Abstract
The Mediator complex is a central component of transcriptional regulation in Eukaryotes. The complex is structurally divided into four modules known as the head, middle, tail and kinase modules, and in Arabidopsis thaliana, comprises 28-34 subunits. Here, we explore the functions of four Arabidopsis Mediator tail subunits, MED2, MED5a/b, MED16, and MED23, by comparing the impact of mutations in each on the Arabidopsis transcriptome. We find that these subunits affect both unique and overlapping sets of genes, providing insight into the functional and structural relationships between them. The mutants primarily exhibit changes in the expression of genes related to biotic and abiotic stress. We find evidence for a tissue specific role for MED23, as well as in the production of alternative transcripts. Together, our data help disentangle the individual contributions of these MED subunits to global gene expression and suggest new avenues for future research into their functions.
Keywords: Mediator, Arabidopsis, transcription regulation, gene expression
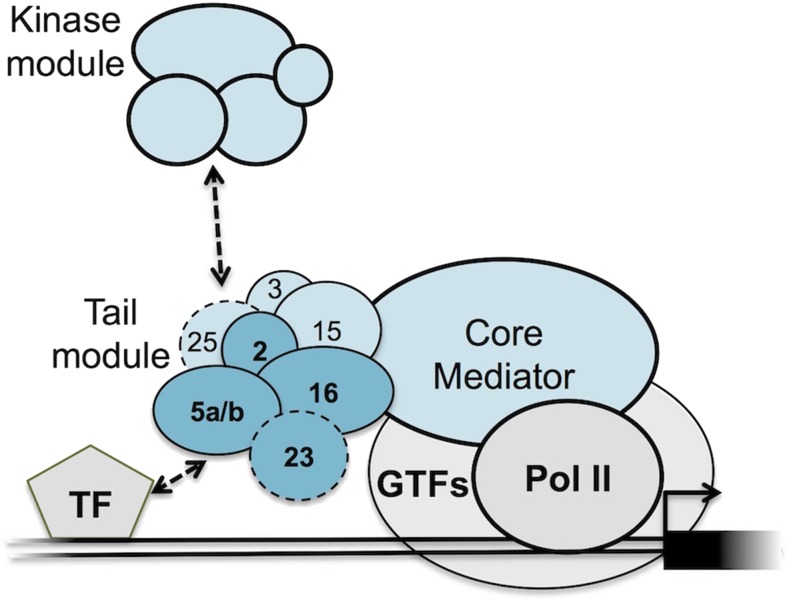
The Mediator complex is an essential co-regulator of eukaryotic transcription, participating in many of the events surrounding transcription initiation (Kelleher et al. 1990; Flanagan et al. 1990; Thompson et al. 1993; Kim et al. 1994; Poss et al. 2013; Allen and Taatjes 2015). Mediator bridges the divide between enhancer-bound transcription factors and promoter-bound RNA Polymerase II (Pol II) to facilitate assembly and function of the preinitiation complex. The individual subunits of the complex have been assigned to four modules, known as the head, middle, tail, and kinase modules, based on their positions within the complex (Figure 1). The head and middle modules contact Pol II, while the tail module primarily interacts with transcription activators (Figure 1; Koh et al. 1998; Myers et al. 1999; Lee et al. 1999; Park et al. 2000; Zhang et al. 2004; Jeronimo et al. 2016; Tsai et al. 2017). The kinase module reversibly associates with the rest of the complex and is thought to play a negative regulatory role by inhibiting interaction of Mediator with Pol II (Elmlund et al. 2006; Knuesel et al. 2009; Tsai et al. 2013). The core Mediator complex has recently been redefined as just the middle and head modules as they are the minimal components required for Mediator to stimulate transcription (Cevher et al. 2014; Plaschka et al. 2015; Jeronimo et al. 2016). Although the core is capable of functioning independently, the majority of evidence suggests that the tail is associated with the core under most circumstances.
Figure 1.
Model of the Arabidopsis Mediator complex. Core Mediator interacts with RNA Pol II and the general transcription factors (GTFs). The tail module (numbered subunits) interacts with DNA-bound transcription factors (TF and the dissociable kinase module. Dark blue subunits are those studied here. The positions of the subunits outlined with dashed lines are not well determined.
Given that the middle and head modules can be recruited to promoters and facilitate preinitiation complex (PIC) assembly independent of the tail module, it appears that a major role of the tail is to increase the probability of Mediator-PIC interactions by recruiting and tethering the complex to promoter-proximal transcription factors (Jeronimo et al. 2016); however, this does not appear to be the only role of the tail and many questions remain regarding its structure and function. The tail is highly flexible and has thus been difficult to visualize using the composite cryo-EM imaging techniques that have recently enabled high resolution structures of core Mediator (Tsai et al. 2017). In addition, many studies of Mediator structure have focused on yeast Mediator complexes, which lack some tail subunits found in humans and plants (Bourbon 2008). Structural, genetic, and functional data from a number of organisms support the existence of two submodules within the tail, one comprising MED2, MED3, and MED15, and another comprising MED5, MED16, and MED23 (Li et al. 1995; Ito et al. 2002; Zhang et al. 2004; Béve et al. 2005; Robinson et al. 2015). Although loss of MED16 results in separation of the rest of the tail from the complex, the free MED2-MED3-MED15 submodule can still be recruited by transcription factors to activate transcription (Zhang et al. 2004; Galdieri et al. 2012). Aside from its role in recruiting Mediator to promoters, the tail module also facilitates reinitiation by helping to maintain a scaffold PIC (Reeves and Hahn 2003). Negative regulation of transcription also occurs through the tail in some instances. CDK8, the enzymatically active subunit of the kinase module, has been shown to phosphorylate both MED2 and MED3, resulting in gene repression (van de Peppel et al. 2005; Gonzalez et al. 2014).
In Arabidopsis, Mediator tail subunits have been shown to be required for the regulation of a variety of processes (reviewed in Yang et al. 2015; Samanta and Thakur 2015a; Dolan et al. 2017). Mediator tail subunits MED16 and MED25 are two of the most extensively studied Arabidopsis MED subunits. MED16/SFR6 was first identified for its role in freezing tolerance and MED25/PFT1 for its role in promoting flowering (Knight 1999; Cerdán and Chory 2003). Since then, both have been shown to function extensively in the regulation of defense-related genes, as well as a number of other processes (Boyce et al. 2003; Knight et al. 2009; Kidd et al. 2009; Elfving et al. 2011; Xu and Li 2011; Wathugala et al. 2012; Chen et al. 2012; Çevik et al. 2012; Sundaravelpandian et al. 2013; Hemsley et al. 2014; Yang et al. 2014; Raya-González et al. 2014; Zhang et al. 2014; Seguela-Arnaud et al. 2015; Zhu et al. 2015; Wang et al. 2016; Muñoz-Parra et al. 2017; Dolan et al. 2017). MED2 has been less well studied, but has been shown to share some functions with MED14 and MED16 in cold-regulated gene expression (Hemsley et al. 2014). MED5a/b also share some functions with MED14, and MED16, but in the regulation of dark induced gene expression (Hemsley et al. 2014). From these studies and others it has become increasingly apparent that normal gene expression requires the concerted action of multiple MED subunits, making it difficult to disentangle the functions of individual subunits (e.g., Figure 4 in Yang et al. 2015). This fact was highlighted by the observation that nine different MED subunits are required for methyl-jasmonate induced expression of PDF1.2 (Wang et al. 2016).
Previously, we showed that MED2, MED16, and MED23 are differentially required for the function of ref4-3, a semi-dominant MED5b mutant that negatively impacts phenylpropanoid accumulation (Dolan et al. 2017). In the present study, we explore the effects of disrupting MED2, MED5a/b, MED16, and MED23 on genome-wide transcription to gain a broader understanding of their roles in gene regulation and their functional relationships to one another. As expected, we find that these subunits have both distinct and overlapping roles in gene regulation. These data lay a foundation for teasing apart the individual contributions of these MED subunits to the expression of different pathways and genes, and more importantly, for understanding how they function as a unit.
Methods
Plant Materials and Growth
Arabidopsis thaliana (ecotype Columbia-0) was grown in Redi-earth Plug and Seedling Mix (Sun Gro Horticulture, Agawam, MA) at a temperature of 23°, under a long-day (16 hr light/8 hr dark) photoperiod with a light intensity of 100 μE m−2 s−1. Seeds were planted nine per 4” x 4” pot and held for two days at 4° before transferring to the growth chamber.
Salk insertion lines were obtained from the Arabidopsis Biological Resource Center (Ohio State University) unless otherwise noted. The insertion lines used in this study include: med5b-1/ref4-6 (SALK_ 037472), med5a-1/rfr1-3 (SALK_011621) (Bonawitz et al. 2012), med2-1 (SALK_023845C) (Hemsley et al. 2014), sfr6-2 (SALK_048091) (Knight et al. 2009). The med2-1, and med23-4 mutants were provided to us by Dr. Tesfaye Mengiste (Department of Botany and Plant Pathology, Purdue University). The med16-1/sfr6-2 mutant was provided by Dr. Zhonglin Mou (Department of Microbiology and Cell Science, University of Florida). Homozygous Salk lines were genotyped as previously described (Dolan et al. 2017).
Calculation of Rosette Area
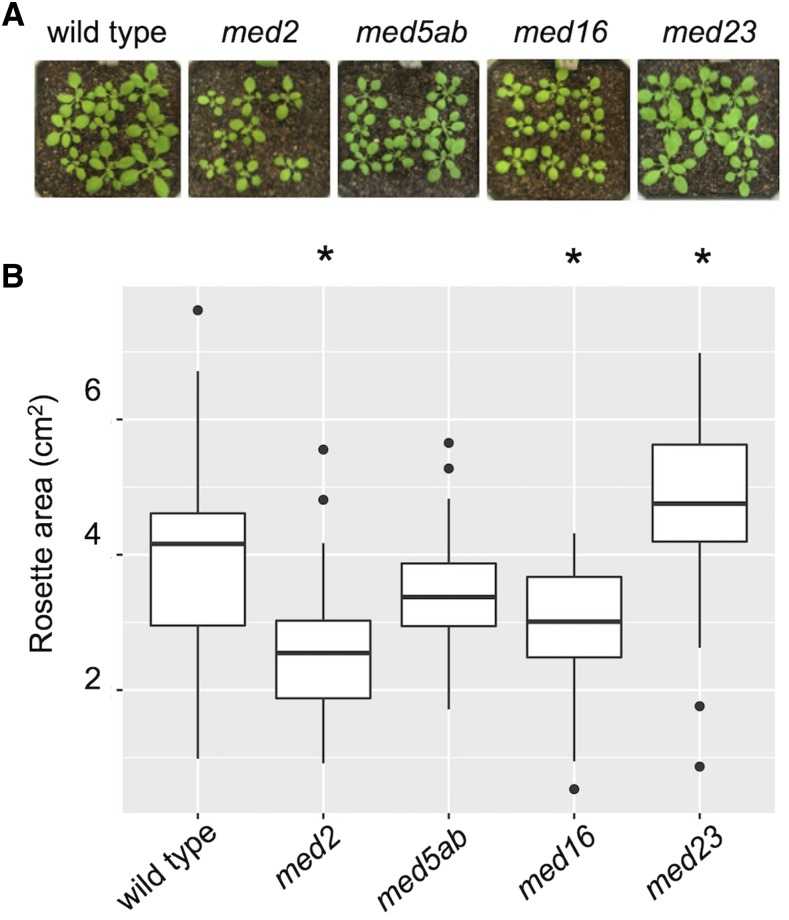
The same plants that were used for RNAseq were used to determine average rosette area. Seventeen days after transfer to the growth chamber, plants were photographed as in Figure 2A. Fiji was used to calculate the visible area of each rosette from the images (Schindelin et al. 2012; Rueden et al. 2017).
Figure 2.
The med mutants grow similar to wild-type plants. (A) A representative subset of the 18-day-old plants used for RNAseq. (B) Rosette areas of wild-type and med mutant plants. Asterisks indicate P < 0.01 compared to wild type (t-test, n = 36-51)
Determination of Flowering Time
Pots were randomized within the growth chamber to minimize positional effects on growth. The number of rosette leaves was counted on the first day that the inflorescence reached or exceeded 1 cm and that day was recorded as the day of flowering.
RNA Extraction and Whole Transcriptome Sequencing
Samples were collected for whole transcriptome sequencing (RNAseq) 18 days after transfer to the growth chamber, 6.5 hr after subjective dawn. For each of the four biological replicates, five whole rosettes were pooled from five different pots with randomized locations and immediately flash frozen in liquid nitrogen. Samples were then stored at -80° until RNA extraction. For RNA extraction, the pooled rosettes were ground to a powder under liquid nitrogen using a chilled mortar and pestle. Approximately 80 mg of ground tissue was then transferred to an Eppendorf tube for RNA extraction using the RNEasy Plant Mini kit from Qiagen (Qiagen, Chatsworth, CA). Total RNA was submitted to the Purdue Genomics Core Facility (Purdue University) for purification of polyA+ RNA, library construction, and sequencing. All samples were dual-barcoded, pooled, and loaded onto 4 sequencing lanes. Paired-end, 100 bp sequencing was performed by an Illumina HiSeq2500 machine run in “rapid” mode (Illumina, San Diego, CA). Read mapping was performed by the Purdue Genomics Core using the TAIR10 genome build and Tophat v. 2.1.0 (Trapnell et al. 2009). Transcriptome data has been deposited with the Gene Expression Omnibus under accession GSE95574.
Statistical Analysis of RNAseq Data
RNAseq data were acquired as described previously (Dolan et al. 2017). Briefly, digital gene expression (counts) for every exon was determined using the HTSeq-count program with the intersection “nonempty” option (Anders et al. 2015). Counts were summarized by gene ID. The edgeR program was used for differential gene expression analysis (Robinson et al. 2010). The analysis began with a count table comprising 33,602 genes. Genes expressed at low levels were filtered out by removing any genes for which there was not at least 1 count per million in at least four of the samples. This resulted in a list of 18,842 expressed genes. The exact test for the negative binomial distribution was then used to identify genes that were differentially expressed in the med mutants compared to wild type (FDR < 0.01) (Robinson and Smyth 2008). The results of these analyses are available in Supplemental File S1.
Gene ontology enrichment was performed using DAVID v6.8 (Huang et al. 2008). All genes that were expressed in our data set were used as the set of background genes for enrichment testing. GO terms were considered enriched if the associated Benjamini-Hochberg adjusted P value was less than 0.05. Where noted, redundant GO terms were removed for the purposes of reporting.
Alternative splicing analysis was performed using the procedure provided in the edgeR package (Robinson et al. 2010). Testing was performed between each med mutant and wild type using the diffSpliceDGE function (Lun et al. 2016). Simes’ method was used to convert exon-level P values to genewise P values. Genes with an FDR < 0.05 were considered as having alternatively spliced transcripts.
The Athena analysis suite was used to identify and test for enrichment of transcription factor binding motifs within 1000 bp of the transcription start site of each gene. Motifs were considered enriched if the associated P value was less than 10E-4 (cutoff recommended by the Athena developers based on a Bonferroni correction).
Data and Reagent Availability
Gene expression data has been deposited with the Gene Expression Omnibus under accession GSE95574. File S1 has been uploaded to the GSA Figshare portal. File S1 contains the results of our differential expression analysis. Arabidopsis MED T-DNA lines are available upon request. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6864170.
Results
The med tail mutants exhibit minor changes in development
We previously isolated homozygous T-DNA lines of MED2, MED5a, MED5b, MED16, and MED23, and showed that full-length transcripts of the genes in which the insertions are located are either abolished or substantially reduced (Dolan et al. 2017). Using the med5a and med5b mutants, we created a med5ab double mutant, as the proteins that they encode appear to be largely interchangeable within the complex (Bonawitz et al. 2012). Under our growth conditions, all of these med mutants develop similarly to wild-type plants (Figure 2A and Dolan et al. 2017), with a few exceptions. The med2 plants fail to stand erect as they get taller, indicating that they have weakened inflorescences (Dolan et al. 2017). In addition, med2 and med16 rosettes are slightly smaller and med23 rosettes are slightly larger than those of wild type (Figure 2B). We also observed that med2 and med5ab plants flower early (discussed in more detail below), whereas med16 is known to be late-flowering (Knight et al. 2008).
med tail mutants have unique effects on the transcriptome
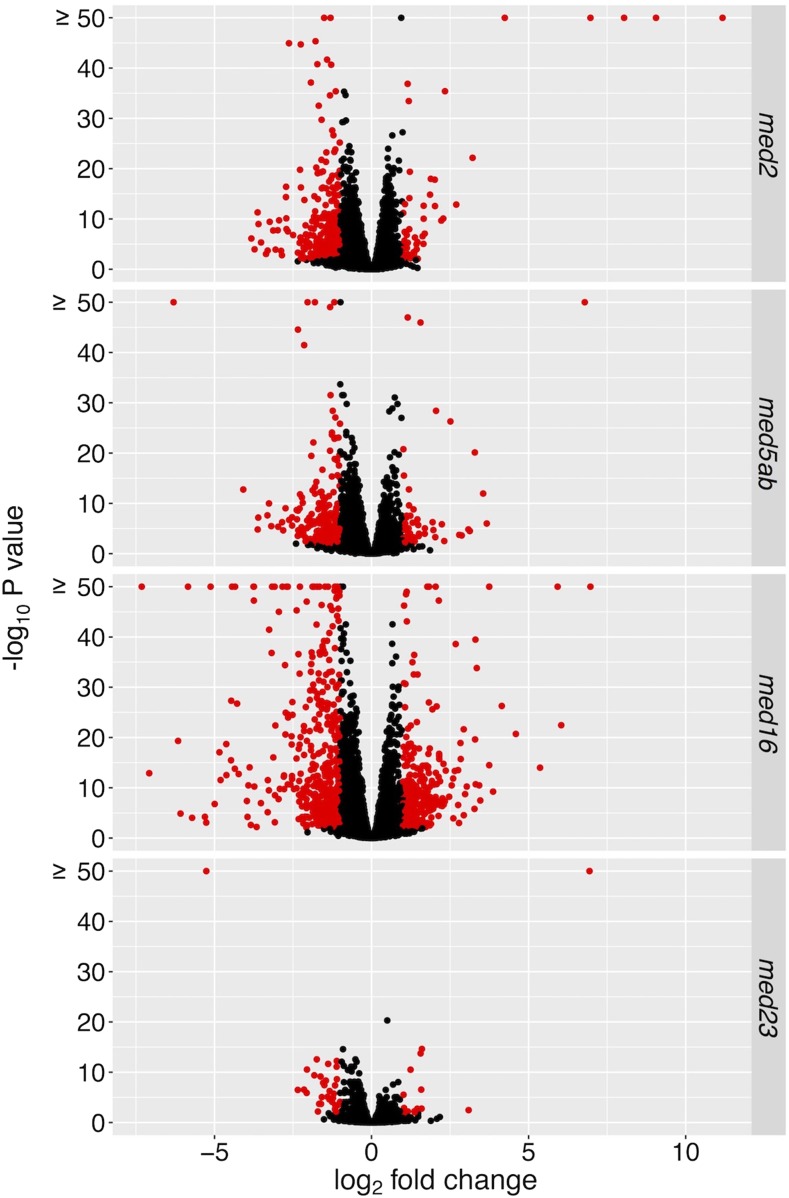
We grew all of the med mutants under a 16 h light, 8 h darkness cycle for 18 days, at which time we collected whole-rosettes for RNA extraction followed by RNAseq analysis (Figure 2 and Dolan et al. 2017). In our previous analysis of the data, we showed that in the med2 and med5ab mutants, significantly more genes are downregulated than upregulated (Dolan et al. 2017). We also showed that many more genes are differentially expressed in med16 than in the other mutants, with a similar number of genes being up- or downregulated, and that in med23 very few genes are differentially expressed (Dolan et al. 2017). As our previous analysis was limited to the role of these genes in the regulation of phenylpropanoid biosynthesis and as suppressors of ref4-3, we sought to more broadly characterize their functions in global gene expression. Here, we analyzed the same data in more detail using a stricter false discovery rate (FDR) of 0.01 and the same twofold change minimum cutoff in order to generate a high-confidence list of differentially expressed genes (DEGs) for each mutant (Figure 3). Using these criteria we found that there were 364 DEGs in med2 (53↑, 311↓), 305 DEGs in med5ab (66↑, 239↓), 768 DEGs in med16 (289↑, 479↓), and 47 DEGs in med23 (15↑, 33↓).
Figure 3.
Volcano plots showing differential gene expression in the med mutants. Genes with an adjusted P value of < 0.01 and a log2 fold-change ≥1 are highlighted in red.
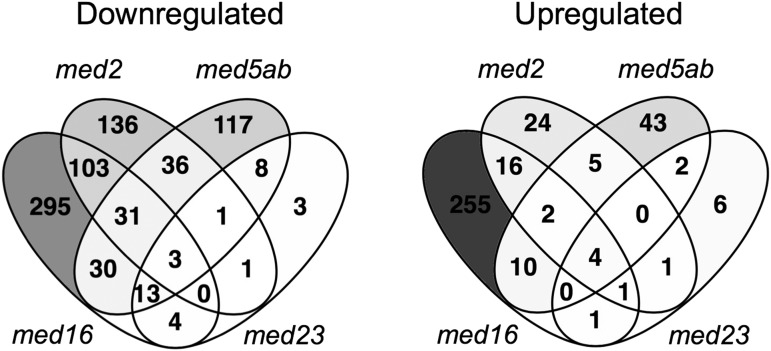
Comparison of the genes that were differentially expressed in each of the four med mutants showed that there were a large number of DEGs unique to each line, except for med23. Most of the DEGs in med23 were also differentially expressed in med5ab and/or med16 (Figure 4). There were also a large number of genes (119) that were shared only by med2 and med16. Only three genes were downregulated in all four mutants. They are DRM2, which encodes an auxin/dormancy associated protein, ERF105, which encodes an ethylene responsive transcription factor, and AT1G35210, which encodes a hypothetical, chloroplast localized protein. Similarly, only four genes were upregulated in all four mutants. They include one gene from the copia-like retrotransposon family (AT5G35935), one gene from the gyspy-like retrotransposon family (AT5G28335), the 5.8S rRNA gene (AT3G41979), and a gene that encodes a defensin-like family protein (AT2G16367). Given that there were so few DEGs in med23, it was not surprising that there was so little overlap between all four mutants. For this reason, we also looked at the DEGs shared just by med2, med5ab, and med16 (Table 1). Among the 31 genes that are downregulated in the three mutants, there is no significant enrichment of any gene ontology (GO) terms. The three mutants share only two upregulated genes, those being MYB47 and SAUR12.
Figure 4.
Overlap in downregulated or upregulated genes between the med mutants. Includes all genes that were differentially expressed compared to wild type (FDR <0.01) with an absolute log2 fold change ≥ 1.
Table 1.
Genes that are differentially expressed in the med2, med5ab and med16 mutants compared to wild type.
| AGI | GENE DESCRIPTION | LOG2 FOLD-CHANGE | ||
|---|---|---|---|---|
| med2 | med5ab | med16 | ||
| UPREGULATED | ||||
| AT1G18710 | MYB DOMAIN PROTEIN 47 (MYB47) | 1.69 | 1.49 | 3.31 |
| AT2G21220 | SAUR-LIKE AUXIN-RESPONSIVE PROTEIN FAMILY (SAUR12) | 1.66 | 1.34 | 1.62 |
| DOWNREGULATED | ||||
| AT1G10070 | BRANCHED-CHAIN AMINO ACID TRANSAMINASE 2 (BCAT-2) | −1.57 | −1.28 | −1.44 |
| AT1G15125 | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | −1.52 | −2.19 | −4.47 |
| AT1G19380 | Protein of unknown function (DUF1195) | −1.31 | −1.12 | −1.77 |
| AT1G21100 | O-methyltransferase family protein | −1.14 | −1.21 | −1.63 |
| AT1G27020 | unknown protein | −1.56 | −1.25 | −1.16 |
| AT1G51820 | Leucine-rich repeat protein kinase family protein | −1.32 | −2.05 | −1.44 |
| AT1G69880 | THIOREDOXIN H-TYPE 8 (TH8) | −3.24 | −1.49 | −3.30 |
| AT1G73330 | DROUGHT-REPRESSED 4 (DR4) | −1.72 | −1.15 | −2.66 |
| AT2G05440 | GLYCINE RICH PROTEIN 9 (GRP9) | −1.47 | −4.08 | −4.35 |
| AT2G26560 | PHOSPHOLIPASE A 2A (PLA2A) | −1.74 | −1.91 | −2.53 |
| AT2G40330 | PYR1-LIKE 6 (PYL6) | −1.59 | −1.05 | −1.80 |
| AT2G43120 | RmlC-like cupins superfamily protein | −1.93 | −1.10 | −3.76 |
| AT3G10020 | unknown protein | −1.09 | −1.14 | −1.51 |
| AT3G22060 | Receptor-like protein kinase-related family protein | −1.59 | −1.26 | −1.23 |
| AT3G26200 | CYP71B22 | −2.25 | −1.17 | −3.74 |
| AT3G43828 | CACTA-like transposase family | −1.72 | −1.25 | −1.42 |
| AT3G48520 | CYP94B3 | −3.83 | −1.92 | −3.97 |
| AT3G49620 | DARK INDUCIBLE 11 (DIN11) | −2.29 | −1.61 | −1.74 |
| AT3G50010 | Cysteine/Histidine-rich C1 domain family protein | −1.07 | −2.29 | −2.67 |
| AT3G51400 | protein of unknown function (DUF241) | −1.80 | −1.01 | −2.26 |
| AT4G11460 | CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE 30 (CRK30) | −1.71 | −1.09 | −1.11 |
| AT4G15210 | BETA-AMYLASE 5 (BAM5) | −1.59 | −3.26 | −4.62 |
| AT4G33467 | unknown protein | −1.98 | −2.74 | −4.47 |
| AT4G35770 | SENESCENCE 1 (SEN1) | −1.54 | −1.76 | −2.15 |
| AT5G14360 | Ubiquitin-like superfamily protein | −1.71 | −1.41 | −1.70 |
| AT5G39890 | Protein of unknown function (DUF1637) | −1.99 | −1.14 | −1.66 |
| AT5G41761 | unknown protein | −1.14 | −1.91 | −4.84 |
| AT5G44420 | PLANT DEFENSIN 1.2 (PDF1.2) | −2.16 | −2.85 | −7.08 |
| AT5G51790 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | −1.85 | −1.08 | −1.27 |
| AT5G56870 | BETA-GALACTOSIDASE 4 (BGAL4) | −1.06 | −1.04 | −1.05 |
| AT5G62360 | Plant invertase/pectin methylesterase inhibitor superfamily protein | −1.04 | −1.92 | −4.44 |
To determine how the expression profiles of the mutants correlate more broadly, we compared the expression of all DEGs that had an FDR < 0.01 in at least one of the mutants (Figure 5A). This approach revealed a positive correlation in the expression profiles of med5ab and med23 (r = 0.61). There was little correlation between the other expression profiles with med5ab and med16 being the most different from one another. Stronger correlations were observed when the comparisons were limited to only those genes that met the FDR cutoff in both mutants (Figure 5B), except in the case of med16 and med5ab, in which many genes were differentially expressed in opposite directions.
Figure 5.
Pairwise comparison of the gene expression profiles of the med mutants. Scatter plots comparing the log2 fold change in expression compared to wild type of genes that are (A) differentially expressed in any of the four mutants (FDR < 0.01) or (B) differentially expressed in both mutants being compared. The Pearson (r) correlation is given for each pair of comparisons.
MED tail mutants affect different biological processes
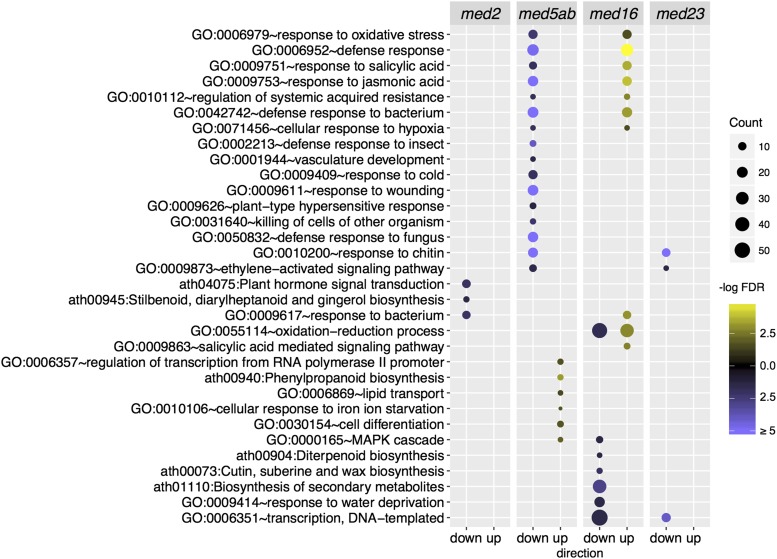
Gene ontology (GO) term enrichment analysis of the DEGs in each of the mutants showed substantial differences in the pathways and processes affected (Figure 6). Defense and cellular stress pathways are upregulated in med16, whereas the same pathways are downregulated in med5ab. Several other defense pathways are downregulated in med5ab that are not affected in the other mutants, as are “vasculature development” and “response to cold”.
Figure 6.
Gene ontology enrichment among genes that are differentially expressed in the med mutants. Enrichment of “Biological process” GO-terms and KEGG pathways. Terms that were largely redundant were removed. Direction indicates the subset of genes with increased or decreased expression in each of the mutants compared to wild type (FDR < 0.01, absolute log2 fold change ≥ 1). The brightness of the circles indicates the significance of the term or pathway (-log FDR) and their size indicates the number of genes that are associated with that term or pathway.
In med16, many genes related to the biosynthesis of secondary metabolites, response to water deprivation, and transcription, are downregulated. Among the 311 genes that are downregulated in med2, only three GO-terms are enriched; they are, “plant hormone signal transduction”, “response to bacterium”, and “stillbenoid, diarylheptanoid and gingerol biosynthesis”. Likewise, only three GO-terms are enriched in the med23 mutant and all are downregulated. They include “response to chitin” and “ethylene-activated signaling pathway”, which are shared with med5ab, and “transcription, DNA-templated”, which is shared with med16.
Hierarchical clustering identifies genes that require different subsets of MED tail subunits for their proper expression
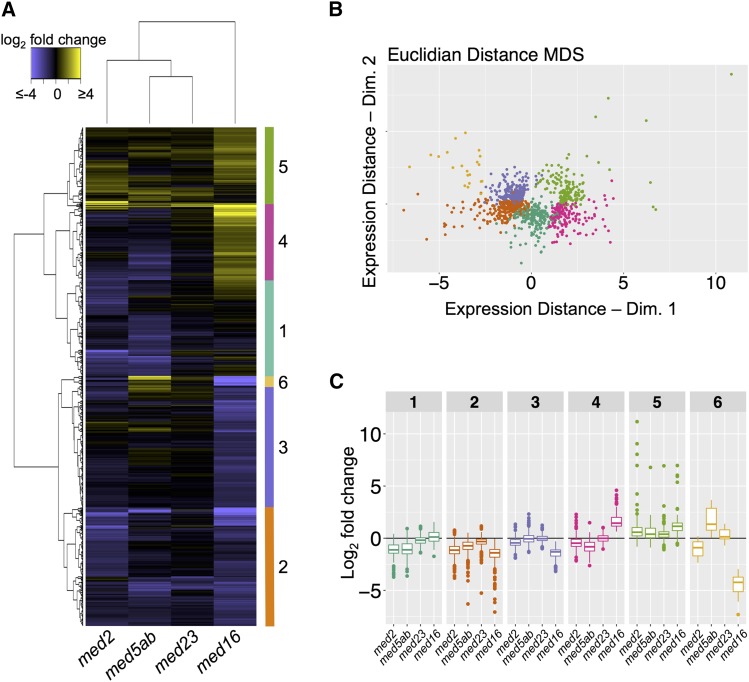
To identify groups of genes that behave similarly or differently in the med mutants, we performed hierarchical clustering using the complete set of 1080 DEGs (Figure 7). Six major gene clusters were identified (Figure 7A and B). Cluster 1 contains genes that are largely downregulated in med2 and med5ab and is enriched for defense-related genes (Figure 7C, Table 2). Cluster 2 contains genes that are downregulated in all of the mutants and is enriched
Figure 7.
Hierarchical clustering of all genes differentially expressed in the med mutants. (A) Hierarchical clustering of log2 fold change expression values. (B) Multidimensional scaling of differentially expressed genes based on their log2 fold change expression values and colored by cluster membership. (C) Boxplots representing the fold-change values according to genotype and cluster membership in A and B.
Table 2. Gene ontology enrichment of gene clusters in Figure 7.
| CLUSTER | CATEGORY | TERM | COUNT | %a | BH PVALb |
|---|---|---|---|---|---|
| TOTAL | |||||
| CLUSTER 1 | GOTERM_BP_DIRECT | GO:0050832∼defense response to fungus | 16 | 7.69 | 1.50E-06 |
| 208 | GOTERM_BP_DIRECT | GO:0042742∼defense response to bacterium | 18 | 8.65 | 2.99E-06 |
| GOTERM_CC_DIRECT | GO:0016021∼integral component of membrane | 72 | 34.62 | 6.94E-05 | |
| GOTERM_BP_DIRECT | GO:0010200∼response to chitin | 11 | 5.29 | 8.34E-05 | |
| GOTERM_CC_DIRECT | GO:0005886∼plasma membrane | 57 | 27.40 | 1.75E-04 | |
| GOTERM_CC_DIRECT | GO:0005576∼extracellular region | 32 | 15.38 | 2.51E-04 | |
| GOTERM_BP_DIRECT | GO:0010112∼regulation of systemic acquired resistance | 5 | 2.40 | 7.60E-04 | |
| GOTERM_BP_DIRECT | GO:0009611∼response to wounding | 12 | 5.77 | 9.69E-04 | |
| KEGG_PATHWAY | ath04626:Plant-pathogen interaction | 9 | 4.33 | 1.13E-03 | |
| GOTERM_BP_DIRECT | GO:0009751∼response to salicylic acid | 10 | 4.81 | 2.43E-03 | |
| GOTERM_BP_DIRECT | GO:0006952∼defense response | 18 | 8.65 | 2.50E-03 | |
| GOTERM_BP_DIRECT | GO:0009753∼response to jasmonic acid | 10 | 4.81 | 2.55E-03 | |
| GOTERM_BP_DIRECT | GO:0009617∼response to bacterium | 8 | 3.85 | 3.21E-03 | |
| GOTERM_MF_DIRECT | GO:0030246∼carbohydrate binding | 11 | 5.29 | 4.38E-03 | |
| GOTERM_BP_DIRECT | GO:0012501∼programmed cell death | 4 | 1.92 | 2.70E-02 | |
| CLUSTER 2 | |||||
| 257 | GOTERM_BP_DIRECT | GO:0009414∼response to water deprivation | 14 | 5.45 | 1.72E-02 |
| KEGG_PATHWAY | ath04075:Plant hormone signal transduction | 9 | 3.50 | 4.09E-02 | |
| CLUSTER 3 | |||||
| 264 | GOTERM_CC_DIRECT | GO:0005576∼extracellular region | 46 | 17.42 | 2.29E-08 |
| GOTERM_MF_DIRECT | GO:0046983∼protein dimerization activity | 17 | 6.44 | 3.72E-07 | |
| GOTERM_MF_DIRECT | GO:0000977∼RNA polymerase II regulatory region sequence-specific DNA binding | 11 | 4.17 | 1.65E-05 | |
| GOTERM_BP_DIRECT | GO:0000165∼MAPK cascade | 8 | 3.03 | 1.10E-04 | |
| GOTERM_BP_DIRECT | GO:0045944∼positive regulation of transcription from RNA polymerase II promoter | 8 | 3.03 | 1.11E-03 | |
| GOTERM_MF_DIRECT | GO:0033946∼xyloglucan-specific endo-beta-1, 4-glucanase activity | 4 | 1.52 | 1.69E-03 | |
| GOTERM_MF_DIRECT | GO:0008794∼arsenate reductase (glutaredoxin) activity | 5 | 1.89 | 1.53E-03 | |
| KEGG_PATHWAY | ath01110:Biosynthesis of secondary metabolites | 23 | 8.71 | 1.25E-02 | |
| GOTERM_MF_DIRECT | GO:0051537∼2 iron, 2 sulfur cluster binding | 7 | 2.65 | 3.16E-03 | |
| GOTERM_MF_DIRECT | GO:0015035∼protein disulfide oxidoreductase activity | 8 | 3.03 | 3.22E-03 | |
| KEGG_PATHWAY | ath00073:Cutin, suberine and wax biosynthesis | 4 | 1.52 | 2.23E-02 | |
| GOTERM_MF_DIRECT | GO:0003700∼transcription factor activity, sequence-specific DNA binding | 30 | 11.36 | 1.21E-02 | |
| CLUSTER 4 | |||||
| 159 | GOTERM_BP_DIRECT | GO:0009753∼response to jasmonic acid | 13 | 8.18 | 9.35E-07 |
| GOTERM_BP_DIRECT | GO:0042742∼defense response to bacterium | 16 | 10.06 | 1.31E-06 | |
| GOTERM_BP_DIRECT | GO:0009617∼response to bacterium | 11 | 6.92 | 1.62E-06 | |
| GOTERM_BP_DIRECT | GO:0006952∼defense response | 20 | 12.58 | 3.13E-06 | |
| GOTERM_BP_DIRECT | GO:0009751∼response to salicylic acid | 9 | 5.66 | 1.76E-03 | |
| GOTERM_BP_DIRECT | GO:0009863∼salicylic acid mediated signaling pathway | 5 | 3.14 | 1.90E-03 | |
| GOTERM_BP_DIRECT | GO:0007165∼signal transduction | 12 | 7.55 | 7.09E-03 | |
| GOTERM_BP_DIRECT | GO:0050832∼defense response to fungus | 9 | 5.66 | 7.76E-03 | |
| GOTERM_BP_DIRECT | GO:0009627∼systemic acquired resistance | 5 | 3.14 | 1.04E-02 | |
| KEGG_PATHWAY | ath01110:Biosynthesis of secondary metabolites | 17 | 10.69 | 1.10E-02 | |
| GOTERM_BP_DIRECT | GO:0055114∼oxidation-reduction process | 21 | 13.21 | 1.13E-02 | |
| GOTERM_BP_DIRECT | GO:0009695∼jasmonic acid biosynthetic process | 4 | 2.52 | 2.24E-02 | |
| GOTERM_BP_DIRECT | GO:0080027∼response to herbivore | 3 | 1.89 | 2.69E-02 | |
| GOTERM_BP_DIRECT | GO:0009611∼response to wounding | 8 | 5.03 | 2.78E-02 | |
| KEGG_PATHWAY | ath00592:alpha-Linolenic acid metabolism | 4 | 2.52 | 2.92E-02 | |
| GOTERM_BP_DIRECT | GO:0002229∼defense response to oomycetes | 4 | 2.52 | 3.13E-02 | |
| GOTERM_CC_DIRECT | GO:0005576∼extracellular region | 23 | 14.47 | 3.74E-02 | |
| GOTERM_BP_DIRECT | GO:0009620∼response to fungus | 5 | 3.14 | 4.44E-02 | |
| CLUSTER 5 | |||||
| 172 | GOTERM_CC_DIRECT | GO:0005576∼extracellular region | 27 | 15.70 | 4.47E-04 |
| KEGG_PATHWAY | ath00940:Phenylpropanoid biosynthesis | 7 | 4.07 | 1.97E-03 | |
| CLUSTER 6 | |||||
| 21 | GOTERM_CC_DIRECT | GO:0005576∼extracellular region | 11 | 52.38 | 9.89E-05 |
| GOTERM_BP_DIRECT | GO:0010584∼pollen exine formation | 3 | 14.29 | 9.12E-03 |
Percentage of genes in input that are represented by a given gene ontology.
Benjamini-Hochberg-corrected P Value.
for genes related to water deprivation and hormone signal transduction (Figure 7C, Table 2. Cluster 3 contains genes that are downregulated in med16 and to some extent, med2 (Figure 7C, Table 2). Cluster 3 genes encode proteins involved in secondary metabolite biosynthesis, transcription regulation, and extracellular processes. Cluster 4 contains genes that are upregulated in med16 and downregulated in med2 and med5ab and is enriched for genes involved in numerous defense pathways (Figure 7C, Table 2). Cluster 5 contains genes that are upregulated in all of the mutants and is enriched for genes involved in phenylpropanoid biosynthesis and extracellular processes (Figure 7C, Table 2). Finally, cluster 6 contains genes that are strongly downregulated in med16 and upregulated in med5ab. These genes encode proteins involved in pollen exine formation and those that are localized to the extracellular region. Together, these data provide a basis for discovering pathways and processes that require the function of individual or multiple MED tail subunits for their regulation.
The med2 and med5ab mutants are early flowering
As previously mentioned, the med16 mutant is late flowering (Knight et al. 2008) and we initially observed that the med2 and med5ab mutants appeared to flower early. When we quantified this phenomenon, we found that med2 plants flowered an average of 2.1 days earlier and with 2.6 fewer rosette leaves than wild type plants (Figure 8A and B). Similarly, med5ab flowered an average of 1.5 days earlier than wild type and with 2.1 fewer rosettes leaves.
Figure 8.
med2 and med5ab are early flowering and have altered expression of flowering-related genes (A) Days after planting and (B) number of leaves at the time that the first inflorescence reached 1 cm. Asterisks indicate P < 0.01 when compared to wild type (t-test, n = 32-35). Boxes indicate the first quartile, the median, and the third quartile. The whiskers indicate the largest and smallest value no more than 1.5 times the interquartile range. Outliers are individually marked. (C) log2 fold change in expression compared to wild-type of flowering-related genes. Asterisks indicate genes with an FDR < 0.01. Genes that were not expressed are indicated in gray.
Consistent with the previously published results, med16 flowered an average of 8.9 days later and with 7.3 more leaves than wild type. Additionally, med23 plants had an average of 1.3 more leaves at the time of flowering.
In the med16 mutant, the late flowering phenotype was attributed to reduced expression of clock components, leading to a reduced expression of flowering genes, namely CO and FT (Knight et al. 2008). Although CO transcripts were not detectable in the samples we analyzed, expression of FT was strongly reduced in med16 (Figure 8C). In addition, expression of FLC, a negative regulator of the floral transition (Michaels and Amasino 1999), was increased in med16. Examination of the major genes involved in flowering did not reveal an obvious cause for the early flowering of med2 and med5ab (Figure 8C). In the case of med2, FLC is substantially upregulated without concomitant downregulation of it targets SOC1 and FT, suggesting that FLC might partially require MED2 for its function in repressing the floral transition. It is also possible that the effect of med2 and med5ab on flowering time is too subtle to be detected at the transcriptional level. In addition, the expression of many flowering and clock genes cycles diurnally, therefore differences in expression might be less apparent at the time we sampled the plants than at other times during the day.
MED23 and MED5a may have tissue-specific functions
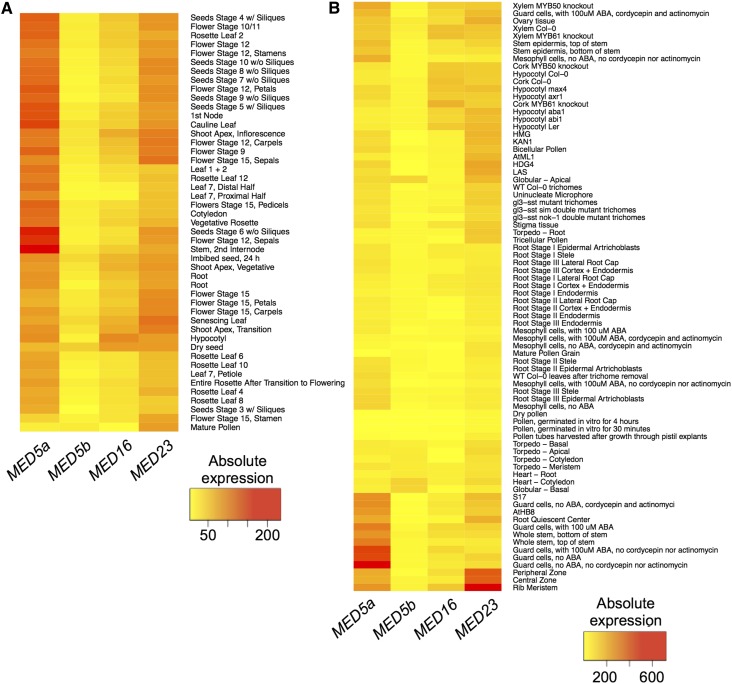
Only nine DEGs were identified in med23, four of which have not been characterized, lending little information as to whether MED23 has any unique functions in transcription regulation (Table 3). To explore whether MED23 might play a more predominant role in other organs or in particular tissues, we used the Arabidopsis eFP browser to compare the expression of the MED23, MED5a, MED5b and MED16 (data for MED2 was not available) during the development of different organs (Figure 9A) and in different tissues (Figure 9B) (Winter et al. 2007). MED23 was expressed in all organs, but was expressed more strongly in seeds, flowers, roots, and shoots than in leaves (Figure 9A). MED23 also showed substantial expression in mature pollen, whereas the other MED genes did not. Most striking, however, was the strong expression of MED23 in the shoot apical meristem (Figure 9B, “Peripheral zone”, “Central zone”, “Rib meristem”). These data suggest that MED23 might have specific functions in meristematic or reproductive development. This hypothesis is strengthened by the observation that the floral specification gene AGAMOUS (AG) is upregulated in med23 (Table 3), and that several other genes involved in embryo, floral, or meristem development are co-expressed with MED23 (Table 4) (ATTED-II v8.0, Aoki et al. 2016). The eFP data also showed that MED5a is more highly expressed than MED5b during most developmental stages, and has a much higher level of expression in guard cells than the other MED subunits we examined.
Table 3.
Genes that are differentially expressed in med23 compared to wild type
| AGI | LOG2 FOLD-CHANGE | BH P VALa | GENE DESCRIPTION |
| AT5G35935 | 6.94 | 1.60E-240 | copia-like retrotransposon family |
| AT4G08093 | 3.09 | 3.38E-03 | expressed protein |
| AT3G30122b | 1.60 | 2.38E-15 | expressed protein |
| AT5G28335 | 1.59 | 1.74E-03 | gypsy-like retrotransposon family |
| AT2G01008b | 1.58 | 2.87E-07 | unknown protein, best match MEE38 |
| AT3G41979 | 1.56 | 1.82E-14 | 5S rRNA |
| AT4G07850 | 1.46 | 1.79E-03 | gypsy-like retrotransposon family |
| AT2G16367 | 1.40 | 6.46E-03 | defensin-like (DEFL) family protein |
| AT3G44765 | 1.37 | 8.90E-03 | other RNA |
| AT2G05914 | 1.37 | 5.86E-03 | Natural antisense transcript overlaps with AT2G05915 |
| AT3G44970b | 1.24 | 3.12E-11 | Cytochrome P450 superfamily protein |
| AT1G30760 | 1.16 | 6.41E-03 | FAD-binding Berberine family protein |
| AT3G22415b | 1.06 | 9.44E-03 | unknown protein |
| AT3G19550b | 1.03 | 1.30E-03 | unknown protein |
| AT4G18960b | 1.02 | 2.98E-06 | AGAMOUS (AG) |
| AT1G23230b | −5.26 | 0.00E+00 | MEDIATOR COMPLEX SUBUNIT 23 (MED23) |
| AT1G35210 | −2.35 | 3.30E-07 | unknown protein |
| AT5G51190 | −2.15 | 2.93E-07 | Integrase-type DNA-binding superfamily protein |
| AT4G17490 | −2.07 | 1.28E-06 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6 (ERF6) |
| AT4G24570 | −2.06 | 2.92E-11 | DICARBOXYLATE CARRIER 2 (DIC2) |
| AT2G25735 | −1.82 | 4.05E-10 | unknown protein |
| AT3G44260 | −1.74 | 2.73E-13 | Polynucleotidyl transferase, ribonuclease H-like |
| AT3G29000 | −1.71 | 6.46E-03 | Calcium-binding EF-hand family protein |
| AT5G27420 | −1.67 | 1.96E-04 | CARBON/NITROGEN INSENSITIVE 1 (CNI1) |
| AT5G04340 | −1.64 | 1.68E-04 | ZINC FINGER OF ARABIDOPSIS THALIANA 6 (ZAT6) |
| AT5G61600 | −1.62 | 7.14E-10 | ETHYLENE RESPONSE FACTOR 104 (ERF104) |
| AT1G07135 | −1.61 | 2.06E-04 | glycine-rich protein |
| AT1G27730 | −1.58 | 8.85E-06 | SALT TOLERANCE ZINC FINGER (STZ) |
| AT5G47230 | −1.54 | 1.29E-08 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5 (ERF5) |
| AT5G56320 | −1.50 | 3.77E-08 | EXPANSIN A14 (EXPA14) |
| AT1G66090 | −1.45 | 4.66E-09 | Disease resistance protein (TIR-NBS class) |
| AT1G74290b | −1.40 | 7.78E-06 | alpha/beta-Hydrolases superfamily protein |
| AT1G53480 | −1.38 | 2.19E-12 | MTO 1 RESPONDING DOWN 1 (MRD1) |
| AT4G23810 | −1.34 | 2.41E-05 | WRKY family transcription factor (WRKY53) |
| AT3G30720 | −1.33 | 5.52E-07 | QUA-QUINE STARCH (QQS) |
| AT5G45340 | −1.26 | 7.79E-05 | CYP707A3 |
| AT2G33830 | −1.23 | 1.29E-05 | Dormancy/auxin associated family protein |
| AT5G23240 | −1.20 | 1.85E-06 | DNAJ heat shock N-terminal domain-containing protein |
| AT3G51860 | −1.17 | 2.28E-03 | CATION EXCHANGER 3 (CAX3) |
| AT2G38470 | −1.16 | 4.15E-08 | WRKY DNA-BINDING PROTEIN 33 (WRKY33) |
| AT2G01010 | −1.15 | 3.81E-04 | 18S rRNA |
| AT5G59820 | −1.13 | 6.84E-03 | C2H2-TYPE ZINC FINGER FAMILY PROTEIN (RHL41) |
| AT3G55980b | −1.11 | 7.92E-12 | SALT-INDUCIBLE ZINC FINGER 1 (SZF1) |
| AT2G47260 | −1.11 | 5.75E-13 | WRKY DNA-BINDING PROTEIN 23 (WRKY23) |
| AT3G16720 | −1.10 | 2.43E-09 | TOXICOS EN LEVADURA 2 (ATL2) |
| AT4G29780 | −1.08 | 3.69E-04 | unkown protein |
| AT5G26920 | −1.02 | 1.96E-04 | CAM-BINDING PROTEIN 60-LIKE G (CBP60G) |
| AT2G24600 | −1.00 | 7.72E-05 | Ankyrin repeat family protein |
Benjamini-Hochberg-corrected P value.
Genes that are differentially expressed in med23 but not the other med mutants.
Figure 9.
Expression of the MED5ab, MED5b, MED16 and MED23 across development of different organs and in different tissues. The (A) “Development” and (B) “Tissue” datasets were retrieved from the Arabidopsis eFP browser.
Table 4.
Top 40 genes coexpressed with MED23 according to mutual rank
| AGI | ALIAS | FUNCTION | MUTUAL RANKa |
| AT1G02080b | transcription | transcription regulators | 6.9 |
| AT1G48090 | calcium-dependent lipid-binding | calcium-dependent lipid-binding family protein | 7.1 |
| AT1G80070b | SUS2 | Pre-mRNA-processing-splicing factor | 7.9 |
| AT5G58410 | HEAT/U-box | HEAT/U-box domain-containing protein | 13 |
| AT4G39850 | PXA1 | peroxisomal ABC transporter 1 | 13.8 |
| AT3G13330 | PA200 | proteasome activating protein 200 | 16.2 |
| AT3G02260b | UMB1 | auxin transport protein (BIG) | 17.2 |
| AT1G50030b,c | TOR | target of rapamycin | 17.3 |
| AT5G23110 | Zinc finger | Zinc finger, C3HC4 type (RING finger) family protein | 20.4 |
| AT4G01290 | chorismate synthase | chorismate synthase | 20.5 |
| AT2G26780 | ARM repeat | ARM repeat superfamily protein | 22.4 |
| AT3G27670 | RST1 | ARM repeat superfamily protein | 22.9 |
| AT2G17930 | Phosphatidylinositol 3- and 4-kinase | Phosphatidylinositol 3- and 4-kinase family protein with FAT domain | 23.8 |
| AT1G20960b | emb1507 | U5 small nuclear ribonucleoprotein helicase, putative | 25.1 |
| AT1G54490b | XRN4 | exoribonuclease 4 | 26.3 |
| AT2G41700 | ABCA1 | ATP-binding cassette A1 | 29.1 |
| AT1G15780 | NRB4/MED15a | Mediator subunit 15a | 29.7 |
| AT5G61140b | helicase | U5 small nuclear ribonucleoprotein helicasea | 30.7 |
| AT5G51340 | Tetratricopeptide repeat (TPR)-like | Tetratricopeptide repeat (TPR)-like superfamily protein | 31.4 |
| AT3G57570 | ARM repeat | ARM repeat superfamily protein | 31.9 |
| AT5G15680 | ARM repeat | ARM repeat superfamily protein | 33 |
| AT4G00450c | MED12 | RNA polymerase II transcription mediators | 33.5 |
| AT3G15880c | WSIP2 | WUS-interacting protein 2 | 34.3 |
| AT3G51050 | FG-GAP repeat | FG-GAP repeat-containing protein | 37.5 |
| AT3G16830c | TPR2 | TOPLESS-related 2 | 40.3 |
| AT3G50590 | Transducin/WD40 repeat-like | Transducin/WD40 repeat-like superfamily protein | 45.2 |
| AT1G72390 | PHL | Phytochrome-dependent late-flowering | 45.5 |
| AT3G60240 | EIF4G | eukaryotic translation initiation factor 4G | 48 |
| AT5G16280 | Tetratricopeptide repeat (TPR)-like | Tetratricopeptide repeat (TPR)-like superfamily protein | 48.6 |
| AT3G08850c | RAPTOR1B | HEAT repeat; WD domain, G-beta repeat protein protein | 49 |
| AT1G55325c | MAB2/MED13 | RNA polymerase II transcription mediators | 49.9 |
| AT5G47010b | UPF1 | RNA helicase, putative | 50.2 |
| AT3G33530 | Transducin | Transducin family protein / WD-40 repeat family protein | 52 |
| AT2G32730 | proteasome | 26S proteasome regulatory complex, Rpn2/Psmd1 subunit | 52.5 |
| AT5G65750 | 2-oxoglutarate dehydrogenase | 2-oxoglutarate dehydrogenase, E1 component | 54 |
| AT3G07160 | GSL10 | glucan synthase-like 10 | 54.7 |
| AT2G28290c | SYD | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 55.1 |
| AT2G33730b | hydrolase | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 55.6 |
| AT5G51660b | CPSF160 | cleavage and polyadenylation specificity factor 160 | 58.8 |
| AT3G50380 | DUF1162 | Protein of unknown function (DUF1162) | 59 |
Based on ATTED-II data set Ath-m.v15-08.
Genes annotated as being involved in RNA processing.
Genes annotated as being involved in embryo, floral or meristem development.
MED tail mutants might affect alternative mRNA processing
Many genes involved in RNA processing are co-expressed with MED23 (Table 4). In humans, MED23 interacts with mRNA processing factors and is required for the alternative splicing and polyadenylation of a significant number of transcripts (Huang et al. 2012). To determine if MED23 or the other MED subunits examined here might be involved in alternative splicing in Arabidopsis, we queried our RNAseq data for differential splicing events using the diffSpliceDGE function in edgeR (Robinson et al. 2010). To detect alternative exon usage, diffSpliceDGE compares the log fold change of individual exons to that of the gene as a whole. Using an FDR cutoff of 0.05, we detected a handful of alternatively spliced (AS) transcripts in each of the mutants, with the most being found in med23 (Figure 10A). The vast majority of these were not differentially expressed at the level of the whole gene. GO-term enrichment analysis of the AS transcripts found in each mutant showed that genes encoding ribosomal proteins, membrane proteins, chloroplast localized proteins, vacuolar proteins, and cell wall proteins were enriched in all four mutants (FDR < 0.05). In addition, the UniProt keyword “alternative splicing” was also enriched in all four lists, indicating that many of these transcripts have previously been shown to be alternatively spliced. Of the approximately 30 alternative splicing events that we examined, all but one occurred at the 5′ or 3′ end of the gene (e.g., Figure 10B), with many occurring within the untranslated region. They also all exhibited relatively small fold-changes, such that they could not be identified from coverage maps by eye. Together, these results suggest that these MED subunits, particularly MED23, might influence alternative RNA processing, either directly or indirectly.
Figure 10.
Alternative splicing occurs in the 5′ and 3′ ends of genes in the med mutants. (A) Number of alternatively spliced transcripts in the med mutants (FDR < 0.05). (B) Two examples of transcripts that are alternatively spliced in med23. Log2 fold change in the expression of individual exons compared to that of the entire gene. Significant exons are highlighted in red.
One of the “alternative splicing” events appeared very different from the rest. AT1G64790 was detected as an alternatively spliced transcript in med2 and med23 because of a large number of reads that mapped to a region spanning the first and second introns of the gene that were not present in wild type (Figure 11). According to the Araport11 annotation of the Arabidopsis genome, this region produces a cluster of 24 nt small RNAs. It has also been shown to be differentially methylated in the C24 and Ler ecotypes, and undergoes transchromosomal methylation in F2 hybrids (Greaves et al. 2014). The derepression of this region suggests that the small RNAs that typically silence this region are not being produced. Mediator has previously been shown to be required for RNA-directed DNA methylation of repeats and transposons (Kim and Chen 2011), and our data suggest that MED2 and MED23 are specifically required for this process at some loci.
Figure 11.
A region that undergoes transcriptional gene silencing is derepressed in med2 and med23. (A) Read coverage and number of intron-spanning reads across (B) a region of chromosome 1, which includes a portion of the ILYTHIA gene, a hypothetical protein and a small RNA. Exons are indicated as black rectangles, UTRs are in gray. Coverage in (A) is from individual wild-type or mutant samples.
Discussion
The Mediator complex is an important hub of transcription regulation. Serving as a platform for the interaction of countless transcription factors, the complex plays an integral role in the development, response, and adaptation of Eukaryotes to their environments. As such, it is somewhat remarkable that, under favorable growth conditions, Arabidopsis is largely robust to perturbation of many Mediator complex subunits. Unlike mice, in which all MED knockouts tested have proved to be embryonic lethal, many of the single Arabidopsis MED mutants studied to date grow well enough in controlled environments that they are fertile (Yin and Wang 2014; Buendía-Monreal and Gillmor 2016). This makes Arabidopsis uniquely suited to studying the effects of disruption of the complex in a developing, multicellular eukaryote. Many studies of Arabidopsis MED mutants have examined the effects of disruption of one or a few MED subunits on a limited number pathways or genes. In the present study, we sought to gain a broader understanding of the function of the Arabidopsis tail module and the relative contributions of its subunits to genome-wide transcription by comparing the effects of mutations in four different MED tail subunits—MED2, MED5a/b, MED16, and MED23—on the transcriptome.
The T-DNA mutants studied here all developed without any major changes in morphology, exhibiting only minor differences in rosette size (Figure 2), enabling our analysis of gene expression to be unencumbered by changes that might arise due to gross differences in developmental programs (Figure 2). We did, however, observe that med16 flowered late, in accordance with previous reports (Knight et al. 2008), and that med2 and med5 flowered early (Figures 8A and 8B). In addition to med16, mutations in eight other MED subunits cause Arabidopsis to flower late (Reviewed in Yang et al. 2015). The med2 and med5ab mutants are unique in that they are the only MED mutants reported to date that cause plants to flower early. Given the large network of genes that impinge on flowering time, and the broad involvement of Mediator in transcriptional regulation, it is not surprising that so many MED mutants affect flowering time. The opposite flowering phenotypes of med2, med5ab and the other med mutants suggests that individual MED subunits can affect the same traits in different ways, likely by affecting the expression of different subsets of genes. Although our gene expression analysis pointed to a potential reason for the early flowering of med2, additional studies will be required to determine the mechanistic cause. At the time that rosettes were sampled for RNAseq analysis, some plants had formed an apical bud. This may explain why genes related related to pollen exine formation appeared to be upregulated in med5ab and downregulated in med16 (Figure 7A, Table 2, Cluster 6)
In the collection of MED mutants we examined, relatively few genes passed our criteria for differential expression (Figure 4). We found that, although the mutants shared many differentially expressed genes, there were also a large number of genes that were uniquely differentially expressed in each mutant. Genes that were upregulated in all four mutants showed an enrichment of genes encoding extracellular proteins, as well as phenylpropanoid related genes (Figure 7A, Cluster 5), consistent with their ability to rescue the phenylpropanoid-deficient mutant ref4-3 (Dolan et al. 2017). Many of the genes that were altered in the mutants were related to abiotic or biotic stress, in which Mediator is known to play a major role (Samanta and Thakur 2015b). The med16 and med5ab mutants were the most different from one another, showing opposite regulation of many of the same genes (Figures 5B and 7A). Conversely, we observed a strong correlation between the gene expression profiles of med5ab and med23 (Figure 5B). This finding is consistent with our previous observation that both med5ab and med23 have higher levels of sinapoylmalate (Dolan et al. 2017) and suggests a broad functional link between the two subunits, possibly mediated by a close physical association within the complex. This close association is also supported by the observation that knocking out med23 in the MED5b mutant ref4-3 strongly and specifically suppresses the transcriptional and phenotypic effects of ref4-3 (Dolan et al. 2017). Our data also suggest that MED2 plays a more general role in gene regulation than some of the other MED subunits, as only a small number of pathways were significantly enriched in the med2 mutant, despite the substantial number of genes that are differentially expressed in that line (Figures 4 and 6).
As we previously reported, the med16 mutant is different from the other med mutants investigated here, in that a large number of genes are upregulated in the mutant, consistent with what has been observed in the yeast (Chen et al. 1993; Covitz et al. 1994; Jiang and Stillman 1995). What was more surprising was that the genes that were upregulated in med16 were associated with defense pathways, including those controlled by salicylic acid and jasmonic acid (Figure 7, Table 2, Cluster 4). MED16 has been extensively reported as being a positive regulator of both SA and JA-mediated defense (Wathugala et al. 2012; Zhang et al. 2012, 2013, Wang et al. 2015, 2016). Given the existence of numerous positive and negative regulators of these pathways, close inspection of the identity and function of these genes will be required to determine how these findings fit with known role of MED16 in defense response pathways. Additionally, many of these genes are downregulated in med5ab (Figure 7, Cluster 4), suggesting a possible antagonistic or epistatic relationship between MED5a/b and MED16 in the expression of defense response genes.
MED23 is one of several subunits that are conserved in metazoans and plants, but not in Saccharomyces (Bourbon 2008). In humans, MED23 plays a variety of important roles, including promoting transcription elongation, alternative splicing, and ubiquitination of histone H2B (Huang et al. 2012; Wang et al. 2013; Yao et al. 2015). Aside from our previous report, the role of MED23 in transcription regulation in plants has yet to be investigated. Our data suggest that MED23 does not play a major role in Arabidopsis rosettes under normal growth conditions. Examination of the expression of MED23 in different organs and tissues, as well as the genes that are co-expressed with MED23, suggested that MED23 might function in reproductive or meristem development. Two of the genes co-expressed with MED23, MED12 and MED13, encode subunits of the Mediator kinase module. MED12 and MED13 play a transient role in early embryo patterning and development, and similar to MED23, are expressed most strongly in the shoot apical meristem (Gillmor et al. 2010). Together, these observations suggest that MED23 might function together with MED12 and MED13 in embryo development, particularly in establishing the shoot apical meristem.
We also discovered evidence of a conserved role for MED23 in alternative splicing, in that a number of RNA processing factors are co-expressed with MED23 and that more alternative transcripts were produced in med23 than in the other mutants (Table 3, Figure 10A). All of the alternative splicing events that we examined occurred at either 5′ or 3′ ends of the genes. GO-term analysis of these genes showed an enrichment of genes encoding membrane proteins or proteins localized to different cellular compartments. Alternative splicing of N- or C-terminal exons can affect where proteins are targeted by changing the inclusion of signal peptides or transmembrane helices (Davis et al. 2006; Dixon et al. 2009; Lamberto et al. 2010; Kriechbaumer et al. 2012; Remy et al. 2013). In addition, alternative UTRs can affect transcript stability and translation efficiency (Reddy et al. 2013). Biochemical validation will be required to determine whether these transcripts truly undergo alternative splicing in the MED mutants, and if so, what consequences they have on protein function or localization. Two major mechanisms have been proposed by which Mediator might affect splicing. In the “recruitment model”, Mediator and Pol II impact splicing by directly interacting with splicing factors to facilitate their recruitment to the transcription machinery (Merkhofer et al. 2014). MED23 has been shown to function in this way in HeLa cells by interacting with and promoting the recruitment of the splicing factor hnRNPL (Huang et al. 2012). Alternatively, Mediator might affect splicing by altering the rate of the transcription elongation, the so-called “kinetic model” of co-transcriptional splicing (Donner et al. 2010; Takahashi et al. 2011; Wang et al. 2013).
In the course of our alternative splicing analysis, we discovered that in med2 and med23 a region that appears to undergo transcriptional gene silencing (TGS) was derepressed (Figure 11, Greaves et al. 2014). Previously, mutation of Arabidopsis MED17, MED18 or MED20a was shown to disrupt TGS at type II loci by reducing the efficiency with which Pol II is recruited, causing reduced production of the long noncoding scaffold RNAs required for the recruitment of Pol V (Kim and Chen 2011). These MED subunits were also shown to be required for TGS at some type I loci, which are not known to require Pol II for silencing, but the mechanism by which they are required is unknown. Whether MED2 and MED23 function similarly remains to be seen.
This study is the first to present a side-by-side comparison of the effects of multiple Arabidopsis med mutants on global gene expression. Importantly, these data begin to unravel the complex network of interactions within Mediator that are required for the regulation of different genes and pathways and they suggest a number of potential avenues for future investigation.
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under Award DE-FG02-07ER15905 and Stanford University’s Global Climate and Energy Project (GCEP).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6864170.
Communicating editor: M. Estelle
Literature Cited
- Allen B. L., Taatjes D. J., 2015. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16: 155–166. 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Okamura Y., Tadaka S., Kinoshita K., Obayashi T., 2016. ATTED-II in 2016: A Plant Coexpression Database Towards Lineage-Specific Coexpression. Plant Cell Physiol. 57: e5 10.1093/pcp/pcv165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béve J., Hu G.-Z., Myers L. C., Balciunas D., Werngren O., et al. , 2005. The structural and functional role of Med5 in the yeast Mediator tail module. J. Biol. Chem. 280: 41366–41372. 10.1074/jbc.M511181200 [DOI] [PubMed] [Google Scholar]
- Bonawitz N. D., Soltau W. L., Blatchley M. R., Powers B. L., Hurlock A. K., et al. , 2012. REF4 and RFR1, subunits of the transcriptional coregulatory complex Mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 287: 5434–5445. 10.1074/jbc.M111.312298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H. M., 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993–4008. 10.1093/nar/gkn349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce J. M., Knight H., Deyholos M., Openshaw M. R., Galbraith D. W., et al. , 2003. The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 34: 395–406. 10.1046/j.1365-313X.2003.01734.x [DOI] [PubMed] [Google Scholar]
- Buendía-Monreal M., Gillmor C. S., 2016. Mediator: A key regulator of plant development. Dev. Biol. 419: 7–18. 10.1016/j.ydbio.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Cerdán P. D., Chory J., 2003. Regulation of flowering time by light quality. Nature 423: 881–885. 10.1038/nature01636 [DOI] [PubMed] [Google Scholar]
- Cevher M. A., Shi Y., Li D., Chait B. T., Malik S., et al. , 2014. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21: 1028–1034. 10.1038/nsmb.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V., Kidd B. N., Zhang P., Hill C., Kiddle S., et al. , 2012. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160: 541–555. 10.1104/pp.112.202697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., et al. , 2012. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. 10.1105/tpc.112.098277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., West R. W., Ma J., Johnson S. L., Gans H., et al. , 1993. TSF1 to TSF6, required for silencing the Saccharomyces cerevisiae GAL genes, are global regulatory genes. Genetics 134: 701–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covitz P. A., Song W., Mitchell A. P., 1994. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics 138: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. J., Hanson K. A., Clark F., Fink J. L., Zhang F., et al. , 2006. Differential Use of Signal Peptides and Membrane Domains Is a Common Occurrence in the Protein Output of Transcriptional Units. PLoS Genet. 2: e46 10.1371/journal.pgen.0020046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. P., Hawkins T., Hussey P. J., Edwards R., 2009. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 60: 1207–1218. 10.1093/jxb/ern365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan W. L., Chapple C., 2017. Conservation and divergence of Mediator structure and function: insights from plants. Plant Cell Physiol. 58: 4–21. 10.1093/pcp/pcw176 [DOI] [PubMed] [Google Scholar]
- Dolan W. L., Dilkes B. P., Stout J. M., Bonawitz N. D., Chapple C., 2017. Mediator Complex Subunits MED2, MED5, MED16, and MED23 Genetically Interact in the Regulation of Phenylpropanoid Biosynthesis. Plant Cell 29: 3269–3285. 10.1105/tpc.17.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A. J., Ebmeier C. C., Taatjes D. J., Espinosa J. M., 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17: 194–201. 10.1038/nsmb.1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., et al. , 2011. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA 108: 8245–8250. 10.1073/pnas.1002981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H., Baraznenok V., Lindahl M., Samuelsen C. O., Koeck P. J. B., et al. , 2006. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. USA 103: 15788–15793. 10.1073/pnas.0607483103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan P. M., Kelleher R. J., Feaver W. J., Lue N. F., LaPointe J. W., et al. , 1990. Resolution of factors required for the initiation of transcription by yeast RNA polymerase II. J. Biol. Chem. 265: 11105–11107. [PubMed] [Google Scholar]
- Galdieri L., Desai P., Vancura A., 2012. Facilitated Assembly of the Preinitiation Complex by Separated Tail and Head/Middle Modules of the Mediator. J. Mol. Biol. 415: 464–474. 10.1016/j.jmb.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor C. S., Park M. Y., Smith M. R., Pepitone R., Kerstetter R. A., et al. , 2010. The MED12–MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122. 10.1242/dev.043174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D., Hamidi N., Del Sol R., Benschop J. J., Nancy T., et al. , 2014. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl. Acad. Sci. USA 111: 2500–2505. 10.1073/pnas.1307525111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves I. K., Groszmann M., Wang A., Peacock W. J., Dennis E. S., 2014. Inheritance of Trans Chromosomal Methylation patterns from Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. USA 111: 2017–2022. 10.1073/pnas.1323656111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P. A., Hurst C. H., Kaliyadasa E., Lamb R., Knight M. R., et al. , 2014. The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484. 10.1105/tpc.113.117796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li W., Yao X., Lin Q.-J., Yin J.-W., et al. , 2012. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell 45: 459–469. 10.1016/j.molcel.2011.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ito M., Okano H. J., Darnell R. B., Roeder R. G., 2002. The TRAP100 component of the TRAP / Mediator complex is essential in broad transcriptional events and development. EMBO J. 21: 3464–3475. 10.1093/emboj/cdf348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C., Langelier M.-F., Bataille A. R., Pascal J. M., Pugh B. F., et al. , 2016. Tail and kinase modules differently regulate core Med/iator recruitment and function in vivo. Mol. Cell 64: 455–466. 10.1016/j.molcel.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. W., Stillman D. J., 1995. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics 140: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher R. J., Flanagan P. M., Kornberg R. D., 1990. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215. 10.1016/0092-8674(90)90685-8 [DOI] [PubMed] [Google Scholar]
- Kidd B. N., Edgar C. I., Kumar K. K., Aitken E. A., Schenk P. M., et al. , 2009. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252. 10.1105/tpc.109.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D., 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608. 10.1016/0092-8674(94)90221-6 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Chen X., 2011. The plant Mediator and its role in noncoding RNA production. Front. Biol. (Beijing) 6: 125–132. 10.1007/s11515-011-1133-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., 1999. The sfr 6 Mutation in Arabidopsis Suppresses Low-Temperature Induction of Genes Dependent on the CRT/DRE Sequence Motif. Plant Cell 11: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Mugford S. G., Ulker B., Gao D., Thorlby G., et al. , 2009. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58: 97–108. 10.1111/j.1365-313X.2008.03763.x [DOI] [PubMed] [Google Scholar]
- Knight H., Thomson A. J. W., McWatters H. G., 2008. Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 148: 293–303. 10.1104/pp.108.123901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M. T., Meyer K. D., Bernecky C., Taatjes D. J., 2009. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23: 439–451. 10.1101/gad.1767009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S. S., Ansari A. Z., Ptashne M., Young R. A., 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1: 895–904. 10.1016/S1097-2765(00)80088-X [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Wang P., Hawes C., Abell B. M., 2012. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 70: 292–302. 10.1111/j.1365-313X.2011.04866.x [DOI] [PubMed] [Google Scholar]
- Lamberto I., Percudani R., Gatti R., Folli C., Petrucco S., 2010. Conserved Alternative Splicing of Arabidopsis Transthyretin-Like Determines Protein Localization and S-Allantoin Synthesis in Peroxisomes. Plant Cell 22: 1564–1574. 10.1105/tpc.109.070102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C., Park J. M., Min S., Han S. J., Kim Y. J., 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19: 2967–2976. 10.1128/MCB.19.4.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bjorklund S., Jiang Y. W., Kim Y. J., Lane W. S., et al. , 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92: 10864–10868. 10.1073/pnas.92.24.10864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A. T. L., Chen Y., Smyth G. K., 2016. It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR, pp. 391–416 in Statistical Genomics. Methods and Protocols, edited by Ewy M., Sean D. Humana Press, New York, NY: 10.1007/978-1-4939-3578-9_19 [DOI] [PubMed] [Google Scholar]
- Merkhofer E. C., Hu P., Johnson T. L., 2014. Introduction to cotranscriptional RNA splicing. Methods Mol. Biol. 1126: 83–96. 10.1007/978-1-62703-980-2_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M., 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Parra E., Pelagio-Flores R., Raya-González J., Salmerón-Barrera G., Ruiz-Herrera L. F., et al. , 2017. Plant-plant interactions influence developmental phase transitions, grain productivity and root system architecture in Arabidopsis via auxin and PFT1/MED25 signalling. Plant Cell Environ. 40: 1887–1899. 10.1111/pce.12993 [DOI] [PubMed] [Google Scholar]
- Myers L. C., Gustafsson C. M., Hayashibara K. C., Brown P. O., Kornberg R. D., 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96: 67–72. 10.1073/pnas.96.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Kim H. S., Han S. J., Hwang M. S., Lee Y. C., et al. , 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20: 8709–8719. 10.1128/MCB.20.23.8709-8719.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T. J. P., van Leenen D., et al. , 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522. 10.1016/j.molcel.2005.06.033 [DOI] [PubMed] [Google Scholar]
- Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., et al. , 2015. Architecture of the RNA polymerase II–Mediator core initiation complex. Nature 518: 376–380. 10.1038/nature14229 [DOI] [PubMed] [Google Scholar]
- Poss Z. C., Ebmeier C. C., Taatjes D. J., 2013. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48: 575–608. 10.3109/10409238.2013.840259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya-González J., Ortiz-Castro R., Ruíz-Herrera L. F., Kazan K., López-Bucio J., 2014. PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis. Plant Physiol. 165: 880–894. 10.1104/pp.114.239806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S. N., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the Alternative Splicing Landscape in Plants. Plant Cell 25: 3657–3683. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves W. M., Hahn S., 2003. Activator-Independent Functions of the Yeast Mediator Sin4 Complex in Preinitiation Complex Formation and Transcription Reinitiation. Mol. Cell. Biol. 23: 349–358. 10.1128/MCB.23.1.349-358.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E., Cabrito T. R., Baster P., Batista R. A., Teixeira M. C., et al. , 2013. A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25: 901–926. 10.1105/tpc.113.110353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Smyth G. K., 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9: 321–332. 10.1093/biostatistics/kxm030 [DOI] [PubMed] [Google Scholar]
- Robinson P. J., Trnka M. J., Pellarin R., Greenberg C. H., Bushnell D. A., et al. , 2015. Molecular architecture of the yeast Mediator complex. eLife 4: e08719 10.7554/eLife.08719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., et al. , 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18: 529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S., Thakur J. K., 2015a Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6: 757 10.3389/fpls.2015.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S., Thakur J., 2015b Role of plant mediator complex in stress response in Elucidation Abiotic Stress Signaling in Plants. Springer, New York: 10.1007/978-1-4939-2540-7_1 [DOI] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguela-Arnaud M., Smith C., Uribe M. C., May S., Fischl H., et al. , 2015. The Mediator complex subunits MED25/PFT1 and MED8 are required for transcriptional responses to changes in cell wall arabinose composition and glucose treatment in Arabidopsis thaliana. BMC Plant Biol. 15: 215 10.1186/s12870-015-0592-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaravelpandian K., Chandrika N. N. P., Schmidt W., 2013. PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol. 197: 151–161. 10.1111/nph.12000 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A. S., et al. , 2011. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104. 10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. M., Koleske A. J., Chao D. M., Young R. A., 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73: 1361–1375. 10.1016/0092-8674(93)90362-T [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.-L., Sato S., Tomomori-Sato C., Conaway R. C., Conaway J. W., et al. , 2013. A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20: 611–619. 10.1038/nsmb.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.-L., Yu X., Gopalan S., Chao T.-C., Zhang Y., et al. , 2017. Mediator structure and rearrangements required for holoenzyme formation. Nature 544: 196–201. 10.1038/nature21393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Du X., Mou Z., 2016. The Mediator Complex Subunits MED14, MED15, and MED16 Are Involved in Defense Signaling Crosstalk in Arabidopsis. Front. Plant Sci. 7: 1947 10.3389/fpls.2016.01947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yao J., Du X., Zhang Y., Sun Y., et al. , 2015. The Arabidopsis Mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol. 169: 856–872. 10.1104/pp.15.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yao X., Huang Y., Hu X., Liu R., et al. , 2013. Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription 4: 39–51. 10.4161/trns.22874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathugala D. L., Hemsley P. A., Moffat C. S., Cremelie P., Knight M. R., et al. , 2012. The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195: 217–230. 10.1111/j.1469-8137.2012.04138.x [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., et al. , 2007. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS One 2: e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Li Y., 2011. Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554. 10.1242/dev.071423 [DOI] [PubMed] [Google Scholar]
- Yang Y., Li L., Qu L.-J., 2015. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 58: 106–118. 10.1111/jipb.12377 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ou B., Zhang J., Si W., Gu H., et al. , 2014. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77: 838–851. 10.1111/tpj.12440 [DOI] [PubMed] [Google Scholar]
- Yao X., Tang Z., Fu X., Yin J., Liang Y., et al. , 2015. The Mediator subunit MED23 couples H2B mono-ubiquitination to transcriptional control and cell fate determination. EMBO J. 34: 2885–2902. 10.15252/embj.201591279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Wang G., 2014. The Mediator complex: a master coordinator of transcription and cell lineage development. Development 141: 977–987. 10.1242/dev.098392 [DOI] [PubMed] [Google Scholar]
- Zhang F., Sumibcay L., Hinnebusch A. G., Swanson M. J., 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24: 6871–6886. 10.1128/MCB.24.15.6871-6886.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z., 2012. The Arabidopsis Mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309. 10.1105/tpc.112.103317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wu H., Wang N., Fan H., Chen C., et al. , 2014. Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol. 203: 770–783. 10.1111/nph.12860 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yao J., Zhang Y., Sun Y., Mou Z., 2013. The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75: 484–497. 10.1111/tpj.12216 [DOI] [PubMed] [Google Scholar]
- Zhu W., Yao X., Liang Y., Liang D., Song L., et al. , 2015. Mediator Med23 deficiency enhances neural differentiation of murine embryonic stem cells through modulating BMP signaling. Development 142: 465–476. 10.1242/dev.112946 [DOI] [PubMed] [Google Scholar]