagen, Valencia, CA). DNA-free total RNA (5 μg) was used for reverse transcription (RT) reaction followed by qPCR with LC480 SYBR Green I master using universal cycling conditions in LightCycler® 480 (Roche, South San Francisco, CA). The primer sequences were obtained from the Universal Probe Library (Roche). The PCR cycling conditions were 45 cycles of 10s at 95°C, 45s at 55°C and 20s at 72°C. Second derivative Cq value determination method was used to compare the fold differences. Cp is the cycle at which the threshold is crossed. For analysis of miRNA expression level, qPCR was performed as above using TaqMan microRNA assays according to the manufacturer’s instructions (Applied Biosystems, Grand Island, NY). All miRNA expression levels are expressed relative to the U6 small nuclear (sn) RNA expression level performed on the same sample. Experiments were performed in triplicate samples.
Western blotting
Whole cell extracts from the cultured cells were isolated using lysis buffer (1% Triton X-100, 20mM Tris–HCl pH 7.5, 150mM NaCl, 1mM EDTA, 1mM EGTA, 2.5mM sodium pyrophosphate, 1 μM β-glycerophosphate, 1mM sodium orthovanadate and 1mg/ml PMSF). Whole cell extracts were then fractionated by SDS-PAGE and transferred to Immobilon protein membrane (Millipore, Billerica, MA). Membranes were incubated successively with the primary and the secondary antibodies, and exposed to the chemiluminescence reagent (Amersham Pharmacia Biotech, Piscataway, NJ) for signal detection.
Gene promoter luciferase assay
To assess whether GRHL2 regulates miR-200 gene promoters, we performed a bioinformatic search for potential promoter regions of miR-200 family using the University of California, Santa Cruz (UCSC) table browser http://genome.ucsc.edu/cgi-bin/hgTables(23). We amplified the miR-200b/a/429 and miR-200c/141 promoter regions from genomic DNAs of SCC4 by PCR amplification and cloned the promoter fragments in pGL3B-Luc reporter plasmid (Promega), expressing firefly luciferase. Primers for the promoter of miR-200b/a/429 (chr1: 1141407-1142388) are as follows: Forward, 5′-GGTGGAAGGTGCCAGAAAAC-3′ and Reverse, 5-TGAGGGTTGCATGGGACT-3′; primers for the promoter of miR-200c/141 (chr12: 6942123-6943102) are: Forward, 5′-CAGCTGGGGTCTCTGGGTAG-3′ and Reverse, 5′-CTTGGGTCAGGCAGCTTCA-3′. The promoter-luciferase constructs were transfected into SCC4 derivatives using Lipofectin® Reagent (Invitrogen), along with pRL-SV40 containing Renilla luciferase cDNA under the control of SV40 enhancer/promoter. Cells were collected after 48-h post-transfection, and the lysates were prepared using dual Luciferase Reporter Assay System (Promega). Firefly and Renilla luciferase activities were measured using a luminometer (Turner Designs, Sunnyvale, CA). Renilla luciferase activity was used as control for the varied transfected efficiency. All experiments were conducted in triplicate.
Chromatin immunoprecipitation assay
We performed chromatin immunoprecipitation (ChIP) assay based on the protocols published in our previous paper (6). The sequences for the PCR primers of gene promoters are as follows, miR-200b (−231 to −34): 5′-CCCACTCCGACCTAGTCCTC-3′ (Forward) and 5′-ACTCG CTGGGAAGCTCAGTA -3′ (Reverse); miR-200c (−190 to −19): 5′- CTGCTTG GACTGCAACCTGG- 3′ (Forward) and 5′- ACCTTGGGTCAGGCAGCTTC-3′ (Reverse); hTERT (−53 to 48): 5′- CCCCTCCCCTTCCTTTCCGC-3′ (Forward) and 5′- GTGGCCGGGGCCAGGGCTT -3′ (Reverse). The promoter regions analyzed in this study for GRHL2 enrichment included the proximal regions near the transcription start sites, which are frequently bound by GRHL2 for regulation of the gene expression (6). All experiments were repeated in triplicate.
Colony formation assay and anchorage-independent growth assay
Cells were plated at low density (200 cells per well in six-well plates) and cultured for 14 days. Cell clones were stained with 0.5% crystal violet to reveal colony forming efficiency and size of each colony. To demonstrate tumorigenic ability in vitro, each well of a six-well culture plates was coated with 2ml of culture medium in soft agar (DMEM/F12, 10% FBS, 0.6% agar). After the bottom layer was solidified, 2ml of top agar–medium mixture (DMEM/F12, 10% FBS, 0.3% agar) containing 1×104 cells was added, and the cultures were incubated with 95% air, 5% CO2 and 100% humidity at 37°C for 2 weeks. Tumor colonies were visualized after staining in 0.005% crystal violet (Sigma-Aldrich). All experiments were conducted in triplicate.
Cell migration/invasion assay
Cells were seeded in the upper chamber of Matrigel-coated 24-well transwell inserts with 8 µm pores (Corning, NY). The bottom chambers were filled with the culture medium (DMEM/F12 with 10% FBS). After 48h, non-invaded cells on the upper surface of the membrane were mechanically removed with a cotton swab, and the invasive cells on the lower membrane surface were fixed with methanol and stained with 1% crystal violet. All experiments were done in triplicate.
Immunohistochemistry
In situ expression of GRHL2 and Keratin1 (K1) were determined in oral mucosal tissue specimens by immunohistochemistry (IHC). Normal human oral mucosa (NHOM) (n = 10) was obtained from UCLA School of Dentistry (Los Angeles, CA); Specimens diagnosed as precancerous lesions (n = 12) or OSCC (n = 26) were obtained from the Oral Pathology Diagnostic Laboratory, UCLA (Los Angeles, CA). IHC was performed with appropriated primary antibodies on a 4-μm thick sections according to the methods described elsewhere (24). GRHL2 staining intensity per each sample was scored by independent evaluators including one oral pathologist, as negative (–), weak (+), moderate (++), strong (+++) or very strong (++++).
Immunofluorescent staining
Cells were cultured in Nunc™ Lab-Tek™ II Chamber Slide™ System (Thermo Scientific, Rochester, NY) to reach 70–80% confluence, fixed in 2% paraformaldehyde for 20min. Cells were permeabilized with 0.2% Triton X-100 in phosphate-buffered saline for 10min, then blocked for 1h in phosphate-buffered saline containing 2% FBS, and incubated overnight at 4°C with primary antibody. After three washes with phosphate-buffered saline, cells were incubated with secondary antibody for 1h. Secondary antibodies used were anti-mouse Alexa 488 (green) or anti-rabbit Alexa 594 (red). Slides were mounted in Prolong Gold w/DAPI (Invitrogen). Images were taken on a Leica inverted microscope.
Tumor xenograft assay
SCC4/EGFP and SCC4/ShGRHL2 cells (5×106) were injected subcutaneously into 8-week-old immunocompromised (nu/nu) mice (Charles River Laboratories, San Diego, CA). Kinetics of tumor growth was monitored by measuring the tumor volume and plotted against days post-injection. After 1, 2, 3, 5 weeks post-injection, some tumors were excised for histological validation by haematoxylin and eosin (H&E) staining. Total of 40 immunocompromised mice were used (2 groups × 4 time points × 5 mice each group). All animal experiments were conducted according to the guidelines and approval of the UCLA Institutional Animal Care and Use Committee.
Laser-captured microdissection
Following histological examination of H & E staining of oral mucosal tissues [i.e. NHOM (n = 8), precancerous oral lesions (n = 5) and OSCC tissues (n = 9)], epithelial layers from the paraffin-embedded tissue samples were excised by laser-captured microdissection (LCM) using Leica (LMD) 7000 system (Leica Microsystems Inc, Richmond, IL) at the California NanoSystems Institute at UCLA (Los Angeles, CA). LCM-derived tissue RNAs were extracted using a high pure RNA paraffin kit (Roche). RT was performed with RNA isolated from the tissue sections using a Superscript II RT kit (Invitrogen) with random hexamer primers (Promega) according to the manufacturer’s instructions. Transcript expression was analyzed by qPCR with the LightCycler® 480 system (Roche). Thermocycling conditions for all PCR reactions included an initial denaturation stage at 95°C for 10min, followed by 50 cycles at 95°C for 15s and finally 60°C for 1min (25). All experiments were repeated in triplicate.
Antibodies
The following primary antibodies were used in this study: GAPDH, ZEB1, E-Cadherin (E-Cad) and p63 from Santa Cruz Biotech. (Santa Cruz, CA); GRHL2 (Abnova, Taiwan); Keratin 1 from Covance (Emeryville, CA); N-Cadherin (N-Cad) from BD Biosciences (San Jose, CA); Fibronectin from Sigma-Aldrich and Oct-4, Nanog and c-Myc from Cell Signaling Technology Inc. (Danvers, MA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Santa Cruz Biotech.
Statistical analysis
Statistical analysis was performed using the Student t test (two-tailed) for the qPCR gene expression and spheroid formation experiments. One-way analysis of variance (ANOVA) and post hoc t tests were applied to analyze the data from LCM experiment. P < 0.05 were considered to be significant. All data are expressed as mean ± SD.
Results
GRHL2 is involved in regulating tumorigenic ability of OSCCs
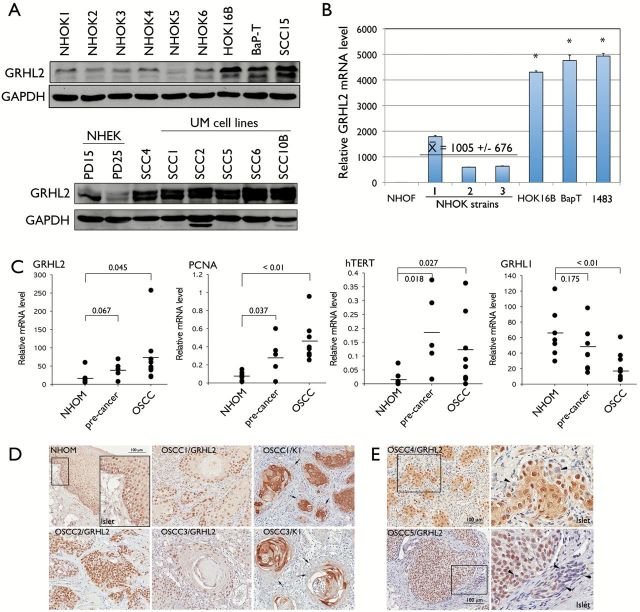
We surveyed the GRHL2 protein expression in rapidly proliferating primary oral keratinocyte cultures (NHOK and NHEK) and OSCC cell lines. Enhanced GRHL2 expression level was found in HPV-immortalized human oral keratinocytes (HOK-16B) and the OSCC cell lines (HOK-16B/BaP-T, SCC4, SCC15, 1483 and UM-OSCC cell lines) compared with those in NHOK and NHEK (Figure 1A and B). To quantify the expression levels of GRHL2 gene in OSCC tumors, GRHL2 mRNA expression was determined by qRT-PCR in oral epithelial tissues dissected from archived specimens by LCM. GRHL2 mRNA level was elevated in OSCCs compared with NHOM and preneoplastic oral epithelia (Figure 1C). We previously identified GRHL2 as a novel transcription regulator of hTERT gene through promoter-magnetic precipitation (PMP) (7). hTERT plays a pivotal role in OSCC formation (26). Expression of hTERT and proliferating cell nuclear antigen (PCNA), both of which are downstream targets of GRHL2 (6), were upregulated in OSCC compared with NHOM, while GRHL1, an isoform of GRHL2, showed an opposite pattern of expression in NHOM, precancer and OSCC, compared with that of GRHL2 (Figure 1C). GRHL2 protein level was also determined in situ in archived histologic specimens of NHOM and OSCCs by IHC. As previously reported (6), GRHL2 was predominantly expressed in the basal layer of NHOM. Its staining intensity was reduced in the differentiated suprabasal layer cells (Figure 1D). GRHL2 was also expressed in neoplastic and pre-neoplastic epithelia. However, higher levels of GRHL2 staining intensity (++++) were detected in OSCCs specimens (18 out of 26) compared with those in the normal and dysplastic samples examined (Supplementary Table S1, available at Carcinogenesis Online). In OSCCs, expression of GRHL2 was not homogenous among tumor cells (Figure 1E). Strong GRHL2 expression was detected in the proliferative cells at the outer border of the invasive islands and in the cells with undifferentiated, basaloid morphology. OSCC tissue specimens were derived from gingiva, buccal mucosa and tongue. Regardless of the intraoral site, GRHL2 staining intensity appears to be elevated in OSCC compared with the normal tissues. The staining patterns for GRHL2 and K1 were mutually exclusive in OSCC tissues, consistent with the negative regulation of K1 expression by GRHL2, as previously reported (6). These data indicate that GRHL2 expression in oral epithelial cells is associated with enhanced cell proliferation and undifferentiated status, and that aberrant GRHL2 overexpression may attribute to oral carcinogenesis.
Figure 1.
GRHL2 expression is enhanced in cells and tissues of OSCCs. (A) Western blotting was performed for GRHL2 using whole cell extracts from six different strains of NHOK, immortalized cells (HOK-16B) and OSCC cell lines (HOK-16B/BaP-T and SCC15). Another blot containing whole cell extracts from NHEK and OSCC cell lines, including SCC4 and those from UM-OSCCs were probed for GRHL2 protein levels. GAPDH was used as loading control. PD refers to population doublings. (B) qRT-PCR was performed with RNA extracted from NHOF, three strains of rapidly proliferating NHOK and the established cell lines (HOK-16B, HOK-16B/BaP-T and 1483). Bars indicate standard deviation and the asterisk (*) indicates statistical significance (P < 0.05), compared with the mean values of the control group (NHOK). (C) Epithelial tissues were excised from NHOM, preneoplastic oral lesions and OSCC tissues by laser-captured microdissection (LCM). mRNA levels of GRHL2, PCNA, hTERT and GRHL1 were determined from each sample by qRT-PCR. P values are shown in each group. (D) IHC was performed with NHOM, preneoplastic oral lesions and OSCC tissues using α-GRHL2 or α-K1 antibody. Representative staining results were shown for various OSCC samples (OSCC1–OSCC5). GRHL2 and K1 staining patterns appear to be mutually exclusive; tumor cells with strong GRHL2 appear to be completely lacking K1 staining (arrows). (E) In some tumors, GRHL2 expression pattern was found heterogeneous within the same tumor island; some cells exhibited strong GHRL2 staining while others completely lacked (arrowheads). Right-side panel shows higher magnification of the view field within the boxed area.
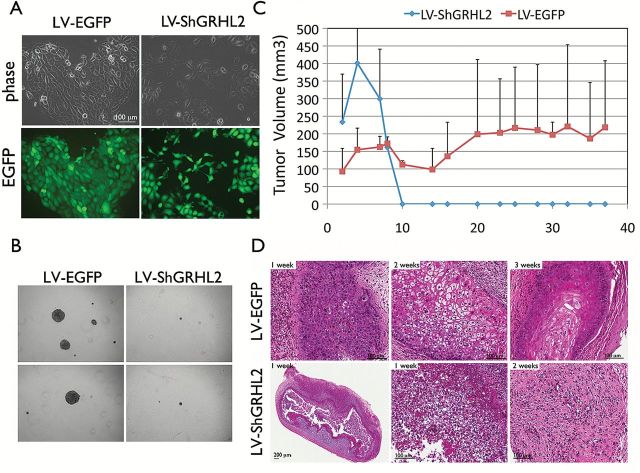
To explore the function of GRHL2 in the transformed phenotype of OSCC cells, we compared the change of phenotype and tumorigenic ability of SCC4 cells in which the endogenous GRHL2 was knocked down by lentiviral vector expressing shRNA against GRHL2 (SCC4/ShGRHL2) and its control cells infected with the control vector (SCC4/EGFP). In the monolayer culture, SCC4/ShGRHL2 cells demonstrated altered morphology, exhibiting mesenchymal spindle shape (Figure 2A). We also measured the longest diameter of cells in SCC4/EGFP and SCC4/ShGRHL2 to quantitate the difference in cell shape, and found that GRHL2 knockdown in cells led to elongation of cells, consistent with the morphological changes (Supplementary Figure S1A, available at Carcinogenesis Online). GRHL2 knockdown in SCC4 also led to reduced cell proliferation rate and colony formation abilities in monolayer culture (Supplementary Figure S1B and S3B, available at Carcinogenesis Online). SCC4/EGFP readily formed colonies in soft agar, reflecting anchorage-independent growth, while GRHL2 knockdown in SCC4 led to almost complete loss of anchorage-independent growth, suggesting that GRHL2 is important for the transformed phenotype in OSCCs in vitro (Figure 2B). Subcutaneous injection of SCC4/EGFP into immunocompromised (nu/nu) mice could induce palpable tumor nodule formation after 1 week post-injection, and the nodules steadily increased in volume over 5-week period (Figure 2C). However, GRHL2 knockdown led to complete loss of palpable tumor nodules within 10 days post-injection. Histology revealed the initially formed nodules by the SCC4/ShGRHL2 cells demonstrated massive cell death within 1 week post-injection and were not detectable after 10 days (Figure 2D). Hence, sustained GRHL2 expression is important for the maintenance of tumorigenic ability of OSCCs.
Figure 2.
GRHL2 is required for the maintenance of transformed phenotype of OSCC cells. (A) Epithelial morphology was lost in SCC4/ShGRHL2 cells compared with the control (SCC4/EGFP) cells. GRHL2 expression was knocked down using lentiviral vector (LV-ShGRHL2) in SCC4. As control, SCC4 was infected with lentiviral vector expressing EGFP (LV-EGFP). (B) Anchorage-independent growth assay was performed with SCC4/EGFP and SCC4/ShGRHL2 in soft agar plates. Growth of colonies in soft agar was visualized under light microscopy. Representative colony sizes are shown here after 14 days post-seeding. (C) GRHL2 is associated with the tumorigenicity of OSCCs in vivo. SCC4/EGFP or SCC4/ShGRHL2 cells were injected subcutaneously at 5×106 per injection in dorsal flank of immunocompromised (nu/nu) mice (n = 5 per group and per time point). Tumor volume was measured once every 2–3 days and plotted against days post-injection. (D) Tumor nodules were harvested at 1, 2 and 3 weeks post-injection and examined the histological change via H & E staining. SCC4/ShGRHL2 tumor regressed rapidly within 10 days post-injection while the control cells (SCC4/EGFP) developed well-differentiated solid tumor.
GRHL2 maintains the stem-like characteristics of OSCCs and upregulates Oct-4 pluripotency gene
In the next experiments, we sought to explain the mechanism underlying the role of GRHL2 in maintaining the tumorigenicity of OSCCs. Recent studies found the pathophysiologic role of self-renewing cells with cancer stem characteristics in long-term maintenance of cancers (27). Phenotypic studies of cancer cells with stem characteristics are aided by tumor spheroid model, in which the tumor initiating cells are enriched in non-adherent tumor spheroids (28). Thus, tumor spheroid formation is a measure of stem-like properties of cancer cells. We initially investigated the effects of GRHL2 on spheroid formation. SCC4/EGFP cells formed tumor spheroids in low-attachment plates, while SCC4/ShGRHL2 cells demonstrated notably reduced number and size of spheroids (Figure 3A, Supplementary Figure S3B, available at Carcinogenesis Online). To distinguish between tumor spheroids and cell aggregation, we allowed spheroid formation from a single cell plated in 96-well plates. Again, SCC4/EGFP readily formed tumor spheroids from single cells, while SCC4/ShGRHL2 cells did not (Figure 3B). We assessed the formation of secondary tumor spheroids by replating the cells derived from the primary tumor spheroids. With GRHL2 knockdown, secondary tumor spheroid formation was diminished compared with those of controls (Figure 3C and D). Overexpression of GRHL2 in SCC9 cells, which do not express endogenous GRHL2, increased the number and size of tumor spheroids compared with the control cells infected with empty vector (SCC9/LXSN) (Supplementary Figure S2, available at Carcinogenesis Online).
Figure 3.
GRHL2 promotes cancer stem-like characteristics of OSCC cells. (A) Tumor spheroid assay was performed with SCC4/EGFP and SCC4/ShGRHL2 cells in suspension culture for 7 days. Shown here are two independent colonies (C8 and C10) of SCC4/ShGRHL2 cells after cell cloning, demonstrating that the altered tumor spheroid formation by GRHL2 knockdown is not clone-specific. (B) SCC4 cells were seeded in limiting dilution in ultra low attachment 96-well plates to form tumor spheroids to rule out the possibility that the tumor spheroids were cell aggregation rather than growth of a single cell. GRHL2 knockdown strongly reduced the size and number of colonies derived from a single cell. Number of spheroids formed in each well of the 96-well plates were counted for each cell type and plotted. P value is shown in comparison with the control group (SCC4/EGFP). (C) To determine self-renewal characteristics of SCC4/EGFP and SCC4/ShGRHL2 cells, the primary tumor spheroids were collected and disbursed into single cells, which were then plated to form secondary tumor spheroids. After the suspension culture for 7 days, tumor spheroid size and number were determined and plotted (D). P values are shown in comparison to the control group (SCC4/EGFP). (E) Expression levels of pluripotency genes, e.g. Nanog, Oct-4, KLF4, Lin28, Sox9 and EZH2, were determined by qRT-PCR in SCC4 cells with or without GRHL2 knockdown. Bars indicate standard deviation and asterisk (*) indicates statistical significance (P < 0.05), compared with the mean values of the control groups (SCC4/EGFP). (F) ChIP assay was performed with α-GRHL2 antibody in SCC4 cells, and binding in Oct-4 proximal promoter region was assessed by PCR. IVL promoter served as a positive control. TATA binding protein (TBP) also served as the positive control, while IgGs were negative controls. (G) Western blot analysis of Oct-4, Nanog and c-Myc expression in SCC4/ShGRHL2 cells compared with the control, SCC4/EGFP. (H) Western blotting was performed with whole cell extracts of SCC9 and NHOF for GRHL2 and Oct-4 after GRHL2 overexpression via retroviral vector (LXSN-GRHL2). Primary NHOK and SCC4 cells were included for comparison. GAPDH was used for loading control. (I) GRHL2 and Oct-4 protein levels were determined in P19 embryonic carcinoma cell line during retinoic acid (RA)-induced differentiation (100nM), and mouse embryoid body differentiation by Western blotting. GAPDH was used as a loading control.
Since our data suggest potential involvement of GRHL2 in stemness and maintenance of cancer phenotype, we then explored whether GRHL2 regulates gene expression of pluripotency factors, e.g. Oct-4, KLF4, Lin28, Nanog, Sox9 and EZH2, many of which are involved in cellular reprogramming. All tested genes, with exception for Lin28, were downregulated in SCC4 with endogenous GRHL2 knockdown (Figure 3E), suggesting that GRHL2 modulates stemness through transcriptional regulation of the reprogramming factors. Among these genes, we investigated the impact of GRHL2 on the expression of Oct-4 due to its importance in both stem cells and cancers (29). In particular, GRHL2 was found to bind to the proximal region of the Oct-4 gene promoter (Figure 3F). When GRHL2 was knocked down in SCC4, protein levels of Oct-4, Nanog and c-Myc were notably reduced (Figure 3G). When GRHL2 was overexpressed in SCC9 and NHOF, there was an increase in the level of Oct-4 protein levels in these two cells (Figure 3H). We also determined the change of GRHL2 expression level in P19 embryonic carcinoma cell line and mouse embryonic stem cells during terminal differentiation induced by retinoic acid treatment or embryoid body formation, respectively. In both cell types, GRHL2 expression level was readily detected in cells with undifferentiated status and lost during retinoic acid-induced differentiation and embryoid body formation, along with the altered expression of Oct-4 (Figure 3I). These data indicate that GRHL2 regulates the stem-like properties and expression of pluripotency genes in OSCC cells.
GRHL2 determines the epithelial phenotype of OSCC cells through regulating miR-200 genes
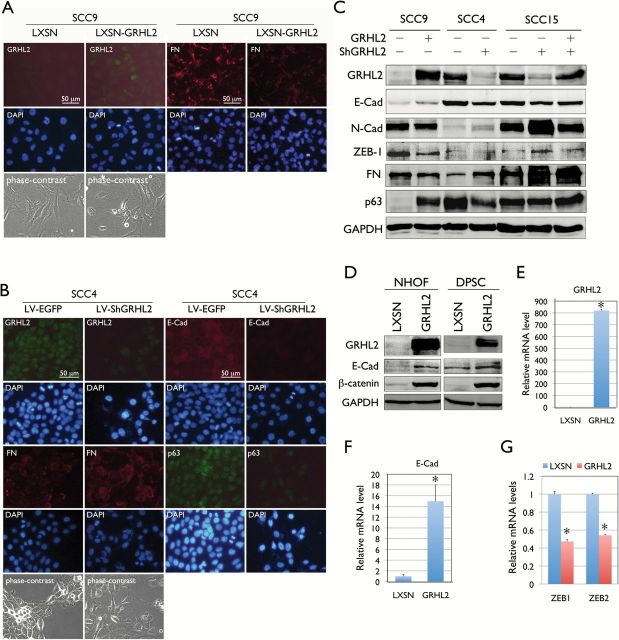
OSCCs develop tissue destruction at the primary site and rarely progress into distant metastasis (15). However, metastatic nature of OSCC is the culprit of the mortality associated with the disease, and it is critical to understand the molecular mechanism of epithelial plasticity underlying the metastatic cell transformation. To demonstrate the role of GRHL2 in epithelial plasticity in OSCCs, we transduced GRHL2 into SCC9, which lack the endogenous expression, and knocked down the GRHL2 expression in SCC4 and SCC15, both of which express high level of the gene. Immunofluorescent staining revealed that ectopic expression of GRHL2 in SCC9 led to reduced expression of the mesenchymal cell marker, e.g. fibronectin (FN) (Figure 4A). Also, knockdown of GRHL2 in SCC4 or SCC15 inhibited expression of the epithelial cell markers, e.g., E-Cad, p63, K14 and β-catenin, while enhanced expression of the mesenchymal cell markers, e.g., FN, ZEB1 and N-Cad (Figure 4B). We also knocked down GRHL2 in SCC4 and SCC15 using a different lentiviral vector (LV2) to generate SCC4/ShGRHL2 and SCC15/ShGRHL2 cells. When GRHL2 was knocked down using the LV2-ShGRHL2 vector, we also observed loss of epithelial markers, e.g. E-Cad and β-catenin and induction of N-Cad and FN (Supplementary Figure S3 and S4, available at Carcinogenesis Online). Ectopic expression of GRHL2 in NHOF and dental pulp stem cell induced epithelial phenotype in cells by inducing expression of E-Cad and β-catenin, both of which make up the adherens junction in epithelial cells (30) (Figure 4D). We also examined the gene expression of E-Cad, ZEB1 and ZEB2 in SCC9 cells transduced with GRHL2 and its control cells by qRT-PCR. Ectopic expression of GRHL2 induced E-Cad expression while suppressed ZEB1 and ZEB2 expression, which are in accordance with the Western blotting results (Figure 4E–G). These data indicate that GRHL2 is an epithelial-specific transcriptional factor that determines epithelial phenotype of OSCC cells.
Figure 4.
GRHL2 regulates epithelial plasticity in OSCC. (A) Immunofluorescent staining was performed in SCC9 cells with or without GRHL2 overexpression using the retroviral vector, LXSN-GRHL2. GRHL2 overexpression and repressed FN expression were confirmed in SCC9/GRHL2 cells after G418 selection compared with the empty vector (LXSN) control. (B) Immunofluorescent staining was performed in SCC4/EGFP and SCC4/ShGRHL2 cells for GRHL2, E-Cad, p63 and FN. GRHL2 knockdown led to loss of E-Cad and p63 levels, while FN was induced. Phase contrast views for each cell group were shown. (C) GRHL2 expression levels were altered in SCC9, SCC4 and SCC15 cell lines by either overexpression (in SCC9) or knockdown (in SCC4 and SCC15). Alteration of GRHL2 level led to corresponding changes in proteins involved in epithelial (e.g. E-Cad and p63) and mesenchymal (e.g. N-Cad, ZEB1 and FN) phenotypes. In SCC15 cells, GRHL2 was re-expressed by retroviral vector (LXSN-GRHL2) infection after lentivirus-mediated GRHL2 knockdown. GRHL2 re-expression partially restored the epithelial phenotype, e.g. loss of N-Cad and ZEB1. (D) Ectopic GRHL2 expression in NHOF and DPSC enhanced the expression of E-Cad and β-catenin, which make up the epithelial adherens complex. (E-G) Expression levels of E-Cad, ZEB1 and ZEB2 were determined by qRT-PCR in SCC9 with or without GRHL2 transduction by retroviral vector (LXSN-GRHL2). Bars indicate standard deviation and asterisk (*) indicates statistical significance (P < 0.05), compared with the mean values of their control cells.
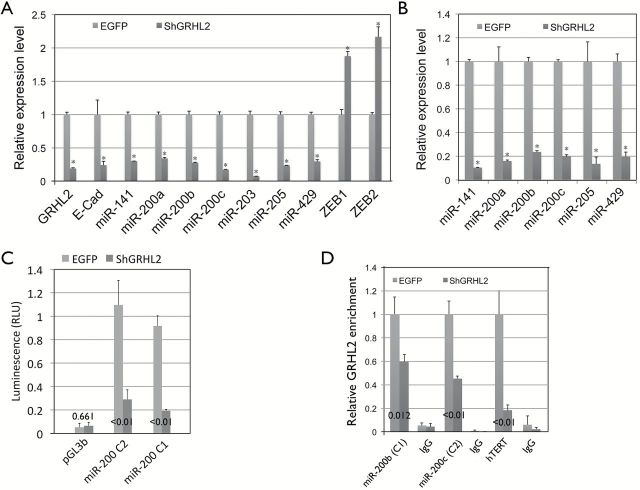
Next, we investigated how GRHL2 regulates epithelial phenotype in OSCC. In breast cancers, miR-200 is associated with the epithelial phenotype and facilitates tumor formation (20). We sought to explore the relationship between GRHL2 and miR-200 family genes, e.g., miR-141, miR-200a, miR-200b, miR-200c and miR-429, which negatively regulate EMT through the negative feedback loop with ZEB proteins (31). miR-200 family genes are also unique mediators of the reprogramming factors Oct-4/Sox2 and induce somatic cell reprogramming at the early stage (32). We first checked the expression levels of the miR-200 genes in SCC4/EGFP and SCC4/ShGRHL2 cells and found that all members of the miR-200 family were downregulated by GRHL2 knockdown, while ZEB1 and ZEB2 expression were elevated (Figure 5A). Likewise, GRHL2 knockdown in HaCaT cells showed drastic loss of miR-200 gene expression (Figure 5B). Promoter-luciferase reporter assay revealed that the loss of GRHL2 in SCC4 resulted in diminution of miR-200 promoter activities (Figure 5C). ChIP assay revealed that knockdown of GRHL2 led to reduced enrichment of GRHL2 at the miR-200 promoters, authenticating the binding event (Figure 5D). These data suggest that GRHL2 regulates the expression of miR-200 family genes through direct promoter DNA binding.
Figure 5.
GRHL2 regulates the expression of miR-200 family genes through direct promoter binding. (A) miR-200 family genes were downregulated in SCC4/ShGRHL2 compared with the control (SCC4/EGFP), as determined by qPCR. In the same samples, knockdown of GRHL2 reduced E-Cad mRNA level and enhanced ZEB1 and ZEB2 mRNA levels. (B) Diminution of miR-200 gene levels was confirmed in HaCaT cells after GRHL2 knockdown by LV-ShGRHL2 lentiviral vector compared with the empty vector control (LV-EGFP). Asterisk (*) indicates P < 0.05 compared with control group (LV-EGFP). (C) miR-200 gene promoter activity was compared in SCC4/EGFP and SCC4/ShGRHL2 cells by promoter-luciferase (firefly) reporter assay. Co-tranfection with Renilla luciferase reporter plasmid driven by SV40 promoter was used for normalization. (D) GRHL2 binding was assessed by ChIP assay using α-GRHL2 antibody or IgG (negative control) in SCC4/EGFP and SCC4/ShGRHL2 cells. Immunoprecipitates were used for amplification of the miR-200 promoter fragments (miR-200b and miR-200c representing the clusters C1 and C2, respectively) near the transcription start sites by qPCR. Enrichment of GRHL2 in hTERT promoter was used as a positive control. P values are shown in comparison to the control group (SCC4/EGFP).
Since GRHL2 knockdown led to phenotypic change in SCC4, e.g. inhibition of cell proliferation and colony formation, reduction in tumor spheroid formation, we tested whether such phenotype could be reversed by overexpression of miR-200 genes. Overexpression of miR-200 cluster 1 (miR-200b, miR-200a and miR-429) and cluster 2 (miR-200c and miR-141) genes in SCC4/ShGRHL2 cells partially rescued the proliferation rate (Figure 6A). GRHL2 knockdown led to loss of E-Cad and p63, and drastic enhancement of N-Cad expression, reminiscent of EMT (Figure 6B). When miR-200 C1 or C2 was overexpressed in these cells, there was partial restoration of protein levels of E-Cad and N-Cad, but p63 expression remained unchanged. In addition, miR-200 expression in SCC4/ShGRHL2 cells led to appearance of epithelial phenotype, e.g. cobble-stone morphology, and reduced cell migration, as well as tumor spheroid forming abilities (Figure 6C–F). Hence, it appears that GRHL2 directly regulates the expression of the miR-200 family genes, which partly mediate the phenotypic effects of GRHL2 on OSCC cells.
Figure 6.
Overexpression of miR-200 family genes partially reverses the phenotypic changes caused by GRHL2 knockdown in OSCC cells. (A) SCC4/ShGRHL2 cells were infected with the retroviral vectors expressing miR-200 cluster 1 (miR-200b, -200a, -429) named MSCV-miR-200C1 or miR-200 cluster 2 (miR-200c and -141) named MSCV-miR-200C2. After selection with puromycin (1 μg/ml), the cells were maintained in culture for 7 days to assess altered cell proliferation rate; cell numbers were plotted against time in culture. (B) Western blotting was performed for GRHL2 and epithelial/mesenchymal markers, e.g. E-Cad, N-Cad and p63, in SCC4/EGFP, SCC4/ShGRHL2/MSCV (empty vector), SCC4/ShGRHL2/miR-200C1 and SCC4/ShGRHL2/miR-200C2. (C) Four days after seeding the same cell numbers across the groups, photomicrographs were taken to reveal the altered cell confluence and changes in intercellular adhesion. (D) Tumor spheroid assay was performed with SCC4/EGFP, SCC4/ShGRHL2/MSCV and SCC4/ShGRHL2/miR-200C1. In addition, colony formation assay (E) and Matrigel invasion assay (F) were performed among the indicated cell groups to test the effects of miR-200 gene overexpression on reversing the phenotypic changes induced by GRHL2 knockdown in SCC4 cells.
Discussion
This study demonstrates the functional role of GRHL2 in the maintenance of the transformed phenotype in OSCCs. GRHL2 expression level is elevated in the OSCC tissue specimens compared with the normal control, as revealed by IHC and qRT-PCR of samples isolated by LCM. However, it should be noted that GRHL2 is also expressed in normal epithelial tissues in order to coordinate epithelial cell proliferation, differentiation and regeneration (6). Although GRHL2 expression is in general elevated in OSCCs, some cells in tumor may lack the gene expression, depending on the status of differentiation and migration. OSCC specimens shown in Figure 1D clearly demonstrate the mutually exclusive expression between GRHL2 and K1, since GRHL2 is associated with undifferentiated status (6). GRHL2 expression was also lacking in other OSCC samples without exhibition of cell differentiation (Figure 1E), presumably due to occurrence of EMT and cell migration within tumor islands. Thus, although GRHL2 is strongly expressed in OSCC compared to the normal counterpart, GRHL2 level and expression pattern are heterogeneous among different OSCC specimens, presumably depending on the level of cell differentiation and tumor behavior.
There is controversy in regards to the role of GRHL2 in cancer. Several reports demonstrated procarcinogenic effects of GRHL2; enhanced GRHL2 expression was positively correlated with poor cancer survival, and GRHL2 promoted breast carcinogenesis through maintaining epithelial phenotype (11,33). On the contrary, other studies noted the inhibitory effects of GRHL2 on EMT as an indicator of its tumor suppressive role and diminution of GRHL2 expression in gastric cancer also as support of its tumor suppressive function (13,14). This discrepancy may reflect the dynamic regulation of GRHL2 expression in cancers under different stages in the disease progression. Clearly, GRHL2 has growth-promoting effects in epithelial cells, and this was evinced in our prior studies in which GRHL2 overexpression in NHOK and NHEK led to drastic increase in replicative potential and repressed terminal differentiation (5,6). Ectopic expression of GRHL2 in NIH3T3 cells induced epithelial phenotype, enhanced cell proliferation and tumorigenic conversion in vivo, indicating its oncogenic effects (8). Hence, it is plausible to postulate that aberrant GRHL2 expression is phasic during cancer progression—it is high during the initial stage of primary tumor development, then temporarily lost during EMT to allow cell migration, and reactivated upon metastatic colonization, during which GRHL2 triggers distant metastasis through mesenchymal–epithelial transition (MET).
The seeming disconnect between GRHL2 and cancer stem cells may stem from the fact that it inhibits EMT, which is generally considered to confer cells with stem-like properties, with migratory and invasive capabilities associated with metastatic behavior (34). However, recent studies show that self-renewal is not linked to the mesenchymal phenotype and that epithelial-type cells are suitable for tumor colonization and proliferation. In prostate cancers, tumor-initiating cells demonstrate epithelial phenotype (35). Also, Korpal et al. showed that overexpression of miR-200 cluster genes led to epithelial phenotypic change and increased metastatic tumors in 4T07 breast cancer cell line, which has very weak metastatic activity (20). Prrx1 is a developmental transcription factor involved in the induction of EMT in embryos and cancers, and metastatic colonization of BT-549 breast cancer cells required the loss of Prrx1 and consequent induction of MET. Interestingly, MET by loss of Prrx1 led to acquisition of stem cell properties in cancer cells, suggesting that EMT and stemness can be uncoupled (36). The induced pluripotent stem cell (iPSC) studies revealed that miR-200 can mediate Oct-4/Sox2 signaling and induce MET and iPSC generation (32). Our current study also demonstrated that EMT and stemness might be uncoupled by GRHL2, which is associated with epithelial characteristics and enhances stemness of OSCCs. Hence, it is likely that GRHL2 knockdown and Prrx1-mediated EMT overlap in molecular signaling, and/or that this new concept of coupling between enhanced stemness and MET is a common requirement for metastatic colonization. This directly contrasts with several other reports showing that EMT is linked with stemness. Some studies even show that artificial induction of EMT in breast cancer cells by overexpression of TGF-β, SNAIL or TWIST led to stem cell characteristics, e.g. altered stem marker expression, mammosphere formation and multilineage differentiation (37).
In conclusion, the current study showed the requirement of GRHL2 in tumorigenic potential of OSCCs and the underlying mechanism, which may involve transcriptional regulation of its target genes, e.g. Oct-4 and miR-200 family members. The significance of these findings lies in part in the regulation of epithelial plasticity during OSCC primary tumor development, dissemination through EMT and metastatic tumor formation by distant colonization. It appears that GRHL2 level in OSCCs modulates the epithelial plasticity during the disease progression, and further research will elucidate how this functional relationship between GRHL2 and epithelial phenotype may be intervened for anticancer therapies.
Supplementary material
Supplementary Table 1 and Figures 1–13 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health/National Institute of Dental and Craniofacial Research (1R56DE024593-01 to M.K.K., 1R03DE024259-01A1 to W.C.); M.K.K. is also supported by the Jack A. Weichman Endowed Fund.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- ChIP
chromatin immunoprecipitation
- EMT
epithelial–mesenchymal transition
- FBS
fetal bovine serum
- GRHL2
grainyhead-like 2
- IHC
immunohistochemistry
- LCM
laser-captured microdissection
- MET
mesenchymal–epithelial transition
- NHEK
normal human epidermal keratinocytes
- NHOF
normal human oral fibroblasts
- NHOM
normal human oral mucosa
- OSCC
oral squamous cell carcinomas
- siRNA
small interfering RNA
References
- 1. Bray S.J., et al. (1991) Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev., 5, 1672–1683. [DOI] [PubMed] [Google Scholar]
- 2. Huang J.D., et al. (1995) Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev., 9, 3177–3189. [DOI] [PubMed] [Google Scholar]
- 3. Stramer B., et al. (2005) Cell biology: master regulators of sealing and healing. Curr. Biol., 15, R425–R427. [DOI] [PubMed] [Google Scholar]
- 4. Narasimha M., et al. (2008) Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J. Cell Sci., 121(Pt 6), 747–752. [DOI] [PubMed] [Google Scholar]
- 5. Chen W., et al. (2010) Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5’-CpG island in normal human keratinocytes. J. Biol. Chem., 285, 40852–40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen W., et al. (2012) Grainyhead-like 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism. Cell Death Dis., 3, e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang X., et al. (2009) Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene, 28, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Werner S., et al. (2013) Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem., 288, 22993–23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanaka Y., et al. (2008) Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J. Hepatol., 49, 746–757. [DOI] [PubMed] [Google Scholar]
- 10. Dompe N., et al. (2011) A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc. Natl. Acad. Sci. USA, 108, E943–E951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiang X., et al. (2012) Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One, 7, e50781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cieply B., et al. (2012) Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res., 72, 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cieply B., et al. (2013) Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res., 73, 6299–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiang J., et al. (2013) Expression and role of grainyhead-like 2 in gastric cancer. Med. Oncol., 30, 714. [DOI] [PubMed] [Google Scholar]
- 15. Garavello W., et al. (2006) Risk factors for distant metastases in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg., 132, 762–766. [DOI] [PubMed] [Google Scholar]
- 16. Kang M.K., et al. (1998) Replicative senescence of normal human oral keratinocytes is associated with the loss of telomerase activity without shortening of telomeres. Cell Growth Differ., 9, 85–95. [PubMed] [Google Scholar]
- 17. Kang M.K., et al. (2000) In vitro replication and differentiation of normal human oral keratinocytes. Exp. Cell Res., 258, 288–297. [DOI] [PubMed] [Google Scholar]
- 18. Li S.L., et al. (1992) Sequential combined tumorigenic effect of HPV-16 and chemical carcinogens. Carcinogenesis, 13, 1981–1987. [DOI] [PubMed] [Google Scholar]
- 19. Kang M.K., et al. (2007) Extension of cell life span using exogenous telomerase. Methods Mol. Biol., 371, 151–165. [DOI] [PubMed] [Google Scholar]
- 20. Korpal M., et al. (2011) Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med., 17, 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gill R.M., et al. (1998) Regulation of expression and activity of distinct pRB, E2F, D-type cyclin, and CKI family members during terminal differentiation of P19 cells. Exp. Cell Res., 244, 157–170. [DOI] [PubMed] [Google Scholar]
- 22. Deshpande A.M., et al. (2012) Cdk2ap2 is a novel regulator for self-renewal of murine embryonic stem cells. Stem Cells Dev., 21, 3010–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu F., et al. (2006) The UCSC Known Genes. Bioinformatics, 22, 1036–1046. [DOI] [PubMed] [Google Scholar]
- 24. Kang M.K., et al. (2007) Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br. J. Cancer, 96, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams D.L., et al. (2008) Immunocytochemistry and laser capture microdissection for real-time quantitative PCR identify hindbrain neurons activated by interaction between leptin and cholecystokinin. J. Histochem. Cytochem., 56, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim H.R., et al. (2001) Elevated expression of hTERT is associated with dysplastic cell transformation during human oral carcinogenesis in situ. Clin. Cancer Res., 7, 3079–3086. [PubMed] [Google Scholar]
- 27. Clevers H. (2011) The cancer stem cell: premises, promises and challenges. Nat. Med., 17, 313–319. [DOI] [PubMed] [Google Scholar]
- 28. Lobo N.A., et al. (2007) The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol., 23, 675–699. [DOI] [PubMed] [Google Scholar]
- 29. Glinsky G.V. (2008) “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J. Clin. Oncol., 26, 2846–2853. [DOI] [PubMed] [Google Scholar]
- 30. Brembeck F.H., et al. (2006) Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev., 16, 51–59. [DOI] [PubMed] [Google Scholar]
- 31. Brabletz S., et al. (2010) The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep., 11, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang G., et al. (2013) Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc. Natl. Acad. Sci. USA, 110, 2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang X., et al. (2013) Bridging cancer biology with the clinic: relative expression of a GRHL2-mediated gene-set pair predicts breast cancer metastasis. PLoS One, 8, e56195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kalluri R., et al. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest., 119, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Celià-Terrassa T., et al. (2012) Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J. Clin. Invest., 122, 1849–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ocaña O.H., et al. (2012) Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell, 22, 709–724. [DOI] [PubMed] [Google Scholar]
- 37. Mani S.A., et al. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell, 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.