Abstract
Microplitis demolitor (Hymenoptera: Braconidae) is a parasitoid used as a biological control agent to control larval-stage Lepidoptera and serves as a model for studying the function and evolution of symbiotic viruses in the genus Bracovirus. Here we present the M. demolitor genome (assembly version 2.0), with a genome size of 241 Mb, and a N50 scaffold and contig size of 1.1 Mb and 14 Kb, respectively. Using RNA-Seq data and manual annotation of genes of viral origin, we produced a high-quality gene set that includes 18,586 eukaryotic and 171 virus-derived protein-coding genes. Bracoviruses are dsDNA viruses with unusual genome architecture, in which the viral genome is integrated into the wasp genome and is comprised of two distinct components: proviral segments that are amplified, circularized, and packaged into virions for export into the wasp’s host via oviposition; and replication genes. This genome assembly revealed that at least two scaffolds contain both nudivirus-like genes and proviral segments, demonstrating that at least some of these components are near each other in the genome on a single chromosome. The updated assembly and annotation are available in several publicly accessible databases; including the National Center for Biotechnology Information and the Ag Data Commons. In addition, all raw sequence data available for M. demolitor have been consolidated and are available for visualization at the i5k Workspace. This whole genome assembly and annotation represents the only genome-scale, annotated assembly from the lineage of parasitoid wasps that has associations with bracoviruses (the ‘microgastroid complex’), providing important baseline knowledge about the architecture of co-opted virus symbiont genomes.
Keywords: symbiosis, Hymenoptera, Braconidae, Microplitis demolitor bracovirus (MdBV), Polydnaviridae
Microplitis demolitor (Hymenoptera: Braconidae) is a parasitoid wasp species that completes its immature stages of development in larval Lepidoptera. It is endemic to Queensland, Australia, and was first introduced to the United States in 1983 for biological control of Helicoverpa zea and select other lepidopteran pests (Shepard et al. 1983; Burke 2016). It has become an important system for studying the function and evolution of mutualistic symbiotic viruses in the genus Bracovirus (family Polydnaviridae). Bracoviruses are produced in wasp ovaries in the nuclei of specialized (calyx) cells, which lyse and release virions into the reproductive tract where they form a paste-like “calyx fluid” (Stoltz and Vinson 1979). During parasitism, the female wasp uses her ovipositor to inject eggs and calyx fluid into the body of the host insect, where the virions immediately infect host cells, particularly host blood or hemocyte cells (Beck et al. 2007; Strand et al. 1992; Strand 1994). The virions carry circular double-stranded DNA molecules that are injected into host cell nuclei where virulence genes are transcribed rapidly after parasitism (Stoltz and Vinson 1979; Strand et al. 1992). The protein products of virulence genes are involved in suppressing the host immune system and altering host development to favor the survival of the wasp egg and larva (Strand and Burke 2014).
Bracoviruses (BVs) evolved approximately 100 million years ago from an ancestral virus in the family Nudiviridae (Bézier et al. 2009; Murphy et al. 2008). Today, all known BVs persist in wasps as integrated proviruses. BV genome architecture is unusual because genes are dispersed in the wasp genome and organized in ways that enable formation of replication-defective virions that wasps use to infect hosts (Bézier et al. 2009; Bézier et al. 2013; Burke et al. 2014; Herniou et al. 2013). The elements of Microplitis demolitor bracovirus (MdBV) within the M. demolitor genome have been described in depth, using the assembly named Mdem1 as a reference (Burke et al. 2014). Although the M. demolitor genome sequence was generated primarily to focus upon MdBV, there are few genomic resources available for braconid wasps and other parasitoids, making the wasp genome useful for researchers in other fields (e.g., Geib et al. 2017; Bewick et al. 2017; Zhou et al. 2015). In this manuscript, we announce the full genome sequence of M. demolitor with an improved assembly and an annotated gene set for both wasp and viral genes. This publicly available genome assembly will continue to facilitate research on bracoviruses but also provide a resource to help address other questions specific to M. demolitor and to enable comparative analyses with other insect species.
Materials and Methods
Wasp samples
Wasp samples were derived from a culture maintained at the University of Georgia as described previously (Burke 2016). DNA was isolated from single and pooled male wasps with a high-salt precipitation method to maintain the integrity of high molecular weight DNA as described in Burke et al. (2014).
Whole genome sequencing and assembly
In addition to the sequencing libraries reported in Burke et al. (2014) (180 bp, 1.5 kb, 5 kb, and 10 kb), a new 20 kb long-insert mate-pair library was constructed from pooled adult male DNA using Illumina’s Nextera Mate-Pairs Sample Prep Kit. All libraries were sequenced for 100 cycles on a HiSeq2000 using TruSeq chemistry. Raw reads were trimmed, filtered, and error-corrected as described in Burke et al. (2014). The Mdem1 assembly was further improved by additional scaffolding with the 20 kb Nextera mate-pair library and use of GapCloser v1.12 to close gaps generated in the scaffolding process with short paired read data (Luo et al. 2012). The genome assembly was screened by NCBI during the whole genome submission process to filter out adapter, vector, and other contaminant sequences. Methods used to generate RNASeq and viral DNA libraries and sequence data used for mapping have been described previously (Bitra et al. 2016; Burke and Strand 2014; Burke et al. 2014; Burke and Strand 2012; Burke 2016).
Automated annotation of the M. demolitor genome
Structural and functional annotation of genes was performed with the NCBI Eukaryotic Genome Annotation Pipeline. This automated pipeline utilized short read transcript evidence from existing RNASeq data for M. demolitor (Burke et al. 2014), in addition to the MdBV proviral segments present in GenBank (Webb et al. 2006), NCBI RefSeq protein sets for Fopius arisanus, Nasonia vitripennis and Apis mellifera and 81,697 protein sequences from GenBank derived from the Insecta. Alignments were used to inform gene model prediction using the NCBI eukaryotic gene prediction tool GNOMON. Details of the annotation process can be accessed at: https://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/. The completeness of the annotated gene set was analyzed by identifying the number of arthropod Benchmark Universal Single-Copy Orthologs (BUSCOs) (Simão et al. 2015). BUSCO v.1.1b1 was run on the RefSeq Gene set at the predicted peptide level (“-m OGS”). BUSCO results were compared to the RefSeq Gene sets for braconid species F. arisanus and D. alloeum as well as Nasonia vitripennis, for which a large portion of the genome is mapped to one of five chromosomes (Werren et al. 2010).
Manual annotation of M. demolitor genes of viral origin
Manual verification or correction of nudivirus-like replication genes and proviral genes was performed using the M. demolitor jBrowse/Apollo instance hosted at the USDA National Agricultural Library i5k Workspace. Protein sequences from the previously published manually curated viral gene set from the Mdem1 assembly were aligned to the genome using a modified version of exonerate v. 2.3.0 in which the gff3 output is compatible with jBrowse for upload as a custom track (available at https://github.com/hotdogee/exonerate-gff3). Exonerate alignments were used as the basis for correction of existing gene models or addition of gene models missing in the Mdem2 annotation. The boundaries of proviral segments and replication units in the Mdem2 assembly were identified by searching for sequence motifs that define these regions, along with use of short read mapping data from existing deep sequencing data from MdBV viral DNA and DNA isolated from ovaries when replication and associated amplification of viral DNA is at its peak (Burke and Strand 2014; Burke et al. 2015). Short read data were filtered with the fastx toolkit to retain reads with a phred score equivalent >30 for >90% of bases within a read. Quality filtered reads from sequenced DNAs were mapped to the Mdem2 assembly using bwa mem v. 0.7.15. Similarly, quality-filtered RNASeq data from infected host cells of Chrysodeixis includens or Trichoplusia ni were mapped to the proviral regions of the Mdem2 assembly with hisat2 v.2.1.0 (Burke et al. 2014, Bitra et al. 2016, Kim et al. 2015). Any reads that did not map to the proviral segments were removed using samtools v.1.3.1 (Li et al. 2009).
Data Availability
All raw sequencing data are available from the NCBI Sequence Read Archive (see Table 1 for accessions). The genome assembly, WGS Project AZMT02, is represented as BioSample SAMN02708865 with identical records in GenBank as accession GCA_000572035.2 and RefSeq as accession GCF_000572035.2 with the name Mdem2. All current annotations are associated with the RefSeq assembly as release 101. An FTP site for data download is at ftp://ftp.ncbi.nlm.nih.gov/genomes/Microplitis_demolitor/. NCBI’s Genome Data Viewer can be accessed at https://www.ncbi.nlm.nih.gov/genome/gdv/?acc=GCF_000806365.1&context=genome and an overview of release 100 annotations can be accessed at https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Microplitis_demolitor/101. Curation of this assembly and consolidated sequence-based resources are hosted by the i5k Workspace (https://i5k.nal.usda.gov/) allowing visualization within jBrowse, manual curation with Apollo and other tools.
Table 1. Raw reads generated for assembly.
| SRA | Library type | Read pairs | Base pairs | Material |
|---|---|---|---|---|
| SRX610757 | 180 bp | 138.9 M | 17.5 Gb | Single adult male |
| SRX641381 | 1.5 kb | 164.3 M | 20.9 Gb | 40 pooled adult male wasps |
| SRX641400 | 5 kb | 53.6 M | 7.1 Gb | 40 pooled adult male wasps |
| SRX641403 | 10 kb | 174.3 M | 22.1 Gb | 100 pooled adult male wasps |
| SRX641469 | 20 kb | 230.2 M | 28.3 Gb | 100 pooled adult male wasps |
| SRX981480 | 5 kb | 37.8 M | 5.1 Gb | 120 pooled adult male wasps |
Results And Discussion
In total, approximately 17.5 Gb of small-insert sequence data were generated from a single male adult wasp for the Mdem1 assembly, along with 129.4 Gb of data generated from larger insert libraries (1.5, 5, 10, and 20 kb insert sizes) for scaffolding purposes (Table 1). The 20 kb library derived data were not included in the previous assembly Mdem1. Assembly of these sequence data with SOAPdenovo resulted in a new assembly (Mdem2) that consisted of 1,794 scaffolds with an N50 size of 1.1 Mb and 27,508 contigs with an N50 of 14.12 kb (Table 2). The assembly was 241.2 Mb in total length, which has very good concordance with the genome size estimated by flow cytometry (241 +/− 6 Mb, Burke et al. 2014). Only 14.6% of the genome assembly was comprised of sequence gaps. The overall G + C nucleotide content was 33.1%. These assembly statistics are a large improvement over the Mdem1 assembly, with approximately 65% fewer scaffolds and an N50 size 3.6x longer (Table 2). Genome assemblies are available for three other braconid wasp species while sequences are available for a fourth (Cotesia vestalis) but have not been scaffolded. The Mdem2 assembly statistics are similar to these other braconids and Nasonia vitripennis (family Pteromalidae) in both genome size and G + C content (Table 2).
Table 2. Summary statistics for the Mdem2 assembly, the previous Mdem1 assembly, and select other parasitoid genomes.
| Species | Assembly | NCBI BioProject | Contig count (N50 kb) | Scaffold count (N50 Mb) | Total length (Mb) | GC (%) |
|---|---|---|---|---|---|---|
| Microplitis demolitor | Mdem2 | PRJNA251518 | 27,508 (14.12) | 1,794 (1.14) | 241.2 | 33.1 |
| Microplitis demolitor | Mdem1 | PRJNA251518/PRJNA195937 | 36,718 (13.54) | 5,174 (0.32) | 250.5 | 33.1 |
| Fopius arisanus | ASM80636v1 | PRJNA258104 | 8,510 (51.90) | 1,042 (0.98) | 153.6 | 39.4 |
| Diachasma alloeum | Dall1.0 | PRJNA306876 | 25,534 (44.93) | 3,968 (0.65) | 388.8 | 39.1 |
| Cotesia vestalis | ASM95615v1 | PRJNA271135 | 9,156 (46.06) | — | 186.1 | 30.6 |
| Nasonia vitripennis | Nvit_2.1 | PRJNA13660 | 25,484 (18.84) | 6,169 (0.71) | 295.8 | 40.6 |
Annotation using the NCBI Eukaryotic Annotation Pipeline yielded 12,755 genes or pseudogenes, including 12,144 containing protein coding regions. A total of 19,597 transcripts were annotated, with a mean of 1.54 (median 1) transcripts per gene (Table 3). Evidence for gene annotations were derived from RNA-Seq data from adult wasp ovaries, venom glands, and teratocytes, and larvae (Table 4) and proteins from related species, or ab initio evidence predicted by GNOMON. A large proportion of transcripts (16,219 of 18,586 (87.2%)) were fully supported with experimental evidence. A total of 526 non-coding genes were identified, including tRNAs, lncRNAs and others. Details of the annotation are presented in Table 3 as well as online at https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Microplitis_demolitor/101/.
Table 3. Gene annotation summary statistics.
| Feature | Count | Mean length (bp) | Median length (bp) | Min length (bp) | Max length (bp) |
|---|---|---|---|---|---|
| Genes | 12,670 | 10,107 | 3,317 | 71 | 539,413 |
| All transcripts | 19,597 | 2,356 | 1,840 | 71 | 44,309 |
| mRNA | 18,586 | 2,417 | 1,897 | 189 | 44,309 |
| misc_RNA | 224 | 3,134 | 2,043 | 106 | 19,088 |
| tRNA | 183 | 74 | 73 | 71 | 84 |
| lncRNA | 604 | 911 | 671 | 91 | 5,617 |
| CDSs | 18,586 | 1,916 | 1,401 | 105 | 43,512 |
| Exons | 78,299 | 365 | 211 | 2 | 14,552 |
| Introns | 64,033 | 1,911 | 183 | 30 | 353,735 |
Table 4. Raw reads from M. demolitor tissues or proviral segments used for annotation.
| Track name | Bioproject/ Sample ID | Nucleic acid | Number of reads | Percent aligned (quality-filtered) reads | Reference |
|---|---|---|---|---|---|
| Venom glands | PRJNA214515/SAMN02319525 | RNA | 136.4 M | 95% | Burke and Strand 2014 |
| Larvae | PRJNA214515/SAMN02319526 | RNA | 95.7 M | 93% | Burke and Strand 2014 |
| Teratocytes | PRJNA214515/SAMN02319527 | RNA | 99.0 M | 88% | Burke and Strand 2014 |
| Ovaries | PRJNA214515/SAMN02319528 | RNA | 103.3 M | 90% | Burke and Strand 2012 |
| Infected C. includens hemocytes | PRJNA285771/SAMN03758721 | RNA | 73.7 M | 2% | Burke et al. 2014 |
| Infected C. includens | PRJNA437008/SAMN08637637- | RNA | 22.0 M | 5% | Bitra et al. 2016 |
| SAMN08637639; | |||||
| SAMN08637643- | |||||
| SAMN08637645 | |||||
| Infected T. ni | PRJNA437008/SAMN08637630, SAMN08637631, SAMN08637636, | RNA | 37.5 M | 4% | Bitra et al. 2016 |
| SAMN08637640- | |||||
| SAMN08637642 | |||||
| Bracovirus DNA | PRJNA319039/SAMN04875661 | DNA | 49.5 M | 99% | Burke 2016; Burke et al. 2014 |
| Ovary DNA | PRJNA319039/SAMN04875652 | DNA | 17.6 M | 99% | Burke 2016; Burke et al. 2015 |
BUSCO analysis revealed that the M. demolitor genome assembly and annotation is very complete, with 97% of all BUSCOs conserved in Insecta identified in the protein-coding gene set (Table 5). Only 1.2% of BUSCOs were present as fragments in the M. demolitor annotation, and 0.7% were missing. These results are very similar to BUSCO analyses of other hymenopteran genomes (Table 5).
Table 5. BUSCO analysis of parasitoid wasp genomes.
| Species | CDS count | NCBI Refseq annotation release | Complete (% of total BUSCOs) | Fragmented (% of total BUSCOs) | Missing (% of total BUSCOs) |
|---|---|---|---|---|---|
| M. demolitor | 18,586 | 101 | 2621 (97) | 34 (1.2) | 20 (0.7) |
| F. arisanus | 18,906 | 100 | 2605 (97) | 37 (1.3) | 33 (1.2) |
| D. alloeum | 19,692 | 100 | 2622 (98) | 31 (1.1) | 22 (0.8) |
| N. vitripennis | 24,846 | 102 | 2585 (96) | 34 (1.2) | 50 (1.8) |
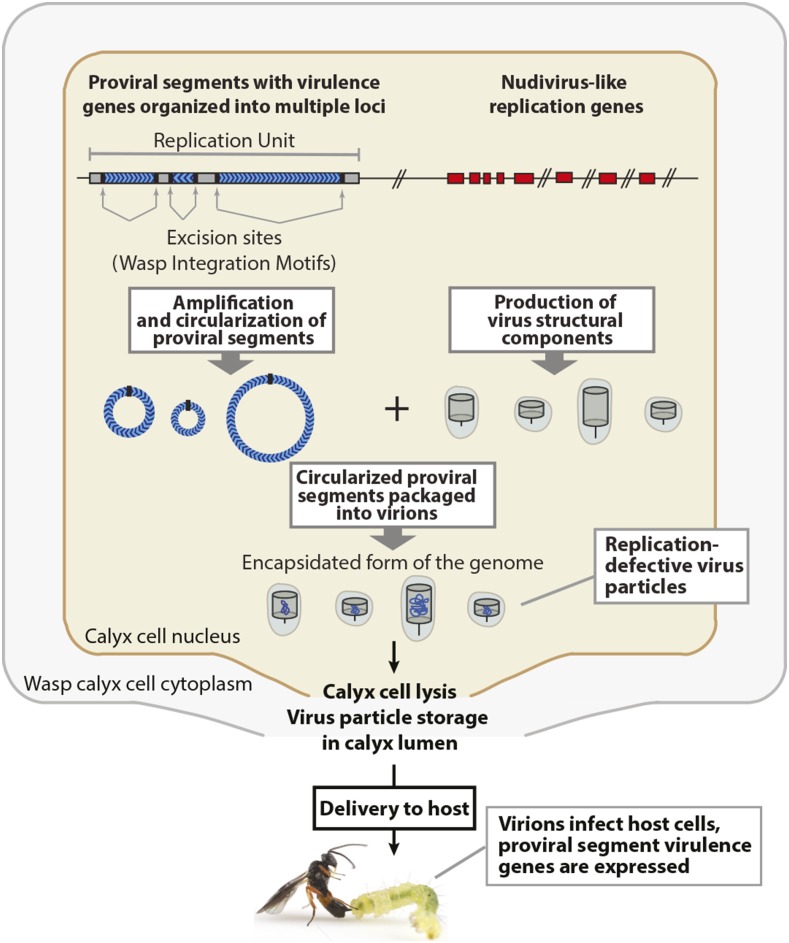
As previously noted, BV genomes are integrated into the genomes of wasps. They also consist of two distinct components: proviral segments and nudivirus-like replication genes (Bézier et al. 2009; Burke et al. 2014, Figure 1). Expression of nudivirus-like replication genes in wasp ovaries results in formation of virions, while proviral segments, bounded by excision motifs targeted by specific nudivirus-like replication genes, are amplified in regions known as Replication Units (RUs), circularized and packaged into virions (Burke et al. 2013; Bézier et al. 2009; Burke et al. 2014; Annaheim and Lanzrein 2007; Savary et al. 1997; Bézier et al. 2013; Burke et al. 2015; Louis et al. 2013). This results in virions that package genes on proviral segments but lack all nudivirus-like replication genes. The genes located on proviral segments are often short and many contain introns (Webb and Strand 2005; Desjardins et al. 2008; Espagne et al. 2004). In contrast, no introns have been described for the nudivirus-like replication genes in bracoviruses (but see below) (Bézier et al. 2009; Burke et al. 2014).
Figure 1.
Schematic showing genome architecture and replication of bracoviruses. Proviral segments (shown as blue chevrons) reside in dispersed locations in the wasp genome and contain virulence genes. Proviral segments specifically amplify in the nuclei of calyx cells as Replication Units prior to segment excision and circularization. Nudivirus-like replication genes also reside in dispersed locations in the wasp genome (as indicated by hash marks) and encode proteins required to make virions. Expression of nudivirus-like replication genes in calyx cell nuclei results in formation of virions that package circularized proviral segments. Mature virions are released into the calyx region of wasp ovaries by lysis of calyx cells. Mature virions are replication-defective because the encapsidated proviral segments do not contain nudivirus-like replication genes. Wasps inject mature virions into host insects, which infect different cell types and express virulence genes that affect host immune defenses and growth.
M. demolitor genes of viral origin were previously described from manual annotation of the Mdem1 assembly (Burke et al. 2014). The genome contained 26 proviral segments that are amplified in eight replication units (Burke et al. 2015; Burke et al. 2014) located at 8 loci on M. demolitor scaffolds. 95 genes were identified in proviral segments, while 76 nudivirus-like replication genes were located on 30 different genome scaffolds. Evidence for these gene models was derived from RNASeq data from wasp cells and tissues as above and also MdBV infected hemocytes (Table 4, Burke et al. 2014). Only a single nudivirus-like replication gene was located on the same scaffold as a proviral segment (HzNVorf93-like and Segment T).
The Mdem2 automatic annotation performed by GNOMON correctly recovered 90% of the M. demolitor viral genes. Eighteen genes that were either missing or incorrectly annotated were manually corrected using alignment with older gene models in the M. demolitor Mdem2 jBrowse/Apollo instance hosted at the i5k Workspace. An additional four gene models (lef-8, lef-9, HzNVorf128-like, and K425_12) were edited to reflect the presence of introns that were previously unidentified.
The architecture of the proviral portion of the M. demolitor genome did not change appreciably between the Mdem1 and Mdem2 assemblies, with proviral segments still located in 8 loci across 9 scaffolds. Coordinates for proviral segments and replication units in the Mdem2 assembly are listed in Table 6. The nudivirus-like replication genes were located on 24 different scaffolds (5 fewer than in Mdem1 assembly). One major difference was that an additional link between proviral segments and nudivirus-like replication genes was identified. Locus 2, containing Segments V, W, E, C and X, was located on a 2.4 Mb scaffold approximately 75 kb away from the nudivirus-like gene p74, and more than 323 kb from several other nudivirus-like replication genes (35a-8 to 35a-14; odv-e66-9 to odv-e66-20; 35a-6 and 35a-7; and helicase). The entire set of proviral segments, replication units, and viral genes are available as gff3 and sequence files at AgDataCommons (http://dx.doi.org/10.15482/USDA.ADC/1432667) and can be uploaded as custom tracks for visualization at the i5k Workspace.
Table 6. Coordinates for boundaries of proviral segments and amplified replication unitsa.
| Segment | Locus | Orientation | Accession number | Segment start | Segment end | Amplification start | Amplification end |
|---|---|---|---|---|---|---|---|
| P | 1 | — | NW_014464280.1 | 204368 | 217086 | 202948 | |
| K1 | 1 | — | NW_014464280.1 | 217246 | 232492 | ||
| K | 1 | — | NW_014464280.1 | >235072 | 245355 | ||
| Q | 1 | — | NW_014464280.1 | 245514 | 261072 | ||
| D | 1 | — | NW_014464280.1 | 261199 | >274917 | >274917 | |
| D | 1 | + | NW_014464327.1 | 325333 | >327729 | >327729 | |
| B | 1 | + | NW_014464327.1 | 315630 | 321681 | ||
| A | 1 | + | NW_014464327.1 | 286830 | 312116 | ||
| L | 1 | + | NW_014464327.1 | 270462 | 285675 | ||
| F | 1 | — | NW_014464327.1 | 259195 | >268430 | ||
| I | 1 | — | NW_014464327.1 | 245419 | >258067* | ||
| M | 1 | + | NW_014464327.1 | 228747 | 244908 | ||
| G | 1 | — | NW_014464327.1 | 215667 | 227866 | 214815 | |
| O | 1 | — | NW_014464327.1 | >209595 | 214086 | 186757 | 214590 |
| V | 2 | + | NW_014463818.1 | >1914991* | 1929656 | 1929927 | |
| W | 2 | + | NW_014463818.1 | 1899259 | >1901856* | ||
| E | 2 | — | NW_014463818.1 | 1890266 | 1898510 | ||
| C | 2 | + | NW_014463818.1 | 1882990 | 1890217 | ||
| X | 2 | — | NW_014463818.1 | 1867002 | 1881299 | 1866484 | |
| N | 3 | — | NW_014463791.1 | 1514968 | 1532949 | 1511956 | 1533620 |
| J | 3 | — | NW_014463791.1 | 1535308 | 1549002 | 1534400 | 1550957 |
| H | 4 | + | NW_014463921.1 | 383509 | 394750 | 382579 | 395765 |
| R | 5 | — | NW_014464373.1 | 36111 | 42534 | 32781 | 43423 |
| S | 6 | — | NW_014463823.1 | 1794688 | 1806656 | 1780975 | 1810556 |
| T | 7 | + | NW_014464188.1 | 421495 | 427196 | 421290 | 429489 |
| U | 8 | + | NW_014463797.1 | 3414939 | 3421513 | 3414736 | 3423057 |
Each proviral segment and its associated locus is listed in a row along with the M. demolitor genome scaffold where it is located. Scaffold accession numbers are indicated along with the coordinates for the boundaries of each proviral segment. Amplification start and end coordinates are listed for each RU that contains one segment. For multi-segment RUs, the amplification start and end coordinates correspond to the outermost segments. “>” signs indicate that gaps in scaffolds or scaffold termini prevent determination of segment or replication unit ends. “*” is similar to “>”, but segment ends are detected in smaller contigs that were not incorporated into scaffolds (e.g., ends of Segments V and W are in NW_014463725.1, Mdem_contig_4120015, while the end of Segment I is in NW_014463324.1, Mdem_contig_4046930).
In addition to updating annotation of the regions of viral origin in the M. demolitor genome, we also consolidated all sequence-based resources we have available for M. demolitor on the jBrowse/Apollo instance of the genome hosted at the i5k Workspace. Genome resources include the most recent genome assembly (Mdem2) and gene sets from NCBI Annotation Release 101. Transcriptome data (in the form of BigWig coverage plots and mapped reads) are available for ovary, teratocyte, venom gland, and larval samples from wasps (Table 4). We have also contributed transcriptome data for all MdBV genes that are expressed in infected host caterpillars. These include the permissive host Chrysodeixis includens and the semipermissive host Trichoplusia ni data from hemocytes and whole body samples (Table 4). Finally, mapped DNA data are available from deep sequencing of DNAs isolated from MdBV virions and M. demolitor ovaries when proviral segment amplification is at its peak (Table 4). These data will facilitate the exploration of the evolution and function of MdBV and other viral symbionts in the future.
The M. demolitor genome described herein represents a high-quality assembly. The assembly of the genome has greatly benefitted from a sequencing strategy in which contigs were built from sequences derived from a single haploid male wasp, followed by scaffolding using sequence data from large-insert libraries. The Mdem1 assembly was also significantly improved with the addition of sequence data derived from a large insert (20kb) mate-pair library used in the Mdem2 assembly. The M. demolitor annotated gene set is similar to related genomes from select other parasitic Hymenoptera in terms of numbers of genes and estimated completeness.
The Mdem2 assembly also provides a more complete picture of the architecture of the MdBV genome in the wasp genome. While proviral segments share no similarity with sequences from pathogenic nudiviruses, prior results showing that the recognition of excision motifs on proviral segments by nudivirus-like integrases strongly suggests that the proviral segments and nudivirus-like replication genes have shared ancestry (Burke et al. 2013). While it is unclear how genome rearrangement of the viral genome was achieved in the wasp genome, the physical location of several nudivirus-like replication genes and proviral segments in neighboring regions of M. demolitor chromosomes provides further evidence for their shared evolutionary history (Strand and Burke 2015). Future assemblies with new long-read sequencing technologies generating chromosome-length scaffolds will provide further insight into the location of viral genome components relative to each other. These data will help to determine whether proviral segment loci and nudivirus-like replication genes are limited to either single or multiple chromosomes in the wasp genome, which will provide information about the events leading to the inception of viral sequences in the wasp genome and their maintenance over time.
Acknowledgments
This work was supported by the National Institutes of Health (F32 AI096552) (GRB), the US National Science Foundation (IOS-12611328) (MRS) and (DEB-1622986) (GRB), the US Department of Agriculture (2009-35302-05250) (MRS JBW) and (2007-35604-17756, 2011-67009-30134) (HMR) the USDA National Institute of Food and Agriculture Hatch project 1013423 (GRB) and the University of Georgia’s Research Foundation, Office for the Vice President for Research, and Agricultural Experiment Station.
Footnotes
Communicating editor: S. L. Celniker
Literature Cited
- Annaheim M., Lanzrein B., 2007. Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers and abundance of proviral and excised segments. J. Gen. Virol. 88: 450–457. 10.1099/vir.0.82396-0 [DOI] [PubMed] [Google Scholar]
- Beck M. H., Inman R. B., Strand M. R., 2007. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 359: 179–189. 10.1016/j.virol.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Bewick A. J., Vogel K. J., Moore A. J., Schmitz R. J., 2017. Evolution of DNA methylation across insects. Mol. Biol. Evol. 34: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bézier A., Annaheim M., Herbinière J., Wetterwald C., Gyapay G., et al. , 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323: 926–930. 10.1126/science.1166788 [DOI] [PubMed] [Google Scholar]
- Bézier A., Louis F., Jancek S., Periquet G., Thézé J., et al. , 2013. Functional endogenous viral elements in the genome of the parasitoid wasp Cotesia congregata: insights into the evolutionary dynamics of bracoviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20130047 10.1098/rstb.2013.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitra K., Burke G. R., Strand M. R., 2016. Permissiveness of lepidopteran hosts is linked to differential expression of bracovirus genes. Virology 492: 259–272. 10.1016/j.virol.2016.02.023 [DOI] [PubMed] [Google Scholar]
- Burke G. R., 2016. Analysis of genetic variation across the encapsidated genome of Microplitis demolitor bracovirus in parasitoid wasps. PLoS One 11: e0158846 10.1371/journal.pone.0158846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke G. R., Simmonds T. J., Thomas S. A., Strand M. R., 2015. Microplitis demolitor bracovirus proviral loci and clustered replication genes exhibit distinct DNA amplification patterns during replication. J. Virol. 89: 9511–9523. 10.1128/JVI.01388-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke G. R., Strand M. R., 2012. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor Bracovirus. J. Virol. 86: 3293–3306. 10.1128/JVI.06434-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke G. R., Strand M. R., 2014. Systematic analysis of a wasp parasitism arsenal. Mol. Ecol. 23: 890–901. 10.1111/mec.12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke G. R., Thomas S. A., Eum J. H., Strand M. R., 2013. Mutualistic polydnaviruses share essential replication gene functions with pathogenic ancestors. PLoS Pathog. 9: e1003348 10.1371/journal.ppat.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke G. R., Walden K. K., Whitfield J. B., Robertson H. M., Strand M. R., 2014. Widespread genome reorganization of an obligate virus mutualist. PLoS Genet. 10: e1004660 10.1371/journal.pgen.1004660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C. A., Gundersen-Rindal D. E., Hostetler J. B., Tallon L. J., Fadrosh D. W., et al. , 2008. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9: R183 10.1186/gb-2008-9-12-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E., Dupuy C., Huguet E., Cattolico L., Provost B., et al. , 2004. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306: 286–289. 10.1126/science.1103066 [DOI] [PubMed] [Google Scholar]
- Geib S. M., Liang G. H., Murphy T. D., Sim S. B., 2017. Whole genome sequencing of the braconid parasitoid wasp Fopius arisanus, an important biocontrol agent of pest tepritid fruit flies. G3 (Bethesda) 7: 2407–2411. 10.1534/g3.117.040741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herniou E. A., Huguet E., Thézé J., Bézier A., Periquet G., et al. , 2013. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20130051 10.1098/rstb.2013.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis F., Bézier A., Periquet G., Ferras C., Drezen J. M., et al. , 2013. The bracovirus genome of the parasitoid wasp Cotesia congregata is amplified within 13 replication units, including sequences not packaged in the particles. J. Virol. 87: 9649–9660. 10.1128/JVI.00886-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18 (erratum: Gigascience 4: 1) 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy N., Banks J. C., Whitfield J. B., Austin A. D., 2008. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol. Phylogenet. Evol. 47: 378–395. 10.1016/j.ympev.2008.01.022 [DOI] [PubMed] [Google Scholar]
- Savary S., Beckage N., Tan F., Periquet G., Drezen J. M., 1997. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastinae) at potential recombinase binding sites. J. Gen. Virol. 78: 3125–3134. 10.1099/0022-1317-78-12-3125 [DOI] [PubMed] [Google Scholar]
- Shepard M., Powell J. E., Jones W. A., 1983. Biology of Microplitis demolitor (Hymenoptera: Braconidae), an imported parasitoid of Heliothis (Lepidoptera: Noctuidae) spp. and the soybean looper, Pseudoplusia includens (Lepidoptera: Noctuidae). Environ. Entomol. 12: 641–645. 10.1093/ee/12.3.641 [DOI] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Stoltz D. B., Vinson S. B., 1979. Viruses and parasitism in insects. Adv. Virus Res. 24: 125–171. 10.1016/S0065-3527(08)60393-0 [DOI] [PubMed] [Google Scholar]
- Strand M. R., 1994. Microplitis demolitor polydnavirus infects and expresses in specific morphotypes of Pseudoplusia includens haemocytes. J. Gen. Virol. 75: 3007–3020. 10.1099/0022-1317-75-11-3007 [DOI] [PubMed] [Google Scholar]
- Strand M. R., Burke G. R., 2014. Polydnaviruses: nature’s genetic engineers. Annu. Rev. Virol. 1: 333–354. 10.1146/annurev-virology-031413-085451 [DOI] [PubMed] [Google Scholar]
- Strand M. R., Burke G. R., 2015. Polydnaviruses: from discovery to current insights. Virology 479–480: 393–402. 10.1016/j.virol.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand M. R., McKenzie D. I., Grassl V., Dover B. A., Aiken J. M., 1992. Persistence and expression of Microplitis demolitor polydnavirus in Pseudoplusia includens. J. Gen. Virol. 73: 1627–1635. 10.1099/0022-1317-73-7-1627 [DOI] [PubMed] [Google Scholar]
- Webb B. A., Strand M. R., Dickey S. E., Beck M. H., Hilgarth R. S., et al. , 2006. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347: 160–174. 10.1016/j.virol.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Webb B. A., Strand M. R., 2005. The biology and genomics of polydnaviruses, pp. 323–360 in Comprehensive Molecular Insect Science. Elsevier, Amsterdam. [Google Scholar]
- Werren J. H., Richards S., Desjardins C. A., Niehuis O., Gadau J., et al. , 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327: 343–348. 10.1126/science.1178028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Rokas A., Berger S. L., Liebig J., Ray A., et al. , 2015. Chemoreceptor evolution in Hymenoptera and its implications for the evolution of eusociality. Genome Biol. Evol. 7: 2407–2416. 10.1093/gbe/evv149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw sequencing data are available from the NCBI Sequence Read Archive (see Table 1 for accessions). The genome assembly, WGS Project AZMT02, is represented as BioSample SAMN02708865 with identical records in GenBank as accession GCA_000572035.2 and RefSeq as accession GCF_000572035.2 with the name Mdem2. All current annotations are associated with the RefSeq assembly as release 101. An FTP site for data download is at ftp://ftp.ncbi.nlm.nih.gov/genomes/Microplitis_demolitor/. NCBI’s Genome Data Viewer can be accessed at https://www.ncbi.nlm.nih.gov/genome/gdv/?acc=GCF_000806365.1&context=genome and an overview of release 100 annotations can be accessed at https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Microplitis_demolitor/101. Curation of this assembly and consolidated sequence-based resources are hosted by the i5k Workspace (https://i5k.nal.usda.gov/) allowing visualization within jBrowse, manual curation with Apollo and other tools.
Table 1. Raw reads generated for assembly.
| SRA | Library type | Read pairs | Base pairs | Material |
|---|---|---|---|---|
| SRX610757 | 180 bp | 138.9 M | 17.5 Gb | Single adult male |
| SRX641381 | 1.5 kb | 164.3 M | 20.9 Gb | 40 pooled adult male wasps |
| SRX641400 | 5 kb | 53.6 M | 7.1 Gb | 40 pooled adult male wasps |
| SRX641403 | 10 kb | 174.3 M | 22.1 Gb | 100 pooled adult male wasps |
| SRX641469 | 20 kb | 230.2 M | 28.3 Gb | 100 pooled adult male wasps |
| SRX981480 | 5 kb | 37.8 M | 5.1 Gb | 120 pooled adult male wasps |