Abstract
Understanding how genomic variation causes differences in observable phenotypes remains a major challenge in biology. It is difficult to trace the sequence of events originating from genomic variants to changes in transcriptional responses or protein modifications. Ideally, one would conduct experiments with individuals that are at either extreme of the trait of interest, but such resources are often not available. Further, advances in genome editing will enable testing of candidate polymorphisms individually and in combination. Here we have created a resource for the study of sleep with 39 inbred lines of Drosophila—the Sleep Inbred Panel (SIP). SIP lines have stable long- and short-sleeping phenotypes developed from naturally occurring polymorphisms. These lines are fully sequenced, enabling more accurate targeting for genome editing and transgenic constructs. This panel facilitates the study of intermediate transcriptional and proteomic correlates of sleep, and supports genome editing studies to verify polymorphisms associated with sleep duration.
Keywords: sleep, Drosophila melanogaster, whole-genome sequence
Genomic studies of wild-derived populations of flies have identified thousands of polymorphisms that affect morphological, physiological, and behavioral complex traits (Ayroles et al. 2009; Jordan et al. 2012; Mackay et al. 2012; Weber et al. 2012; Chow et al. 2013; Harbison et al. 2013; Ellis et al. 2014; Vaisnav et al. 2014; Arya et al. 2015; Ayroles et al. 2015; Dembeck et al. 2015a; Dembeck et al. 2015b; Gaertner et al. 2015; Garlapow et al. 2015; Ivanov et al. 2015; Montgomery et al. 2015; Morgante et al. 2015; Morozova et al. 2015; Shorter et al. 2015; Unckless et al. 2015; Zwarts et al. 2015; Chow et al. 2016; He et al. 2016; Hunter et al. 2016; Vonesch et al. 2016; Harbison et al. 2017; Lobell et al. 2017; Wu et al. 2018). A challenging next step is to demonstrate how the polymorphisms associated with a trait influence phenotype (Mackay et al. 2009; Albert and Kruglyak 2015). One potential approach is to measure the phenotypic, transcriptional, and proteomic impact of perturbing candidate polymorphisms, a strategy that has become possible with the advent of genome editing (Bassett and Liu 2014; Albert and Kruglyak 2015). Such perturbations are best made in consistent genetic backgrounds, where one can accurately estimate enhancing and suppressing epistatic effects (Yamamoto et al. 2008; Swarup et al. 2012; Mackay 2014). Here we developed a 39-line panel of inbred flies having extreme long and short sleep duration, which we refer to as the Sleep Inbred Panel (SIP). Because the SIP lines have extreme differences in phenotype, advanced intercross population designs developed from two or more strains could be employed to identify context-dependent pleiotropic loci or genetic modifiers (Lawson et al. 2011; Huang et al. 2012; Kislukhin et al. 2013; Swarup et al. 2013; King et al. 2014; Najarro et al. 2015; Shorter et al. 2015; Chow et al. 2016; Chandler et al. 2017). The SIP is therefore a useful tool for the design of genome modifications, the identification of phenotypic, transcriptional, and proteomic correlates, and the understanding of context-dependent effects.
Materials and Methods
Construction of the Sleep Inbred Panel
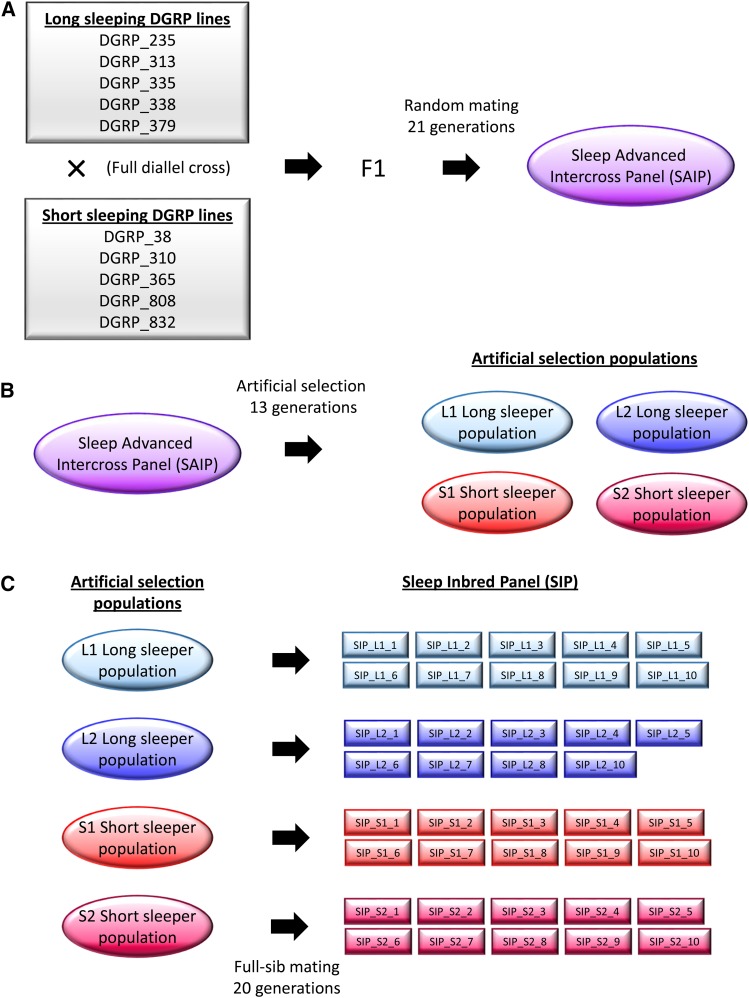
The process for construction of the Sleep Inbred Panel is outlined in Figure 1 and involves three major steps. The first two steps were done previously (Harbison et al. 2017), but we outline them here briefly. The first step was the construction of an outbred population of flies, the Sleep Advanced Intercross Population (SAIP) using ten lines from the Drosophila Genetic Reference Panel (DGRP) (Mackay et al. 2012; Huang et al. 2014) with the most extreme night sleep phenotypes in both sexes (Figure 1A). The five lines with shortest average night sleep were DGRP_38, DGRP_310, DGRP_365, DGRP_808, and DGRP_832 (Harbison et al. 2013). The five lines with the longest average night sleep were DGRP_235, DGRP_313, DGRP_335, DGRP_338, and DGRP_379 (Harbison et al. 2013). All ten lines were crossed in a full diallel design, resulting in 100 crosses. We randomly assigned two virgin females and two males from the F1 of each cross into 20 bottles, with 10 males and 10 females in each bottle. At each subsequent generation, we randomly mixed 20 virgin females and 20 males from each bottle to propagate the next generation. Each generation of random mating had a census population size of 800. We continued this random mating scheme for 21 generations, resulting in the SAIP (Harbison et al. 2017).
Figure 1.
Construction of the Sleep Inbred Panel. The steps used to construct the SIP are indicated. (A) Step 1 is the construction of the Sleep Advanced Intercross Population (SAIP) using 10 long- and short-sleeping inbred lines. The F1 of a full diallel cross was randomly mated for 21 generations to produce the outbred SAIP. (B) Step 2 is an artificial selection protocol applied to the SAIP to produce two replicate long- and two replicate short-sleeping populations of flies. (C) Step 3 is the construction of the SIP via 20 generations of full-sib mating.
The second step was to select for long and short night sleep duration (Figure 1B). To do this we split the SAIP into four populations by seeding four bottles with 25 randomly chosen flies of each sex. Two populations were selected for long night sleep (L1 and L2), and two populations were selected for short night sleep (S1 and S2) using the following artificial selection procedure each generation. We measured sleep and activity over a 5-day period in 100 virgins of each sex from each population. The 25 males and females with the highest (lowest) night sleep within each population were chosen as parents for the next generation of long (short) sleepers. We repeated this procedure for 13 generations. This resulted in two short-sleeper populations with average night sleep of 111.9 ± 10.74 min. and 54.8 ± 5.66 min. for replicate 1 and replicate 2, respectively, and two long-sleeper populations with average night sleep of 685.0 ± 3.35 min. and 678.5 ± 3.46 min. for replicate 1 and 2, respectively (Harbison et al. 2017). After Generation 13, the flies were maintained for each population/replicate via random mating for 17 generations.
The third step was the creation of the Sleep Inbred Panel (Figure 1C). At generation 51, the long- and short-sleeping selected populations were used to create inbred lines. We created 10 lines each from the L1, S1, and S2 populations, and 9 lines from the L2 population (39 lines total). Each line was created using a single male and a single female from one of the populations to start the line; one male and one female from the progeny were used to propagate the line to the next generation. Full-sib mating continued in this manner for 20 generations. Inbred stocks were maintained past generation 20 by random mating.
Rearing and assay conditions
For culturing and sleep assays, flies were reared in a single incubator under standard conditions (25°, 60% humidity, 12:12 hr light:dark cycle) on standard Drosophila medium (https://bdsc.indiana.edu/information/recipes/bloomfood.html). Prior to sleep assays, male and female flies were collected as virgins and aged to 4 – 7 days in same-sex vials of 20 flies each to standardize mating status and social exposure (Ganguly-Fitzgerald et al. 2006; Isaac et al. 2010). For sleep assays, lines were randomly assigned to one of four blocks: three blocks had ten lines, and one block had nine lines. Sleep assays were replicated twice for each SIP line. The first replicate was measured in the generation immediately following the inbreeding procedure, while the second replicate was measured two generations later. We did not observe any differences in night sleep among replicate measures (Table S1). A total of 32 flies/sex/line were measured.
Sleep phenotyping
We measured sleep and activity in the SIP in the rearing and assay conditions stipulated above. Individual virgin males and females were placed into Drosophila Activity Monitoring System (DAM2) monitors (Trikinetics, Waltham, MA) under CO2 anesthesia. Activity counts were recorded for the subsequent seven days; the first day of data were discarded as the flies were recovering from CO2 and acclimating to the monitor tubes. At the end of the seven-day period, each fly was visually examined; data from flies that did not survive the duration of the monitoring period was discarded. We used a C# program (R. Sean Barnes) to calculate the sleep duration, the number of sleep bouts, and the average sleep bout length during the day or night; the waking activity, which is the number of activity counts divided by the number of minutes spent awake in a 24-hour period; and the sleep latency, which is the amount of time before the first night sleep bout.
Phenotypic data analysis
Lines of the SIP originate from four different selection populations: L1 and L2, which were two replicate populations selected for long sleep; and S1 and S2, the two replicate populations selected for short sleep. We first analyzed the sleep parameters for their differences among selection scheme and replicate population within selection scheme using the ANOVA model Y = µ + Sel + Reppop(Sel) + Sex + Rep + Sex×Reppop(Sel) + Rep×Reppop(Sel) + Sex×Reppop×Rep(Sel) + ε, where Sel is selection scheme, Reppop is replicate population, Rep is phenotypic replicate, and ε is the error term. There were significant differences in sleep phenotypes among selection schemes and replicate populations. Next, we compared the mean sleep of each SIP line with the mean of its progenitor population (i.e., the artificially selected population from which each SIP line was derived) using the ANOVA model Y = µ + Sex + Line + Rep + Sex×Line + Sex×Rep + Line×Rep + Line×Sex×Rep + ε, where Rep and ε are as defined above. We used post-hoc Tukey comparisons to determine which lines were significantly different from the progenitor.
DNA extraction and sequencing
Two replicates of thirty female flies were flash-frozen from each line. DNA was extracted using a cell lysis solution [1.58 g of Tris-HCl (Quality Biological, Gaithersburg, MD), 37.22 g EDTA disodium salt (Quality Biological, Gaithersburg, MD) and filled to 1 liter with RNase/DNase-free water, adjusting the pH to 8.0 with 10 M NaOH (Sigma Aldrich, St. Louis, MO) when necessary]. Flies were homogenized using an Omni Bead Ruptor (Omni International, Kennesaw, GA). The solution was incubated with 10% SDS (Thermo Fisher Scientific, Waltham, MA) and 20 mg/mL Proteinase K (Thermo Fisher Scientific, Waltham, MA) at 65° for 1 hr. The lysate was RNase A treated (20 mg/ml) (Thermo Fisher Scientific, Waltham, MA) by mixing and incubating at 37° for 15 min. Ammonium Acetate (Quality Biological, Gaithersburg, MD) solution was added to samples chilled on ice for 5 min to precipitate proteins. 100% isopropanol (VWR International, Radnor, PA) was added and mixed to precipitate the DNA; samples were incubated for 1 hr at -20°. The DNA pellet was washed with 75% ethanol (NIH Supply Center, Gaithersburg, MD), then re-hydrated in RNase/DNase-free water. DNA samples were then purified using phenol-chloroform extraction. We diluted each DNA sample with 10mM Tris (Quality Biological, Gaithersburg, MD), 1mM EDTA, pH 7.8 to bring sample volume to 200 µL. Next, 200 µL of phenol:chloroform:isoamyl alcohol (25:24:1) (Sigma Aldrich, St. Louis, MO) was added to each sample. We then centrifuged samples and transferred the aqueous phase to a new 1.5mL tube. We added 200uL of chloroform (NIH Supply Center, Gaithersburg, MD) to each tube, centrifuged samples and transferred the upper aqueous layer to a new 1.5mL tube. Next, DNA precipitation was initiated by adding 20 μL of sodium acetate (NaOAc) (Sigma Aldrich, St. Louis, MO), 500 μL of ethanol, and 1 μL of glycogen. Samples were then placed on ice, centrifuged at maximum speed for 30 min, and then the supernatant was discarded. We washed the pellet with 500 uL of ethanol and centrifuged samples for 5 min. Afterward, we removed the supernatant and dissolved the pellet in 25 μL sterile 10mM Tris, 0.1mM EDTA, pH 7.8. The samples were heated for 2 min at 55°. We measured DNA concentration and quality with Nanodrop 8000 (Thermo Fisher Scientific, Waltham, MA).
Tru-Seq PCR-Free Library Method
For all lines save one, two micrograms of genomic DNA were sheared to ∼550 bp using a Covaris E220 with settings: duty cycle 10%; intensity 175; cycles/burst 200; and time 80s. Only one microgram of DNA was available for line SIP_L2_2, so the DNA was sheared to ∼350 bp using a Covaris E220 with settings: duty cycle 10%; intensity 3; cycles/burst 200; and time 60s. Libraries were constructed using the Tru-Seq DNA PCR-Free LT Sample Prep Kit (Illumina, San Diego, CA) according to the manufacturer’s protocol. The libraries were pooled and run on an Illumina HiSeq 2500 with version 3 sequencing reagents to generate a minimum of 10 million paired-end 251-base reads per library (Illumina, San Diego, CA), resulting in 30-50X genome coverage on average (Figure S1). The HiSeq data were processed using RTA1.18.64 and CASAVA 1.8.2.
Sequence processing, alignment, and variant calls
All sequence reads were aligned to D. melanogaster assembly BDGP Release 6, UCSC version dm6 (obtained from UCSC Genome Browser FTP site). Alignments were performed using two programs: BWA-MEM version 0.7.12 (Li 2013) and Novoalign version 3.02.07 (Novocraft Technologies, Selangor, Malaysia), using the -t 400 option to optimize alignment speed. PCR duplicates were removed from all aligned read sets using samtools version 0.1.17 (Li et al. 2009). Read groups were added to BWA alignments, which were then realigned around known indels from the set of DGRP Freeze 2 polymorphisms (Huang et al. 2014) using GATK version 2.8.1 (Van der Auwera et al. 2013). Confirmation of sex for each sample was performed by calculating the ratio of the average read depth on the X chromosome to the average read depth on chromosome 2L. The ratio of average read depth on the X chromosome to that of chromosome 2L was greater than 0.96 for every line except for SIP_L2_2, which had a ratio of 0.50. Thus, SIP_L2_2 DNA likely originated from male flies (both sexes were collected for DNA). All variants were called by running LoFreq version 2.1.2 (Wilm et al. 2012), run with the default parameter statement “lofreq call-parallel–pp-threads 8 -f dm6.fa -o lofreq.out.vcf reads.bam”, where “dm6.fa” is the D. melanogaster 6.0 reference sequence file, “lofreq.out.vcf” is the output file, and “reads.bam” is the BAM file aligned reads (either BWA or Novoalign). The call-parallel feature of LoFreq was invoked to call all variants, rare or common. Allele counts for all single nucleotide variant sites were determined using the “bamcounts” command of the bardCNV package (http://github.com/nhansen/BardCNV) with the option -minqual 20 to filter reads for a minimum phred quality of 20 (Table S2). Counts of reads spanning indels were performed by first widening indel variants to their narrowest unambiguous region, then tallying reads with and without the indel using the perl module Bio::SamTools. Confidence intervals with the highest posterior density interval for the estimated read allele proportions were calculated in R using the CRAN “binom” package’s “binom.bayes” function (https://CRAN.R-project.org/doc/FAQ/R-FAQ.html). We plotted LoFreq quality score distributions for known DGRP founder alleles and novel predictions (Figure S2). Using this plot, we set a quality score threshold of 1000 for the novel predicted calls; variants less than this threshold were annotated as low scoring in the final .vcf file’s “FILTER” field. We grouped variant calls into the following categories: 1) DGRP_SNP, SNP calls that match SNPs (chromosome arm, position, and alternate allele) called as present in one of the 10 DGRP founder lines (Huang et al. 2014); 2) DGRP_UNGENOTYPED_SNP, DGRP SNPs that had a missing entry for at least one of the 10 DGRP founder lines (Huang et al. 2014); 3) DGRP_FILTERED_SNP, SNPs that were part of the original 6,149,822 variants found in the DGRP but due to low quality scores did not make the final list of 4,438,427 (Huang et al. 2014); 4) UNMAPPED_IN_DM3, variants that fell on the Het, U, 4, M, and Y chromosomes of the D. melanogaster 5.0 sequence (dm3) and were not part of the 4,438,427 DGRP variants; 5) DENOVO_SNP, SNPs perfectly associated with one DGRP founder haplotype and not previously known (see the Hidden Markov Model analysis below); 6) SELECTED_DENOVO: non-DGRP SNPs that were detected only on one HMM-predicted founder haplotype, but only within one selected population (e.g., L1, L2, S1, or S2); 7) PUTATIVE_FALSE_POSITIVE_SNP, variants that did not meet de novo SNP criteria and did not fall into any other category; and 8) SNPs removed due to a LoFreq quality score less than 1000. We annotated variant calls with SnpEff version 4.3t (Cingolani et al. 2012).
Mapping of founder genotypes
To predict which of the original 10 DGRP founder haplotypes are present at any genomic locus in each of the 39 SIP lines, we utilized the Hidden Markov Model (HMM) of King et al. (King et al. 2012). Our version of the model considered all DGRP polymorphic sites that are informative in the 10 founder lines and were detected as variant by LoFreq in at least one of the 39 inbred lines. We constructed 55 states: 10 homozygous states, in which both line’s homologous chromosomes derive from the same DGRP founder, and 45 heterozygous states, in which two different DGRP founders’ haplotypes are present in the line. Initiation and emission probabilities were set as in King et al. and transition probabilities were calculated from an empirically-derived tabulation of recombination rates (Comeron et al. 2012) as reported by the program RRC, version 2.3 [Fiston-Lavier AS and Petrov DA. Drosophila melanogaster Recombination Rate Calculator: http://petrov.stanford.edu/cgi-bin/recombination-rates_updateR5.pl]. Recombination rates at positions between those tabulated (Comeron et al. 2012) were estimated by linear interpolation.
To implement the HMM, we altered the Perl script made available by King et al. (King et al. 2012) to (a) read allele counts for inbred lines and genotypes of DGRP founders from tab-delimited files rather than a mysql database; (b) allow for an arbitrary number of founder lines (10 for this study); and (c) read and correctly utilize the recombination rates reported by the program RRC to calculate transition probabilities.
Data availability
The DNA sequences have been deposited in the Sequence Read Archive under ID code SRP126512; BioProject PRJNA421951. Supplementary tables S1-S9, figures S1-S4, and File S1 have been deposited on figshare. A text file of variant calls and confidence intervals using both BWA and Novoalign sequence alignments (Files S2 and S3, respectively) and a list of annotated variants in .vcf format have been provided (Files S4 and S5, respectively) on figshare. The scripts used to conduct the HMM analysis are on github: http://github.com/nhansen/SleepInbredPanel. Sleep Inbred Panel lines are available from the Bloomington Drosophila Stock Center (Flybase stock numbers FBst0076271 – FBst0076309; Bloomington Drosophila stock center numbers 76271-76309) (Bloomington, IN). Supplemental material available at Figshare: https://doi.org/10.25387/g3.6789968.
Results and Discussion
Construction of the Sleep Inbred Panel
The Sleep Inbred Panel (SIP) is the result of 21 generations of outbreeding, 13 generations of artificial selection for extreme sleep duration, 17 generations of post-selection maintenance, and 20 generations of subsequent inbreeding. In a previous study, we constructed the Sleep Advanced Intercross Population (SAIP) by crossing 10 long- and short- sleeping lines of the DGRP in a full diallel cross and then allowing the progeny to mate randomly for 21 generations (Figure 1A) (Harbison et al. 2017). The SAIP was used to conduct an artificial selection experiment in which two populations were selected for long night sleep (L1, L2), and two populations were selected for short night sleep (S1, S2) (Figure 1B) (Harbison et al. 2017). Here, we have preserved the differences in sleep duration observed in the previous study by creating inbred lines from these four artificially-selected populations (Figure 1C). Each line was seeded with a single male and virgin female from one of the four selection populations. Each generation thereafter, a single male and virgin female were used to propagate each line. This full-sib inbreeding continued for 20 generations. Thirty-nine inbred lines were created: 10 lines from the L1 long sleeper population, 9 lines from the L2 long sleeper population, 10 lines from the S1 short sleeper population, and 10 lines from the S2 short sleeper population. We refer to this collection of inbred lines as the Sleep Inbred Panel (SIP).
SIP lines have extreme sleep duration phenotypes
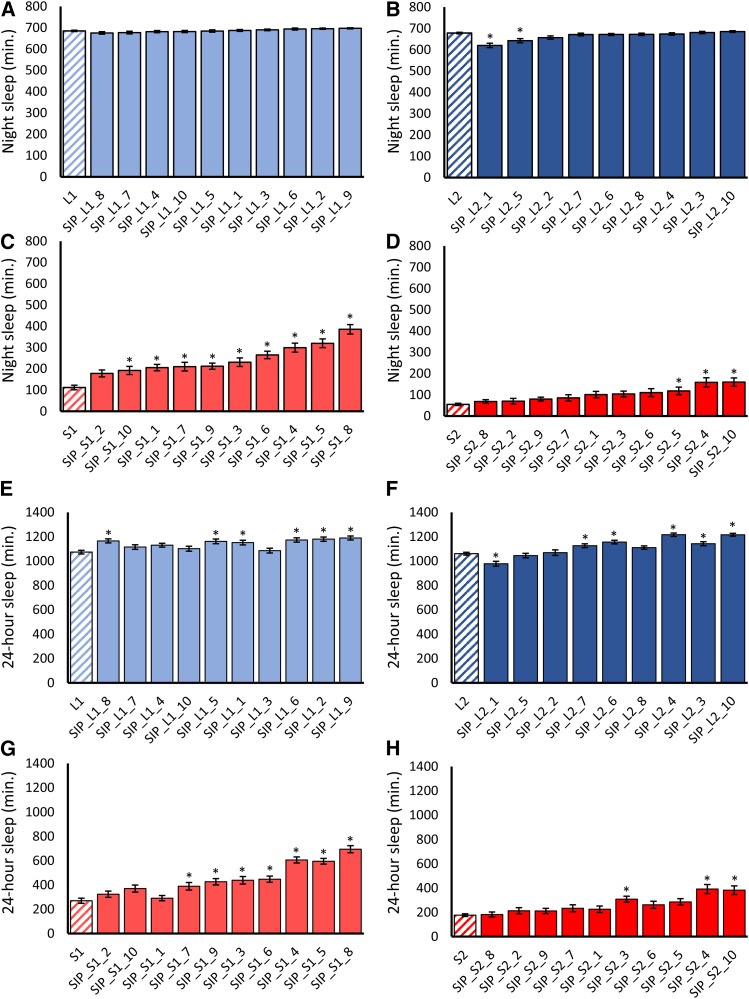
Average night sleep duration in lines of the SIP ranged from 68.61 ± 8.55 min. to 697.14 ± 2.66 min. (Table S3). Differences in night sleep were evident depending upon the direction of selection in the progenitor population (Pselection = 0.0220, four-way nested ANOVA model), and varied among replicate populations selected in the same direction (Preplicatepopulation(selection) = 0.0159, four-way nested ANOVA model) (Table S1). To determine whether we had captured the extreme night sleep phenotypes present in the artificially-selected populations, we compared mean night sleep in the SIP with the mean night sleep in the progenitor artificially selected populations (Harbison et al. 2017). Night sleep in long-sleeper lines was equivalent to that of the L1 and L2 progenitor populations (Figure 2A and 2B; Table S4), except for two L2-derived lines that had significantly reduced sleep (Figure 2B). Night sleep was significantly increased in every line derived from the S1 short sleeper population except for SIP_S1_2 (PLine = 0.0098, 3-way ANOVA model; Figure 2C), however. This result indicated that either inbreeding had not completely captured the short sleep in the S1 population, or that some of the extreme short sleep phenotypes were lost during the 17-generation maintenance period, possibly due to natural selection against short sleep. In contrast, night sleep in lines derived from the S2 population was equivalent to the S2 progenitor population, with the exception of three lines (PLine = ns, 3-way ANOVA; Figure 2D). Similar results were observed if 24-hour sleep duration was considered (Figure 2E-H), though there were more differences among lines (Table S4; see Figure S3 for day sleep phenotypes). Furthermore, night, day, and 24-hour sleep were stable across three generations—that is, replicate 1 and replicate 2 of the sleep measurements (Prep = ns for these three phenotypes) (Table S1). Thus, we largely preserved the extreme long- and short-sleeping night sleep phenotypes that we observed in the original selection populations; interestingly, significant differences from the original selection population means tended to be increases in sleep. Inbred lines derived from the S2 population had 24-hour average sleep duration that was as low if not lower than that of previously identified single-gene mutations and wild-derived inbred lines. Females of SIP_S2_1, SIP_S2_2, SIP_S2_4, SIP_S2_5, SIP_S2_6, SIP_S2_7, SIP_S2_8, and SIP_S2_9 had mean 24-hour sleep times below 250 min, and males of SIP_S2_1, SIP_S2_2, and SIP_S2_8 had mean 24-hour sleep under 300 min (Figure 2H). These short sleep times rival flies with single-gene mutations in Shaker (247 ± 22 min for females and 297 ± 34 for males) (Cirelli et al. 2005), insomniac (317 min for males) (Stavropoulos and Young 2011), and sleepless (Koh et al. 2008). Remarkably, all S2-derived short-sleeping lines had night sleep that was significantly lower than the shortest-sleeping line of the DGRP, DGRP_38, and all but two had shorter 24-hour sleep (Figure S4) (Harbison et al. 2013). The S1-derived lines SIP_S1_1 and SIP_S1_2 had shorter 24-hour sleep as well (Figure S4). Although night sleep in all but one of the long-sleeper lines was the same as the longest-sleeping line of the DGRP (Figure S4), DGRP_335, 24-hour sleep in DGRP_335 was significantly longer than all of the SIP lines (Figure S4). This is likely due to the fact that the selection procedure targeted only night sleep (Harbison et al. 2017); while day and night sleep share some genetic architecture, day sleep is not completely correlated with night sleep (Harbison et al. 2009; Harbison et al. 2013). In addition, we found other sleep traits with significant differences between long- and short-sleep selection schemes, which included day sleep duration (PSelection = 0.0057, four-way nested ANOVA), sleep latency (PSelection = 0.0386, four-way nested ANOVA), and average night bout length (marginally significant PSelection = 0.0553, four-way nested ANOVA). The differences in these sleep parameters between the long and short sleepers reflected correlated responses that we observed in the progenitor populations (Harbison et al. 2017). Stable extreme long and short sleeping phenotypes can therefore be constructed from naturally-occurring variants.
Figure 2.
Night sleep and 24-hour sleep in the Sleep Inbred Panel. Mean night sleep ± SE is plotted for each line of the SIP as compared to the mean of its corresponding progenitor population. SIP lines are ordered from the shortest night sleep to the longest. Solid colors indicate SIP line; diagonal bars indicate the mean of the progenitor population as reported in (Harbison et al. 2017). (A-D) Night sleep in SIP lines derived from the (A) L1 population; (B) L2 population; (C) S1 population; and (D) S2 population. (E-H) 24-hour sleep in SIP lines derived from the (E) L1 population; (F) L2 population; (G) S1 population; and (H) S2 population. Asterisks indicate lines with sleep duration significantly different (P < 0.05) from the progenitor population.
Short sleeper lines of the SIP have more variable day-to-day sleep
We previously noted strong negative correlations between the variability in sleep among individual flies and both night and day sleep duration (Harbison et al. 2013; Harbison et al. 2017); specifically, we found that shorter sleep times were associated with increased variability in sleep duration among flies. We calculated the variability in sleep parameters among individual flies of the SIP as the coefficient of environmental variation, or CVE (Table S5) (Mackay and Lyman 2005). None of the CVE traits were significantly different by selection scheme, suggesting that long sleepers and short sleepers had the same overall inter-individual variability, though night sleep CVE, day sleep CVE, 24-hour sleep CVE, and day bout number CVE were close to significance (Table S1 and S6). However, when we examined daily fluctuations in sleep using the standard deviation of each sleep trait (σ) to represent intra-individual differences (Knutson et al. 2007; Mezick et al. 2009; Buman et al. 2011; Angulo-Barroso et al. 2013; Dillon et al. 2014), we found that night and 24-hour sleep σ and night bout number σ were increased in lines derived from short-sleeping populations, and reduced in lines derived from the long-sleeping populations (P = 0.0398, 0.0428, and 0.0312, respectively, 3-way ANOVA) (Tables S1, S7, and S8). Short sleepers, therefore, have more daily fluctuations in sleep than long sleepers, and their sleep also tends to differ from individual to individual (Harbison et al. 2013; Wu et al. 2018). We speculate that the short sleepers may have greater sensitivity to small environmental fluctuations, and that this may result in more variable sleep.
Genomic architecture of the SIP
We extracted DNA from female flies and sequenced a minimum of 10 million 251 bp paired-end reads per SIP line, producing 30-50X genome coverage on average (Figure S1). We counted polymorphic variants and small indels known to be segregating in the 10 DGRP lines used to create the SAIP (Mackay et al. 2012; Huang et al. 2014). In addition, we searched for potential de novo variants using LoFreq (Wilm et al. 2012). LoFreq detected 1,451,085 (BWA alignment) and 1,298,672 (Novoalign alignment) variants. Results were similar for each sample’s BWA and Novoalign alignment sets, with less than 3% difference among the variants called for the X, 2L, 2R, 3L, 3R, and 4 chromosome arms, while differences between the two alignments were 20% and 22.8% for the mitochondrial genome and the Y chromosome, respectively (Table S2). Most of these variants were known DGRP SNPs (80.9% BWA and 65% Novoalign) (Table S9). We used the distribution of the LoFreq quality scores for the known DGRP SNPs to find a quality score threshold (1000) for the remaining SNPs (Figure S2). We eliminated 247,228 BWA SNPs and 432,612 Novoalign SNPs having quality scores less than 1000 from the final set of variants. We found 2,810 putative novel variants using the BWA alignment and 1,197 with Novoalign that appear to have arisen in the 10 DGRP founders. Furthermore, we found 183 novel variants (BWA) and 114 variants (Novoalign) that were restricted to one artificial population only (i.e., L1, L2, S1, or S2). The numbers of novel variants were reasonable given a recent study of the accumulation of mutations over 60 generations in a single DGRP line (Huang et al. 2016); in that study, the spontaneous mutation rate was estimated as 6.96 × 10−9 for the X chromosome and 6.25 × 10−9 for the autosomes, giving 1,456 de novo mutations. The remaining SNPs mapped to the 4, M, or Y chromosomes or regions not well defined in the D. melanogaster version 5.0 sequence used to call variants in the DGRP. We therefore consider it likely that these variants are part of the 10 DGRP founder genomes. Thus, nearly all the variants that we found map to the DGRP founder lines.
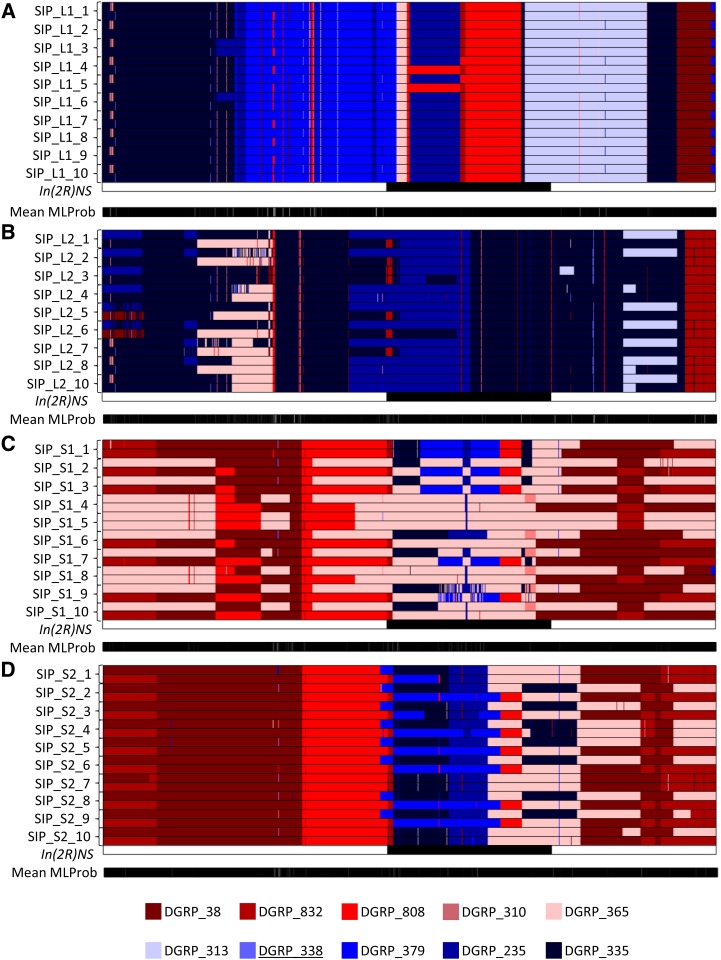
We used a Hidden Markov Model (King et al. 2012) to infer the distribution of the 10 founder DGRP lines along the chromosomes of each SIP line. The model performed well on our data, predicting founder states with posterior probabilities of at least 0.95 on 93.8% of our model’s sites. The fact that founder states were confidently predicted by the HMM suggested that contamination by other genotypes at any stage of the experiment (initial crosses, selection, post-selection maintenance, and inbreeding) was very unlikely. We plotted the inferred genotypes along each chromosome arm of the SIP (chromosome 2R, Figure 3; remaining major chromosome arms, File S1). Figure 3 shows how the founder lines combined to make chromosome 2R in the lines of the SIP. As expected, the greater contribution of the long-sleeping DGRP lines (shaded in hues of blue) can be seen in the L1 and L2 SIP lines (Figure 3A and 3B), while the shorter-sleeping DGRP lines (shaded in reds) contributed more to the S1 and S2 SIP lines (Figure 3C and 3D). The figure also shows the location of an inversion, In(2R)NS, which was heterozygous in one of the founder lines, DGRP_338 (Huang et al. 2014). This inversion does not appear to be present in any of the SIP lines. In addition, the average posterior probabilities are plotted along chromosome length. Brief switches of founder genotype tended to be associated with lower posterior probabilities. Thus, with the HMM model, the overall contribution of each of the original 10 founder DGRP lines can be observed.
Figure 3.
Plot of predicted DGRP founder haplotypes on chromosome 2R from Hidden Markov Model. In each plot, the predicted founder haplotypes are plotted along the length of the chromosome. Short-sleeping founder genotypes are coded in shades of red per the legend at the bottom of the figure; long-sleeping founder lines are coded in shades of blue. The location of chromosomal inversions is indicated by black bars for inversions that were present in the 10 DGRP founder lines. Underlined DGRP lines listed in the legend are heterozygous for the indicated inversion. The average posterior probability is given as a bar underneath the chromosome schematic graded from a probability of 1.0 (black) to 0.0 (white). (A), Lines derived from L1; (B), Lines derived from L2; (C), Lines derived from S1; (D), Lines derived from S2.
The contribution of these founder lines enabled us to compare the homozygosity of the SIP lines to that of the original DGRP founders. While the predicted founder haplotype for a given SNP was often heterozygous (i.e., DGRP_38 and DGRP_832), the SNP alleles themselves were often homozygous. When we compared the actual allelic proportions of each variant of the SIP to the predicted founder alleles to assess homozygosity, the SIP lines were between 1.64% less to 2.14% more homozygous than the genotypes predicted by the HMM model (Table S10).
Here we have developed a panel of 39 inbred long- and short- sleeping lines, a resource that will be useful for developing phenotypic correlates, perturbing genomic variants, and assessing changes in gene expression and protein abundance. These lines are available through the Bloomington Drosophila Stock Center, and the sequences are available through the NCBI Sequence Read Archive.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, the National Heart Lung and Blood Institute. We would like to thank K. Cook and S. Zheng of the Bloomington Drosophila Stock Center and G. Millburn of Flybase-Cambridge for SIP curation, the members of the NISC Consortium for genome sequence data and helpful discussions, and W. Huang for technical assistance. This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
S.T.H. and Y.L.S.N. conceived of the experiments; Y.L.S.N conducted the experiments; S.T.H. and Y.L.S.N. analyzed the data; N.F.H. conducted the sequence analysis; and S.T.H., Y.L.S.N, and N.F.H. wrote the paper.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6789968.
Communicating editor: S. Macdonald
Literature Cited
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Angulo-Barroso R. M., Peirano P., Algarin C., Kaciroti N., Lozoff B., 2013. Motor activity and intra-individual variability according to sleep-wake states in preschool-aged children with iron-deficiency anemia in infancy. Early Hum. Dev. 89: 1025–1031. 10.1016/j.earlhumdev.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya G. H., Magwire M. M., Huang W., Serrano Negron Y. L., Mackay T. F., et al. , 2015. The genetic basis for variation in olfactory behavior in Drosophila melanogaster. Chem. Senses 40: 233–243. 10.1093/chemse/bjv001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Buchanan S. M., O’Leary C., Skutt-Kakaria K., Grenier J. K., et al. , 2015. Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc. Natl. Acad. Sci. USA 112: 6706–6711. 10.1073/pnas.1503830112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. 10.1038/ng.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Liu J.-L., 2014. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genomics 41: 7–19. 10.1016/j.jgg.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Buman M. P., Hekler E. B., Bliwise D. L., King A. C., 2011. Exercise effects on night-to-night fluctuations in self-rated sleep among older adults with sleep complaints. J. Sleep Res. 20: 28–37. 10.1111/j.1365-2869.2010.00866.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler C. H., Chari S., Kowalski A., Choi L., Tack D., et al. , 2017. How well do you know your mutation? Complex effects of genetic background on expressivity, complementation, and ordering of allelic effects. PLoS Genet. 13: e1007075 10.1371/journal.pgen.1007075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y., Kelsey K. J. P., Wolfner M. F., Clark A. G., 2016. Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Hum. Mol. Genet. 25: 651–659. 10.1093/hmg/ddv502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y., Wolfner M. F., Clark A. G., 2013. Large neurological component to genetic differences underlying biased sperm use in Drosophila. Genetics 193: 177–185. 10.1534/genetics.112.146357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–90. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Bushey D., Hill S., Huber R., Kreber R., et al. , 2005. Reduced sleep in Drosophila Shaker mutants. Nature 434: 1087–1092. 10.1038/nature03486 [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembeck L. M., Boroczky K., Huang W., Schal C., Anholt R. R., et al. , 2015a Genetic architecture of natural variation in cuticular hydrocarbon composition in Drosophila melanogaster. eLife 4 10.7554/eLife.09861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembeck L. M., Huang W., Magwire M. M., Lawrence F., Lyman R. F., et al. , 2015b Genetic architecture of abdominal pigmentation in Drosophila melanogaster. PLoS Genet. 11: e1005163 10.1371/journal.pgen.1005163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon H. R., Lichstein K. L., Dautovich N. D., Taylor D. J., Riedel B. W., et al. , 2014. Variability in self-reported normal sleep across the adult age span. J. Gerontol. B Psychol. Sci. Soc. Sci. 70: 46–56. 10.1093/geronb/gbu035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L. L., Huang W., Quinn A. M., Ahuja A., Alfrejd B., et al. , 2014. Intrapopulation genome size variation in D. melanogaster reflects life history variation and plasticity. PLoS Genet. 10: e1004522 10.1371/journal.pgen.1004522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B. E., Ruedi E. A., McCoy L. J., Moore J. M., Wolfner M. F., et al. , 2015. Heritable variation in courtship patterns in Drosophila melanogaster. G3 (Bethesda) 5: 531–539. 10.1534/g3.114.014811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I., Donlea J., Shaw P. J., 2006. Waking experience affects sleep need in Drosophila. Science 313: 1775–1781. 10.1126/science.1130408 [DOI] [PubMed] [Google Scholar]
- Garlapow M. E., Huang W., Yarboro M. T., Peterson K. R., Mackay T. F., 2015. Quantitative genetics of food intake in Drosophila melanogaster. PLoS One 10: e0138129 10.1371/journal.pone.0138129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., Carbone M. A., Ayroles J. F., Stone E. A., Lyman R. F., et al. , 2009. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat. Genet. 41: 371–375. 10.1038/ng.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., McCoy L. J., Mackay T. F., 2013. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14: 281 10.1186/1471-2164-14-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., Serrano Negron Y. L., Hansen N. F., Lobell A. S., 2017. Selection for long and short sleep duration in Drosophila melanogaster reveals the complex genetic network underlying natural variation in sleep. PLoS Genet. 13: e1007098 10.1371/journal.pgen.1007098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhou S., St. Armour G. E., Mackay T. F., Anholt R. R., 2016. Epistatic partners of neurogenic genes modulate Drosophila olfactory behavior. Genes Brain Behav. 15: 280–290. 10.1111/gbb.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Lyman R. F., Lyman R. A., Carbone M. A., Harbison S. T., et al. , 2016. Spontaneous mutations and the origin and maintenance of quantitative genetic variation. eLife 5: e14625 10.7554/eLife.14625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Richards S., Carbone M. A., Zhu D., Anholt R. R., et al. , 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. USA 109: 15553–15559. 10.1073/pnas.1213423109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C. M., Huang W., Mackay T. F., Singh N. D., 2016. The Genetic Architecture of Natural Variation in Recombination Rate in Drosophila melanogaster. PLoS Genet. 12: e1005951 10.1371/journal.pgen.1005951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac R. E., Li C., Leedale A. E., Shirras A. D., 2010. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc. Biol. Sci. 277: 65–70. 10.1098/rspb.2009.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D. K., Escott-Price V., Zhiehm M., Magwire M. M., Mackay T. F., et al. , 2015. Longevity GWAS using the Drosophila Genetic Reference Panel. J Gerontol A Biol. Med. Sci. 70: 1470–1478. 10.1093/gerona/glv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan K. W., Craver K. L., Magwire M. M., Cubilla C. E., Mackay T. F., et al. , 2012. Genome-wide association for sensitivity to chronic oxidative stress in Drosophila melanogaster. PLoS One 7: e38722 10.1371/journal.pone.0038722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Kislukhin G., Walters K. N., Long A. D., 2014. Using Drosophila melanogaster to identify chemotherapy toxicity genes. Genetics 198: 31–43. 10.1534/genetics.114.161968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Macdonald S. J., Long A. D., 2012. Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 191: 935–949. 10.1534/genetics.112.138537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislukhin G., King E. G., Walters K. N., Macdonald S. J., Long A. D., 2013. The genetic architecture of methotrexate toxicity is similar in Drosophila melanogaster and humans. G3 (Bethesda) 3: 1301–1310. 10.1534/g3.113.006619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson K. L., Rathouz P. J., Yan L. L., Liu K., Lauderdale D. S., 2007. Intra-individual daily and yearly variability in actigraphically recorded sleep measures: the CARDIA study. Sleep 30: 793–796. 10.1093/sleep/30.6.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Joiner W. J., Wu M. N., Yue Z., Smith C. J., et al. , 2008. Identification of SLEEPLESS, a sleep-promoting factor. Science 321: 372–376. 10.1126/science.1155942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson H. A., Cady J. E., Partridge C., Wolf J. B., Semenkovich C. F., et al. , 2011. Genetic effects at pleiotropic loci are context-dependent with consequences for the maintenance of genetic variation in populations. PLoS Genet. 7: e1002256 10.1371/journal.pgen.1002256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell A. S., Kaspari R. R., Serrano Negron Y. L., Harbison S. T., 2017. The Genetic Architecture of Ovariole Number in Drosophila melanogaster: Genes with Major, Quantitative, and Pleiotropic Effects. G3 (Bethesda) 7: 2391–2403. 10.1534/g3.117.042390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2014. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Lyman R. F., 2005. Drosophila bristles and the nature of quantitative genetic variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1513–1527. 10.1098/rstb.2005.1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. 10.1038/nrg2612 [DOI] [PubMed] [Google Scholar]
- Mezick E. J., Matthews K. A., Hall M., Kamarck T. W., Buysse D. J., et al. , 2009. Intra-individual variability in sleep duration and fragmentation: Associations with stress. Psychoneuroendocrinology 34: 1346–1354. 10.1016/j.psyneuen.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery S. L., Vorojeikina D., Huang W., Mackay T. F., Anholt R. R., et al. , 2015. Genome-wide association analysis of tolerance to methylmercury toxicity in Drosophila implicates myogenic and neuromuscular developmental pathways. PLoS One 9: e110375 10.1371/journal.pone.0110375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante F., Sorensen P., Sorensen D. A., Maltecca C., Mackay T. F., 2015. Genetic architecture of micro-environmental plasticity in Drosophila melanogaster. Sci. Rep. 5: 9785 10.1038/srep09785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova T. V., Huang W., Pray V. A., Whitham T., Anholt R. R., et al. , 2015. Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult drosophila. BMC Genomics 16: 865 10.1186/s12864-015-2064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarro M. A., Hackett J. L., Smith B. R., Highfill C. A., King E. G., et al. , 2015. Identifying loci contributing to natural variation in xenobiotic resistance in Drosophila. PLoS Genet. 11: e1005663 10.1371/journal.pgen.1005663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Couch C., Huang W., Carbone M. A., Peiffer J., et al. , 2015. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. USA 112: E3555–E3563. 10.1073/pnas.1510104112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N., Young M. W., 2011. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72: 964–976. 10.1016/j.neuron.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Harbison S. T., Hahn L. E., Morozova T. V., Yamamoto A., et al. , 2012. Extensive epistasis for olfactory behavior, sleep and waking activity in Drosophila melanogaster. Genet. Res. 94: 9–20. 10.1017/S001667231200002X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Huang W., Mackay T. F., Anholt R. R., 2013. Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior. Proc. Natl. Acad. Sci. USA 110: 1017–1022. 10.1073/pnas.1220168110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Rottschaefer S. M., Lazzaro B. P., 2015. A genome-wide association study for nutritional indices in Drosophila. G3 (Bethesda) 5: 417–425. 10.1534/g3.114.016477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisnav M., Xing C., Ku H.-C., Hwang D., Stojadinovic S., et al. , 2014. Genome-wide association analysis of radiation resistance in Drosophila melanogaster. PLoS One 9: e104858 10.1371/journal.pone.0104858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., et al. , 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11 10 11–33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonesch S. C., Lamparter D., Mackay T. F., Bergmann S., Hafen E., 2016. Genome-wide analysis reveals novel regulators of growth in Drosophila melanogaster. PLoS Genet. 12: e1005616 10.1371/journal.pgen.1005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. L., Khan G. F., Magwire M. M., Tabor C. L., Mackay T. F., 2012. Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster. PLoS One 7: e34745 10.1371/journal.pone.0034745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A., Aw P. P., Bertrand D., Yeo G. H., Ong S. H., et al. , 2012. LoFreq: a sequence quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40: 11189–11201. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. J., Kumar S., Serrano Negron Y. L., Harbison S. T., 2018. Genotype influences day-to-day variability in sleep in Drosophila melanogaster. Sleep 41: zsx205 10.1093/sleep/zsx205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Zwarts L., Callaerts P., Norga K., Mackay T. F., et al. , 2008. Neurogenetic networks for startle-induced locomotion in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 105: 12393–12398. 10.1073/pnas.0804889105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwarts L., Vanden Broeck L., Cappuyns E., Ayroles J. F., Magwire M. M., et al. , 2015. The genetic basis of natural variation in mushroom body size in Drosophila melanogaster. Nat. Commun. 6: 10115 10.1038/ncomms10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DNA sequences have been deposited in the Sequence Read Archive under ID code SRP126512; BioProject PRJNA421951. Supplementary tables S1-S9, figures S1-S4, and File S1 have been deposited on figshare. A text file of variant calls and confidence intervals using both BWA and Novoalign sequence alignments (Files S2 and S3, respectively) and a list of annotated variants in .vcf format have been provided (Files S4 and S5, respectively) on figshare. The scripts used to conduct the HMM analysis are on github: http://github.com/nhansen/SleepInbredPanel. Sleep Inbred Panel lines are available from the Bloomington Drosophila Stock Center (Flybase stock numbers FBst0076271 – FBst0076309; Bloomington Drosophila stock center numbers 76271-76309) (Bloomington, IN). Supplemental material available at Figshare: https://doi.org/10.25387/g3.6789968.