Abstract
Poly-ADP-ribosylation has been proposed to be a reversible protein modification, participating in diverse cellular functions including DNA repair, chromatin remodeling, genetic stability, mitosis, and cell death. Poly-ADP-ribosylation is initiated by the transfer of the ADP-ribose moiety of NAD+ primarily to the carboxyl groups of glutamate and aspartate and amino group of lysine residues in target proteins, followed by elongation of poly(ADP-ribose) (PAR) chains via α-O-glycosidic (C-1″-C-2′) ribose-ribose bonds. PAR consists of polymers of ADP-ribose (up to 200 units) with branching via α-O-glycosidic (C-1‴-C-2″) ribose-ribose bonds. Further, the pyrophosphate group of each ADP-ribose has two negative charges. Therefore, in proteins modified by PAR, a complex structure with negative charges may lead to dynamic changes of functions. PAR formation is catalyzed by poly(ADP-ribose) polymerases (PARPs) and terminated by several types of enzymes with PAR-degrading activities; poly(ADP-ribose) glycohydrolase (PARG), ADP-ribosyl-acceptor hydrolase (ARH) 3, ARH1, and macrodomain-containing proteins. PARG has been thought to be primarily responsible for PAR degradation. In 2006, ARH3 was cloned and identified as another type of PAR-degrading protein. Although PAR-degrading activity of ARH3 is less than that of PARG, different mechanisms of PAR recognition and the cellular localization of ARH3 appear to be responsible for unique cellular roles of ARH3 involving PAR. In the present review, we focused on our findings regarding structure, biological properties, and cellular functions of ARH3. In addition, we describe the current knowledge of poly-ADP-ribosylation and cell death pathways regulated PARP1, PARG, and ARH3.
Keywords: Poly(ADP-ribose), Poly(ADP-ribose) polymerase 1, ADP-ribose-acceptor hydrolase 3, Parthanatos, Poly(ADP-ribose) glycohydrolase, Apoptosis-inducing factor
1. INTRODUCTION; ADP-RIBOSYL-ACCEPTOR HYDROLASE FAMILY
The ADP-ribosyl-acceptor hydrolase (ARH) family is composed of three 39-kDa proteins, ARH1-3 [1]. ARH1, the founding member of the ARH family, was initially isolated from turkey erythrocytes in 1988, and subsequently from human, rat, and mouse tissues [2, 3]. ARH1 is a cytoplasmic protein, which is widely expressed in tissues. ARH1 is a mono-ADP-ribosyl-acceptor hydrolase responsible for hydrolyzing the ADP-ribosyl-(arginine) protein bond, regenerating the unmodified acceptor [2]. NAD+: arginine mono-ADP-ribosyltransferases (ARTs) catalyze a post-translational modification of proteins, involving the transfer of the ADP-ribose moiety of NAD+ to the arginine residues of target proteins [4, 5]. The reaction is catalyzed by mammalian and bacterial toxin ARTs and the products of both enzyme families are hydrolyzed by ARH1. Therefore, the ARTs and ARH1 constitute a reversible mono-ADP-ribosylation cycle involving target proteins, which affects cellular functions. Using ARH1−/− mice and cells, we have found that ARH1 has crucial roles in tumorigenesis and in counteracting intoxication by cholera toxin [6, 7]. Detailed biological properties and cellular functions of ARH1 have been reviewed [1, 8].
Based on the structure of ARH1, human and mouse gene databases suggested the presence of ARH2 and ARH3 as ARH1-related proteins [9]. ARH2 and ARH3 were cloned and characterized in 2006 [10, 11]. Although ARH3 shows significant structural similarities to ARH1, substrates of ARH3 differ from those of ARH1. To date, ARH3 has been identified as hydrolyzing poly(ADP-ribose) (PAR) and O-acetyl-ADP-ribose (OAADPr), an NAD+-dependent sirtuin reaction product, but not mono-ADP-ribosylated arginine, the primary substrate of ARH1 [10–13]. ARH2 has not been reported to have any enzymatic activity, because of the substitution of amino acids critical for enzymatic activity, which are conserved in both ARH1 and ARH3 [11, 12].
Through PAR and OAADPr hydrolysis, ARH3 may have a role in these two signal transduction pathways and is expected to affect diverse cellular functions. So far, we have found that ARH3 regulates two cellular functions related to PAR degradation [14–16]. First, ARH3 serves as a suppressor of PAR-mediated cell death, called by parthanatos, under oxidative stress induced by exposure to hydrogen peroxide [16]. Second, ARH3 is primarily responsible for PAR hydrolysis in the mitochondrial matrix [14, 15]. Thus, PAR-degrading activity of ARH3 may be critical in the cellular response to stress. In the present review, we focused on our findings regarding structure, biological properties, and cellular functions of ARH3.
2. EXPRESSION AND CELLULAR DISTRIBUTION OF ARH3
ARH3 is conserved among many eukaryotes including insect, fish, chicken, and mammals. In humans, the ARH3 gene is located on chromosome 1 and its transcript contains 5 exons. In human and mouse, ARH3 is ubiquitously expressed in human and mouse tissues and cells [11]. It is highly expressed in brain, thymus, spleen, kidney, and ovary, while less so in heart and muscle. Intracellularly ARH3 is localized in the nucleus, cytoplasm, and mitochondria [11, 14, 16]. Like ARH1, the majority of ARH3 is found in the cytoplasm (65%) [16]. As ARH3 contains a mitochondrial-targeting sequence near its N-terminus, ARH3 is also distributed in the mitochondrial matrix (25%). In addition, ARH3 is also found in the nucleus (10%), although ARH3 does not contain a nuclear localization signal. Mechanisms by which ARH3 is localized in the nucleus have not been elucidated. ARH3 expression in the nucleus might depend on cell type; ARH3 expression was detected in nuclei of mouse brain and cultured mouse embryonic fibroblasts (MEFs), but not HepG2 cells [11, 16].
3. ENZYMATIC ACTIVITY OF ARH3
Human ARH3 has 41% similarity to human ARH1 [11]. ARH1 and ARH3 recognize the ADP-ribose moiety in their substrates; both ARH1 and ARH3 are inhibited best by ADP-ribose, and to a lesser extent by NAD+ and its derivatives including ribose 5-phospate, AMP, and ADP [12, 17]. However, despite the amino acid sequence similarity and the similar substrate recognition, ARH1 and ARH3 prefer different substrates. ARH3 hydrolyzes PAR and OAADPr. ARH3 recognizes the α-O-glycosidic bond formed by a carbon at the C-1″ position of ADP-ribose in PAR and OAADPr, whereas ARH1 hydrolyzes the α-N-glycosidic bond formed between the carbon at the C-1″ position of ADP-ribose and the guanidino group of arginine as well as the linkage in PAR and OAADPr [2, 10–13]. ARH3 does not hydrolyze ADP-ribose attached to arginine, cysteine, diphthamide, or asparagine [11], suggesting that the ADP-ribosyl-amino acid bond might not be a substrate for ARH3. ARH1 has a weak hydrolytic activity toward the α-O-glycosidic bond in PAR and OAADPr; ADP-ribosylated arginine is a much better substrate, indicating that ARH1 also appears to recognize the α-O-glycosidic bond [12]. The structural basis for the difference in the substrate specificity between ARH1 and ARH3 has not been determined.
ARH1 and ARH3 require Mg2+ for maximal activity [2, 11]. In addition to Mg2+, ARH1 in some species such as rat and mouse shows a thiol dependence [3]. In rat ARH1, whose activity depends on DTT, cysteine at position 108 is the critical determinant of thiol dependence [18]. The replacement of cysteine at a position 108 with serine resulted in loss of thiol dependence, while the mutant ARH1 retained maximal activity similar to wild type ARH1. In human ARH1, replacement of serine at position 103 with cysteine resulted in a DTT requirement for maximal activity. Similar to human ARH1, ARH3 does not contain the cysteine residue responsible for thiol dependence [11]. In agreement, ARH3 activity did not require thiol [11].
3.1. PAR Degradation
ARH3 was shown to hydrolyze PAR, resulting in release of ADP-ribose [10–11]. PAR is a polymer composed of ADP-ribose (up to 200 units) in α-O-glycosidic (C-1″-C-2′) ribose-ribose bonds with branching via α-O-glycosidic (C-1‴-C-2″) ribose-ribose bonds at intervals of 50 units of ADP-ribose, in linear PAR chains [19, 20]. ARH3 hydrolyzes α-O-glycosidic (C-1″-C-2′) ribose-ribose bonds in PAR. Crystallization and structure analysis of ARH3 revealed that it is a monomeric protein with two-fold symmetry, formed by 19 α-helices [10]. Some of the α-helices in ARH3 form a cavity necessary to align two Mg2+ ions and to bind one ADP-ribose moiety in PAR with a Kd = 1.6 ± 0.1 μM. ARH3 appears to dock at the terminal ADP-ribose moiety of PAR chains. One of the Mg2+ ions coordinates the 2′-hydroxyl group of the ribose moiety of the second ADP-ribose, resulting in an increased reactivity of the C-1″ atom of the second ADP-ribose to nucleophilic attack, leading to release of the terminal ADP-ribose moiety. Removal of Mg2+ ions in the solvent as well as replacement of amino acids residues critical for arranging two Mg2+ ions resulted in loss of function [10, 11].
3.2. OAADPr Degradation
In addition to PAR-degrading activity, ARH3 hydrolyzes the α-O-glycosidic bond in OAADPr, generating acetate and ADP-ribose [12, 13]. Some sirtuin subtypes possess deacetylase activity, which removes the acetyl group from acetylated-lysine residues with its transfer to the ADP-ribose of NAD+, and release of nicotinamide, resulting in formation of OAADPr [21]. OAADPr itself has diverse physiological functions including regulation of oocyte maturation, gene silencing, energy metabolism, and gating of the TRPM2 cation channel [22–25]. The acetyl group of OAADPr is reversibly interconverted among the hydroxyl groups attached to C-1″, 2″, and 3″ of ribose in OAADPr in a pH-dependent equilibrium [13, 26]. ARH3 only recognized the acetyl group attached to a carbon at the C-1″ position of ribose in OAADPr [13]. At pH 5, 3″-OAADPr isomer, an acetyl group attached to a carbon at C-3″ of ADP-ribose, predominated, but ARH3 did not display OAADPr hydrolase activity. At higher pH, the 1″-OAADPR isomer predominates and ARH3 activity was evident. In addition, 2″- and 3″-N-acetyl-ADP-ribose analogs of OAADPr did not inhibit ARH3 activity. When OAADPr was hydrolyzed by ARH3 in H2O18, O18 was incorporated into ADP-ribose. If nucleophilic attack of H2O18 occurred at 2″- or 3″-OAADPr isomer, O18 should be transferred to acetate. Thus, ARH3 catalyzes the nucleophilic attack of H2O at C-1″ of ribose in OAADPr, cleaving an O-glycosidic bond between ADP-ribose and the acetyl group. The evidence also strongly indicates that ARH3 recognizes the O-glycosidic bond formed by the carbon at the C-1″ position of ADP-ribose, degrading PAR as well as OAADPr.
4. PARP1 MEDIATES CELL DEATH, PARTHANA-TOS, FOLLOWING DNA DAMAGE
Poly-ADP-ribosylation results from the synthesis of ADP-ribose chains attached to proteins and is involved in diverse cellular functions including DNA repair, mitosis, transcription, and cell death [27–29]. PAR itself has an important role in the induction of cell death after DNA damage [30–32]. PAR is synthesized by poly(ADP-ribose) polymerases (PARPs) [33–35]. PARP1 is an abundant nuclear protein, which serves as a sensor of DNA lesions. Enzymatic activity of PARP1 is low in unstimulated cells. Once PARP1 is activated in response to DNA damage by ionizing irradiation, DNA alkylation, and oxidative stress, PAR synthesis is increased rapidly by as much as 500-fold [36]. PAR synthesized by PARP1 auto-modifies PARP1 itself. PAR is also transferred to a variety of nuclear acceptor proteins.
Based on the sequence of the catalytic domain of PARP1, a human database search revealed 17 homologs of PARP proteins [37]. PARP1, PARP2, and PARP3 appear to be DNA damage-inducible PARP isoforms, which are essential for multiple DNA repair systems; single-strand DNA break repair via the base excision repair (BER) and double-strand DNA break repair via homologous recombination (HR) and non-homologous end joining (NHEJ) repair [38–42]. Following DNA-damaging stimuli, these PARP isoforms catalyze PAR synthesis. PARP1 activity accounts for 90% of total PAR production during DNA damage, while the remainder is accounted for by PARP2 [38]. Several DNA repair proteins and scaffold proteins possess PAR-binding domains, including PAR-binding linear motifs, PAR-binding zinc fingers, WWE domains and macrodomains, which are involved in the interaction with PAR [43, 44]. After PARP1 binds to damaged DNA, PAR attached to PARP1 recruits DNA repair machinery to DNA lesions through PAR-binding domains. In addition, PARP1 modifies histones [45, 46]. Electrical repulsion and steric hindrance of PAR attached to histones relax chromatin condensation, leading to chromatin remodeling, which the DNA-repair machinery can access. Thus, the activation of PARP1 by moderate DNA damage results in DNA repair and genetic stability.
In contrast, when DNA damage is beyond the capacity of DNA repair in cells, massive PARP1 activation results in cell death, termed by parthanatos. Parthanatos, PARP1-dependent cell death, was designed to distinguish it from necrosis and apoptosis, although parthanatos shares morphological features with these forms of cell death [30]. Parthanatos is seen in neuronal cells in Parkinson’s disease, animal models of brain ischemia/reperfusion, and NMDA excitotoxicity [31, 47–49]. Since this type of cell death was discovered in 2002, research has been conducted to elucidate intracellular mechanisms underlying the induction of parthanatos. The initial step is export of PAR to the cytoplasm [32]. PAR synthesized by PARP1 in the nucleus translocates to the cytoplasm. After export from the nucleus, PAR interacts with apoptosis-inducing factor (AIF) on the surface of mitochondrial membranes, resulting in AIF release [30, 49, 50]. Owing to the presence of a nuclear localization signal in AIF, the protein released from mitochondria in turn translocates to the nucleus, then inducing large-scale DNA fragmentation and cell death [51]. In addition, recent proteomics studies demonstrate that hexokinase 1, an essential enzyme for initiation of glycolysis by converting glucose to glucose-6-phosphate, contains a PAR-binding motif and also plays an important role in the induction of parthanatos [52, 53]. Hexokinase 1 is localized mainly in the outer mitochondrial membrane through an interaction with the mitochondrial voltage-dependent anion channels (VDAC) [52]. The interaction of hexokinase 1 through its PAR-binding motif with PAR synthesized by PARP1 results in the suppression of hexokinase 1 activity and/or its mislocalization by dissociating it from VDAC, leading to inhibition of glycolysis and ATP loss, resulting in cell death [52, 53]. Thus, PARP1 activation, which is correlated with the extent of DNA damage, can determine whether the cell undergoes DNA repair or death and PAR serves as a death signal transducer between the nucleus and mitochondria.
5. PARG IS PRIMARILY RESPONSIBLE FOR PAR DEGRADATION
Poly(ADP-ribose) glycohydrolase (PARG), which hydrolyzes the α-O-glycosidic ribose-ribose bond in PAR, was purified from calf thymus extracts in 1971. The cDNA encoding bovine PARG was cloned in 1997 [54, 55]. In the process of transcription, PARG mRNA undergoes alternative splicing, generating PARG isoforms of different size, cellular localization, and activity [56, 57]. PARG is composed of 18 exons with separate N-terminal regulatory domain, mitochondrial-targeting sequence, and C-terminal catalytic domain [56]. The regulatory domain contains two nuclear localization signals and a nuclear export signal in exons 1 and 3, respectively, whereas the catalytic domain is essential for PARG activity. In humans, full-length 111-kDa PARG is predominantly localized in the nucleus, because of the nuclear localization signal in exon 1. Human 102-kDa and 99-kDa PARG are translated from the alternative start codon in exon 2 and exon 3, respectively, which removes the nuclear localization signal, resulting in cytoplasmic localization [56]. In addition, small 60-kDa and 55-kDa PARG isoforms are translated from the start codon in exon 1a and exon 4, respectively [57]. 60-kDa PARG isoform is localized in the cytoplasm, whereas 55-kDa PARG isoform is found in mitochondria due to the mitochondrial targeting sequence in exon 4. However, 60-kDa and 55-kDa PARG isoforms are catalytically inactive, because of the absence of exon 5, which is critical for PARG activity [15]. 111-kDa, 102-kDa, and 99-kDa PARG have been reported to shuttle between nucleus and cytoplasm in response to DNA damage [58, 59]. Once DNA damage occurs,102-kDa and 99-kDa PARG isoforms are recruited to DNA damage sites in PAR- and PCNA-dependent mechanisms [58], whereas 111-kDa PARG accumulates in DNA-damage sites, and translocates to the cytoplasm [59]. Relocation of nuclear and cytoplasmic PARG isoforms suggests that these PARG isoforms can degrade PAR in different cellular compartments.
Although still controversial, PARG is believed to have endo- and exoglycosidase activities toward PAR [60–64]. In vitro experiments revealed that PARG preferentially hydrolyzes long and linear PAR, with small and branched PAR less favored as substrates [60, 65, 66]. Hatakeyama et al. indicated that PARG activity toward large PAR (greater than 20 ADP-ribose moieties) proceeds in a biphasic manner [65]. In the early phase, PARG degrades large polymers rapidly, generating small PAR and ADP-ribose. In the late phase, PARG degrades the small PAR at a 20-times slower rate than the initial rate. Km and Vmax for large PAR composed of 18, 45, and 126 ADP-ribose moieties are 0.39, 0.2, 0.11 μM and 110, 123, and 123 μmol/min/mg protein, respectively, while those for small PAR composed of less than 20 ADP-ribose moieties are <10 μM and <45 μmol/min/mg, respectively; the Km for large PAR is lower than that for the small form and large PAR is digested by PARG at a faster rate than the small form [65]. The significant difference of Km values might be related to the observation that PARG requires a certain number of ADP-ribose moieties in the PAR polymer and/or high-ordered structure of PAR for binding. In addition, nuclear PARG purified from human placenta degrades PAR bound to acceptor proteins, such as Histone H1 and PARP1, preferentially to protein-free PAR [67]. Therefore, the different kinetics toward polymer sizes of PAR and preferential degradation of protein-bound PAR suggest that PARG through its endo-glycosidase activity degrades long PAR chains attached to acceptor proteins, generating protein-free small PAR fragments. With exo-glycosidase activity, PARG degrades small PAR polymers, generating ADP-ribose. Thereby, PARG can hydrolyze PAR efficiently through both endo- and exo-glycosidase activities, but can also produce protein-free small PAR fragments, which serve as signaling molecules for the induction of parthanatos.
Genetic disruption of all PARG isoforms resulted in early embryonic lethality, because of excessive PAR accumulation in embryos [68]. Similarly, a Drosophila mutant lacking the catalytic domain of PARG, when grown at 25°C, showed lethality at the larval stage [69]. When cultured at 29°C, some mutants were viable, but exhibited extensive PAR accumulation in the central nervous system, neurodegeneration with reduced locomotor activity, and short life-span. Thus, PARG has an essential role in survival and the development of neuronal cells. In contrast, transgenic mice expressing the targeted deletion of exon 2 and 3 in the PARG gene (PARGΔexon2-3/Δexon2-3), which express 60-kDa cytoplasmic PAR isoforms, but not the 110-kDa nuclear PARG isoform, are viable, but show increased sensitivity to alkylating agents and ionizing radiation, impaired DNA-repair response, and genomic instability [70]. However, there is conflicting evidence regarding PARG effects on cell death. PARGΔexon2-3/Δexon2-3 showed reduced sensitivity to renal and intestinal injury induced by ischemia/reperfusion [71, 72]. Furthermore, administration of PARG inhibitors and depletion of PARG proteins by siRNA or shRNA protected against cell death in animal models of stroke and hydrogen peroxide exposure [16, 73–76]. Dual effects of PARG on cell death may be explained by exo- and endo-glycosidic activities of PARG, which can generate ADP-ribose as well as small, protein-free PAR fragments, acting as a signaling molecule for the induction of parthanatos.
6. CELLULAR FUNCTIONS OF ARH3
6.1. ARH3 Inhibited PARP1-Dependent Cell Death, Parthanatos, through PAR Hydrolysis in the Nucleus and Cytoplasm
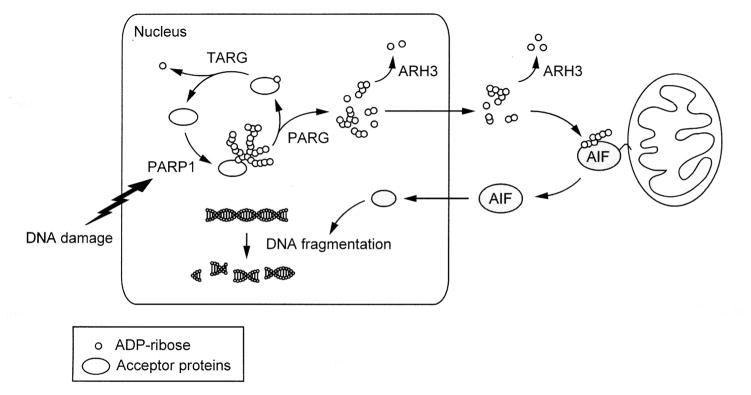
ARH3 has a cytoprotective effect against parthanatos, PARP1-mediated cell death, following exposure to hydrogen peroxide, by catalyzing PAR hydrolysis in the nucleus and cytoplasm (Fig. 1) [16]. ARH3−/− MEFs were more susceptible to hydrogen peroxide-induced cell death than ARH3+/+ MEFs [16]. Cell death observed in ARH3−/− MEFs was not blocked by a caspase inhibitor, zVAD-fmk, and was accompanied by nuclear shrinkage and phosphatidylserine exposure on the external leaflet of the plasma membrane. After exposure to hydrogen peroxide, ARH3−/− MEFs displayed within 20 min elevated PAR accumulation in nuclei, followed by its translocation to cytoplasm within 1 h and then to mitochondria after 2 h. ARH3 is located primarily in the cytoplasm and regulates cytoplasmic PAR content. Moreover, as PAR content was increased in nuclei of ARH3−/− MEFs, ARH3 also regulates nuclear PAR. In ARH3−/− MEFs, increased PAR content in the cytoplasm triggered AIF release from mitochondria and its accumulation in nuclei, resulting in large-scale DNA fragmentation and cell death. In ARH3−/− MEFs, stable expression of PARP1 shRNA prevented PAR accumulation in nuclei, its translocation to the cytoplasm and mitochondria, which then reduced the cytotoxic effect of hydrogen peroxide.
Fig. 1. ARH3 participates in PAR degradation in the nucleus and cytoplasm, thereby inhibiting parthanatos.
Upon DNA damage, activated PARP1 synthesizes PAR from NAD+, resulting in the poly-ADP-ribosylation of nuclear acceptor proteins including PARP1 itself. 110-kDa nuclear PARG, through its exo- and endo-glycosidase activities, degrades PAR polymers, generating ADP-ribose monomers and small PAR fragments, respectively. The initial ADP-ribose attached to acceptor proteins is degraded by macrodomain-containing proteins such as TARG1 (terminal ADP-ribose glycohydrolase; C6orf130) which are responsible for release of an ADP-ribose monomer attached to protein [78]. Small PAR fragments then pass through nuclear membranes, although the detailed mechanisms are still unknown. In the cytoplasm, PAR binds to the PAR-binding site AIF, releasing AIF from mitochondrial membranes. Released AIF is translocated to the nucleus, resulting in DNA fragmentation and cell death. ARH3 is located in the nucleus and cytoplasm, where ARH3 degrades PAR, preventing AIF release from mitochondria.
After genotoxic stimuli, most PAR synthesized by PARP1 is attached to PARP1 itself and other nuclear acceptor proteins. Each ADP-ribose has a pyrophosphate moiety with two negative charges. Therefore, PAR is a large, bulky structure with high negative charges. PAR attached to nuclear proteins is presumed to be unable to pass through nuclear pores. One of the key steps in parthanatos is export of PAR from the nucleus [32]. Through its endoglycosidase activity, PARG may perform a necessary step for PAR export by generating small protein-free PAR fragments from poly-ADP-ribosylated proteins [16]. Partial knockdown of PARG proteins by shRNA in ARH3−/− MEFs reduced sensitivity to hydrogen peroxide-induced parthanatos. This effect resulted from an inability to generate in nuclei protein-free PAR fragments from poly-ADP-ribosylated proteins such as PARP1. ARH3−/− MEFs expressing PARG shRNA showed elevated accumulation of PAR content in nuclei, but retained poly-ADP-ribosylation on PARP1, thereby reducing PAR translocation to the cytoplasm and PAR-mediated AIF release from mitochondria.
A single PARG gene gives rise to multiple isoforms with different localizations and activities [56–57]. Despite the fact that PARG−/− mice show early embryonic lethality, because of robust PAR accumulation, PARGΔexon2-3/Δexon2-3 mice lacking the nuclear 110-kDa PARG isoform, but expressing the cytoplasmic 60-kDa PARG isoform, are viable and fertile [70]. Consistent with this hypothesis, PARGΔexon2-3/Δexon2-3 mice were protected from renal and intestinal injuries after ischemia/reperfusion, while these mice were more susceptible to alkylating agents and ionizing radiation [70]. Therefore, the nuclear PARG isoform might be responsible for generating protein-free PAR fragments from nuclear acceptor proteins in response to certain genotoxic stimuli. In contrast, cytoplasmic PARG isoforms affects cell survival, probably through cytoplasmic PAR degradation.
Based on our data, ARH3 may be critical to cell survival under stress conditions when excessive PAR is synthesized (Fig. 1). The exact values of these kinetics for ARH3 have not been determined. According to Oka et al., 2 μM ARH3 produced as much ADP-ribose as 1 nM PARG, with 900 ng [14C] poly(ADP-ribosylated)PARP as substrate, suggesting ARH3 activity was considerably less than that of PARG [11]. Unlike PARG−/− mice, ARH3−/− mice are healthy and fertile [16]. ARH3−/− MEFs did not show detectable PAR accumulation under basal conditions [16]. These results indicate that ARH3 is not a major regulator of PAR metabolism and PAR-mediated cellular responses under basal conditions. In contrast, PARG is indispensable in PAR metabolism under both basal and DNA-damage conditions. However, PARG preferentially degrades long PAR chains; its activity toward small PAR chains is relatively low [65]. Indeed, the Km value of PARG for small PAR is larger than for large form. In addition, PARG degrades protein-bound PAR more efficiently than protein-free PAR [67]. PARG may generate protein-free small PAR fragments from poly-ADP-ribosylated acceptor proteins under conditions in which excessive PAR is generated in response to DNA damage. These small PAR molecules are exported to the cytoplasm, where they participate in the induction of parthanatos. Based on our findings that ARH3 is mainly localized in the cytoplasm as well as nucleus and mitochondria and PAR export to the cytoplasm was seen in ARH3−/− MEFs after exposure to hydrogen peroxide, ARH3 might act as a scavenger of small, protein-free PAR fragments, which are not degraded effectively by PARG. Thus, sequential actions of PARG and ARH3 have an essential role in regulating PAR localization in the nucleus and cytoplasm and PAR-dependent AIF release from mitochondria, leading to parthanatos.
6.2. ARH3 is Primarily Responsible for Hydrolysis of PAR in the Mitochondrial Matrix
ARH3 was predicted to localize in mitochondria due to its N-terminal mitochondrial targeting sequence; subcellular fractionation analysis confirmed ARH3 distribution in mitochondria [14, 16]. To assess ARH3 activity in mitochondria, the catalytic domain of PARP1 fused with a mitochondrial targeting sequence (mito-PARP1cd) was used to induce PAR synthesis confined to the mitochondrial matrix [14, 15]. Overexpressed ARH3 effectively degraded mito-PARP1cd-induced PAR in the mitochondrial matrix of HEK cells. ARH3−/− MEFs accumulated more mito-PARP1cd-induced PAR content in mitochondria than did ARH3+/+ cells [15]. In addition, in the presence of PJ34, a PARP inhibitor, ARH3−/− MEFs showed reduced turnover of mito-PARP1cd-induced PAR in mitochondria compared to ARH3+/+ MEFs [15]. A splicing event in the PARG gene generated a small 55-kDa PARG isoform, with a mitochondrial-targeting sequence [15, 57]. FLAG-tagged 55-kDa PARG isoform was localized in mitochondria, but failed to hydrolyze mitochondrial PAR. As the 55-kDa PARG isoform does not possess exon 5, which is required for PARG activity, it is presumably an inactive isoform. These results indicate that ARH3, but not the mitochondrial 55-kDa PARG isoform, is primarily responsible for PAR hydrolysis in the mitochondrial matrix. Although some reports describe the presence of PAR in mitochondria, neither a PARP isoform nor poly-ADP-ribosylated acceptor proteins have been identified in mitochondria [77]. Thus, roles of PAR and ARH3 in PAR metabolism in mitochondria are not clear.
CONCLUSION
Using ARH3−/− MEFs, we have elucidated two cellular functions involving the PAR hydrolytic activity of ARH3 [1, 14–16]. Through its PAR-degrading activity, ARH3 regulates PAR metabolism in the nucleus, cytoplasm, and mitochondria. Of importance, ARH3 prevents parthanatos caused by exposure to hydrogen peroxide [16]. Parthanatos participates in neuronal cell death in Parkinson’s disease and stroke [31, 47–49]. Elucidation of the detailed mechanism underlying parthanatos may lead to new therapeutic targets for these diseases. Mouse model studies using ARH3−/− mice will be needed further to confirm the role of ARH3 in neuronal cell death regulated by parthanatos.
Although ARH3 degrades OAADPr as well as PAR, cellular functions related to the OAADPr-degrading activity of ARH3 have not been found. Some sirtuin subtypes possess deacetylase activity, which removes the acetyl group from acetylated-lysine residues, transferring it to the ADP-ribose of NAD+ with the release of nicotinamide, thereby generating OAADPr [21]. Localization of ARH3 overlaps with that of some sirtuin subtypes with deacetylase activity. Therefore, ARH3 might regulate these sirtuin activities through OAADPr metabolism. In addition, OAADPr itself has diverse physiological functions including regulation of oocyte maturation, gene silencing, energy metabolism, and gating of TRPM2 cation channel [22–25]. Thus, ARH3 may have physiological and pathological roles related to its OAADPr-degrading activity.
Acknowledgments
This study was supported by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute and a Grant-in-Aid for Scientific Research (15K18871) from the Ministry of Education, Science, Sports and Culture of Japan.
LIST OF ABBREVIATIONS
- ARH
ADP-ribosyl-acceptor hydrolase
- ART
ADP-ribosyltransferase
- AIF
Apoptosis-inducing factor
- BER
Base excision repair
- mito-PARP1cd
Catalytic domain of PARP1 with a mitochondrial targeting sequence
- HR
Homologous recombination
- MEF
Mouse embryonic fibroblast
- NHEJ
Non-homologous end joining
- OAADPr
O-acetyl-ADP-ribose
- PAR
Poly(ADP-ribose)
- PARG
Poly(ADP-ribose) glycohydrolase
- PARP
Poly(ADP-ribose) polymerase
- PARGΔexon2-3/Δexon2-3
Targeted deletion of exon 2 and 3 in the PARG gene
- TARG1
Terminal ADP-ribose protein glycohydrolase
- VDAC
Voltage-dependent anion channels
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Mashimo M, Kato J, Moss J. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair (Amst) 2014;23:88–94. doi: 10.1016/j.dnarep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moss J, Tsai SC, Adamik R, Chen HC, Stanley SJ. Purification and characterization of ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1988;27(15):5819–5823. doi: 10.1021/bi00415a063. [DOI] [PubMed] [Google Scholar]
- 3.Moss J, Stanley SJ, Nightingale MS, Murtagh JJ, Jr, Monaco L, Mishima K, Chen HC, Williamson KC, Tsai SC. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J Biol Chem. 1992;267(15):10481–10488. [PubMed] [Google Scholar]
- 4.Okazaki IJ, Zolkiewska A, Takada T, Moss J. Characterization of mammalian ADP-ribosylation cycles. Biochimie. 1995;77(5):319–325. doi: 10.1016/0300-9084(96)88141-7. [DOI] [PubMed] [Google Scholar]
- 5.Moss J, Zolkiewska A, Okazaki I. ADP-ribosylarginine hydrolases and ADP-ribosyltransferases. Partners in ADP-ribosylation cycles. Adv Exp Med Biol. 1997;419:25–33. doi: 10.1007/978-1-4419-8632-0_3. [DOI] [PubMed] [Google Scholar]
- 6.Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27(15):5534–5543. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato J, Zhu J, Liu C, Stylianou M, Hoffmann V, Lizak MJ, Glasgow CG, Moss J. ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis. Cancer Res. 2011;71(15):5327–5335. doi: 10.1158/0008-5472.CAN-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takada T, Okazaki IJ, Moss J. ADP-ribosylarginine hydrolases. Mol Cell Biochem. 1994;138(1–2):119–122. doi: 10.1007/BF00928452. [DOI] [PubMed] [Google Scholar]
- 9.Glowacki G, Braren R, Firner K, Nissen M, Kuhl M, Reche P, Bazan F, Cetkovic-Cvrlje M, Leiter E, Haag F, Koch-Nolte F. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11(7):1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller-Dieckmann C, Kernstock S, Lisurek M, von Kries JP, Haag F, Weiss MS, Koch-Nolte F. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci U S A. 2006;103(41):15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281(2):705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 12.Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc Natl Acad Sci U S A. 2006;103(45):16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J, Denu JM, Moss J. Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J Biol Chem. 2011;286(24):21110–21117. doi: 10.1074/jbc.M111.237636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28(2):814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niere M, Mashimo M, Agledal L, Dolle C, Kasamatsu A, Kato J, Moss J, Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J Biol Chem. 2012;287(20):16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashimo M, Kato J, Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc Natl Acad Sci U S A. 2013;110(47):18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss J, Oppenheimer NJ, West RE, Jr, Stanley SJ. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1986;25(19):5408–5414. doi: 10.1021/bi00367a010. [DOI] [PubMed] [Google Scholar]
- 18.Takada T, Iida K, Moss J. Cloning and site-directed mutagenesis of human ADP-ribosylarginine hydrolase. J Biol Chem. 1993;268(24):17837–17843. [PubMed] [Google Scholar]
- 19.Miwa M, Saikawa N, Yamaizumi Z, Nishimura S, Sugimura T. Structure of poly(adenosine diphosphate ribose): identification of 2′-[1″-ribosyl-2″-(or 3″-)(1‴-ribosyl)]adenosine-5′,5″,5‴-tris(phosphate) as a branch linkage. Proc Natl Acad Sci U S A. 1979;76(2):595–599. doi: 10.1073/pnas.76.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juarez-Salinas H, Levi V, Jacobson EL, Jacobson MK. Poly(ADP-ribose) has a branched structure in vivo. J Biol Chem. 1982;257(2):607–609. [PubMed] [Google Scholar]
- 21.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 22.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121(4):515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 23.Borra MT, O’Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277(15):12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 24.Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud AL, Scharenberg AM, Denu JM. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J Biol Chem. 2006;281(20):14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- 25.Tong L, Lee S, Denu JM. Hydrolase regulates NAD+ metabolites and modulates cellular redox. J Biol Chem. 2009;284(17):11256–11266. doi: 10.1074/jbc.M809790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson MD, Denu JM. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277(21):18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 27.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 28.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13(7):411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7(7):517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 30.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 31.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103(48):18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103(48):18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alkhatib HM, Chen DF, Cherney B, Bhatia K, Notario V, Giri C, Stein G, Slattery E, Roeder RG, Smulson ME. Cloning and expression of cDNA for human poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A. 1987;84(5):1224–1228. doi: 10.1073/pnas.84.5.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurosaki T, Ushiro H, Mitsuuchi Y, Suzuki S, Matsuda M, Matsuda Y, Katunuma N, Kangawa K, Matsuo H, Hirose T, et al. Primary structure of human poly(ADP-ribose) synthetase as deduced from cDNA sequence. J Biol Chem. 1987;262(33):15990–15997. [PubMed] [Google Scholar]
- 35.Uchida K, Morita T, Sato T, Ogura T, Yamashita R, Noguchi S, Suzuki H, Nyunoya H, Miwa M, Sugimura T. Nucleotide sequence of a full-length cDNA for human fibroblast poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1987;148(2):617–622. doi: 10.1016/0006-291x(87)90921-1. [DOI] [PubMed] [Google Scholar]
- 36.Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 37.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35(4):208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274(25):17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 39.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A. 2011;108(7):2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41(1):33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Helleday T, Bryant HE, Schultz N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 2005;4(9):1176–1178. doi: 10.4161/cc.4.9.2031. [DOI] [PubMed] [Google Scholar]
- 42.Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39(25):7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 43.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36(22):6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280(15):3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 45.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79(11):3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255(16):7616–7620. [PubMed] [Google Scholar]
- 47.Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, West AB, Koehler RC, Poirier GG, Dawson TM, Dawson VL. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17(6):692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y, Karuppagounder SS, Shin JH, Lee YI, Ko HS, Swing D, Jiang H, Kang SU, Lee BD, Kang HC, Kim D, Tessarollo L, Dawson VL, Dawson TM. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16(10):1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1(5) doi: 10.1042/AN20090046. pii: e00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25(5):259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, Wheeler D, Redpath P, Li J, Romero G, Migaud M, Van Houten B, Poirier GG, Sobol RW. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8(6):1819–1831. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppa-gounder SS, Gagne JP, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci U S A. 2014;111(28):10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miwa M, Sugimura T. Splitting of the ribose-ribose linkage of poly(adenosine diphosphate-robose) by a calf thymus extract. J Biol Chem. 1971;246(20):6362–6364. [PubMed] [Google Scholar]
- 55.Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272(18):11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 56.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297(2):521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 57.Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res. 2007;313(13):2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H, Schreiber V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011;39(12):5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haince JF, Ouellet ME, McDonald D, Hendzel MJ, Poirier GG. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim Biophys Acta. 2006;1763(2):226–237. doi: 10.1016/j.bbamcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 60.Braun SA, Panzeter PL, Collinge MA, Althaus FR. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur J Biochem. 1994;220(2):369–375. doi: 10.1111/j.1432-1033.1994.tb18633.x. [DOI] [PubMed] [Google Scholar]
- 61.Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim Biophys Acta. 1994;1219(2):342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 62.Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, Lafite P, Ahel M, Mitchison TJ, Ahel I, Leys D. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat Commun. 2013;4:2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ikejima M, Gill DM. Poly(ADP-ribose) degradation by glycohydrolase starts with an endonucleolytic incision. J Biol Chem. 1988;263(23):11037–11040. [PubMed] [Google Scholar]
- 64.Miwa M, Tanaka M, Matsushima T, Sugimura T. Purification and properties of glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose) J Biol Chem. 1974;249(11):3475–3482. [PubMed] [Google Scholar]
- 65.Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) J Biol Chem. 1986;261(32):14902–14911. [PubMed] [Google Scholar]
- 66.Malanga M, Althaus FR. Poly(ADP-ribose) molecules formed during DNA repair in vivo. J Biol Chem. 1994;269(26):17691–17696. [PubMed] [Google Scholar]
- 67.Uchida K, Suzuki H, Maruta H, Abe H, Aoki K, Miwa M, Tanuma S. Preferential degradation of protein-bound (ADP-ribose)n by nuclear poly(ADP-ribose) glycohydrolase from human placenta. J Biol Chem. 1993;268(5):3194–3200. [PubMed] [Google Scholar]
- 68.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101(51):17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101(1):82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24(16):7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cuzzocrea S, Di Paola R, Mazzon E, Cortes U, Genovese T, Muia C, Li W, Xu W, Li JH, Zhang J, Wang ZQ. PARG activity mediates intestinal injury induced by splanchnic artery occlusion and reperfusion. FASEB J. 2005;19(6):558–566. doi: 10.1096/fj.04-3117com. [DOI] [PubMed] [Google Scholar]
- 72.Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, Wang ZQ, Thiemermann C. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolase are protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 2005;16(3):712–719. doi: 10.1681/ASN.2004080677. [DOI] [PubMed] [Google Scholar]
- 73.Blenn C, Althaus FR, Malanga M. Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. Biochem J. 2006;396(3):419–429. doi: 10.1042/BJ20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erdelyi K, Bai P, Kovacs I, Szabo E, Mocsar G, Kakuk A, Szabo C, Gergely P, Virag L. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 2009;23(10):3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Genovese T, Di Paola R, Catalano P, Li JH, Xu W, Massuda E, Caputi AP, Zhang J, Cuzzocrea S. Treatment with a novel poly(ADP-ribose) glycohydrolase inhibitor reduces development of septic shock-like syndrome induced by zymosan in mice. Crit Care Med. 2004;32(6):1365–1374. doi: 10.1097/01.ccm.0000127775.70867.0c. [DOI] [PubMed] [Google Scholar]
- 76.Lu XC, Massuda E, Lin Q, Li W, Li JH, Zhang J. Post-treatment with a novel PARG inhibitor reduces infarct in cerebral ischemia in the rat. Brain Res. 2003;978(1–2):99–103. doi: 10.1016/s0006-8993(03)02774-4. [DOI] [PubMed] [Google Scholar]
- 77.Scovassi AI. Mitochondrial poly(ADP-ribosylation): from old data to new perspectives. FASEB J. 2004;18(13):1487–1488. doi: 10.1096/fj.04-1841rev. [DOI] [PubMed] [Google Scholar]
- 78.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32(9):1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]