CONSPECTUS:
Glycosylation is one of the most common protein modifications, and it is essential for mammalian cell survival. It often determines protein folding and trafficking, and regulates nearly every extracellular activity, including cell−cell communication and cell−matrix interactions. Aberrant protein glycosylation events are hallmarks of human diseases such as cancer and infectious diseases. Therefore, glycoproteins can serve as effective biomarkers for disease detection and targets for drug and vaccine development. Despite the importance of glycoproteins, global analysis of protein glycosylation (either glycoproteins or glycans) in complex biological samples has been a daunting task, and here we mainly focus on glycoprotein analysis using mass spectrometry (MS)-based bottom-up proteomics. Although the emergence of MS-based proteomics has provided a great opportunity to analyze glycoproteins globally, the low abundance of many glycoproteins and the heterogeneity of glycans dramatically increase the technical difficulties. In order to overcome these obstacles, considerable progress has been made in recent years, which has contributed to comprehensive analysis of glycoproteins. In our lab, we developed effective MSbased chemical and enzymatic methods to (1) globally analyze glycoproteins in complex biological samples, (2) target glycoproteins specifically on the surface of human cells, (3) systematically quantify glycoprotein and surface glycoprotein dynamics (the abundance changes of glycoproteins as a function of time), and (4) selectively characterize glycoproteins with a particular and important glycan.
In this Account, we first briefly describe the glycopeptide/protein enrichment methods in the literature and then discuss the developments of boronic acid-based methods to enrich glycopeptides for large-scale analysis of protein glycosylation. Boronic acids can form reversible covalent interactions with sugars, but the low binding affinity of normal boronic acid-based methods prevents us from capturing glycoproteins with low abundance, which often contain more valuable information. We enhanced the boronic acid−glycan interactions by using a boronic acid derivative (benzoboroxole) and conjugating it onto a dendrimer to allow synergistic interactions between the boronic acid derivative and sugars. The new method is capable of globally analyzing protein glycosylation with site and glycan structure information, especially for those with low abundance.
In the next part, we discuss the combination of metabolic labeling, click chemistry and enzymatic reactions, and MS-based proteomics as a very powerful approach for surface glycoproteome analysis in human cells. The methods enable us to specifically identify surface glycoproteins and to quantify their abundance changes and dynamics together with quantitative proteomics. The last section of this Account focuses on chemical and enzymatic methods to study glycoproteins containing a particular and important glycan (the Tn antigen, i.e., O-GalNAc). Although not comprehensive, this Account provides an overview of chemical and enzymatic methods to characterize protein glycosylation in combination with MS-based proteomics. These methods will have extensive applications in the fields of biology and biomedicine, which will lead to a better understanding of glycoprotein functions and the molecular mechanisms of diseases. Eventually, glycoproteins will be identified as effective biomarkers for disease detection and drug targets for disease treatment.
Graphical Abstract
1. INTRODUCTION
Protein glycosylation is essential for mammalian cell survival and plays critical roles in many cellular events.1,2 In contrast to DNA and proteins, glycans are not synthesized by following a sequence template but rather are produced by enzymes in cells. Many enzymes are responsible for protein glycosylation and glycan structure construction. Protein glycosylation is highly diverse, which is reflected in not only glycan structures but also the linkages between glycans and proteins. Glycoproteins contain a wealth of valuable information regarding the developmental and disease status of cells. Aberrant protein glycosylation is related to human diseases, including cancer and infectious diseases.3,4 Therefore, glycoproteins may serve as effective biomarkers and drug targets.5
Global and site-specific analysis of glycoproteins is vital to advance glycoscience research and to extract valuable information from glycoproteins for disease detection and treatment.6–11 Because of the heterogeneity of glycans, the low abundances of many glycoproteins, and the dynamic nature of glycosylation, it is extremely challenging to comprehensively analyze glycoproteins in complex biological samples. Innovative and effective analysis methods are urgently needed to advance glycoscience research in order to deepen our understanding of glycoprotein functions and unravel the molecular mechanisms of diseases. Furthermore, global analysis of glycoproteins, particularly those with low abundance, will lead to the discovery of glycoproteins as effective disease biomarkers and the identification of drug targets. This Account mainly summarizes recent mass spectrometry (MS)-based chemical and enzymatic methods developed in our lab for comprehensive analysis of protein glycosylation.
2. EFFECTIVE MS-BASED CHEMICAL METHODS FOR GLOBAL ANALYSIS OF GLYCOPROTEINS
2.1. Glycopeptide Enrichment for Global Analysis
Because the abundances of many glycoproteins are very low, effective enrichment is essential for global characterization of glycoproteins prior to MS analysis. There have been many reports about the development of enrichment methods, and several existing methods have been widely used.11–18 For example, lectins have been extensively applied to enrich glycopeptides/proteins. Zielinska et al.16 used lectin enrichment to analyze the N-glycoproteome in mouse tissues. Although lectins are commonly used, they cannot enrich all glycopeptides because of the limited ability of each lectin to recognize specific glycans.
Hydrophilic interaction liquid chromatography (HILIC) has also been used for glycopeptide/protein enrichment.19,20 However, because HILIC-based methods are dependent only on the hydrophilicity difference, hydrophilic non-glycopeptides cannot be distinguished from glycopeptides, rendering the enrichment specificity suboptimal. Another popular and beautiful method is hydrazide chemistry-based enrichment.13 Glycans are first oxidized to generate aldehyde groups, and then hydrazide chemistry is performed to catch glycopeptides. In recent years, two very elegant methods to enrich and analyze glycoproteins were reported. Woo et al.17 developed the highly innovative isotope-targeted glycoproteomics (IsoTaG) method, which combines metabolic labeling and isotopic recording to analyze glycoproteins. Glycopeptides tagged with the probe display a unique isotopic pattern in full MS, which facilitates the glycopeptide sequencing, and software is employed to recognize the pattern. Sun et al.18 developed another innovative method, termed solid-phase extraction of N-linked glycans and glycositecontaining peptides (NGAG) by combining chemical and enzymatic reactions. This method targets N-glycopeptides, and a total of 2044 unique N-glycopeptides were identified in mammalian cells.
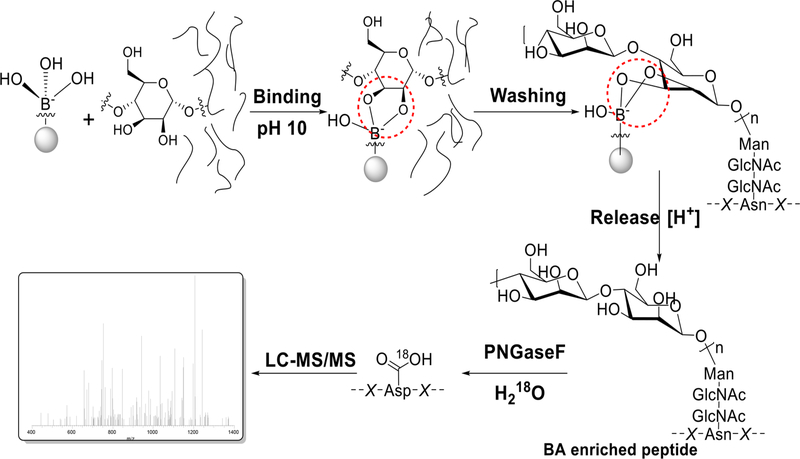
The reversible interactions between boronic acids and sugars have been extensively studied,21,22 and boronic acid-based probes have been explored for sugar detection. Boronic acidbased methods hold great potential to enrich glycopeptides/ proteins.23,24 Because of the common feature of glycans, i.e., one glycan contains multiple hydroxyl groups, boronic acid-based methods are universal to enrich glycopeptides with diverse glycans (Figure 1). By the use of magnetic beads conjugated with phenylboronic acid, glycopeptides were captured under basic conditions. After the removal of non-glycopeptides, intact glycopeptides were released in an acidic solution. This method allowed us to identify the highest number of protein Nglycosylation sites in yeast at that time.24 It was also applied to quantify protein N-glycosylation changes in yeast25 and to investigate the regulation of protein secretion by glycosylation.26 In comparison with the method combining metabolic labeling and click chemistry, the boronic acid-based method had higher glycoproteome coverage while glycoprotein quantification with the other showed more dynamic results because metabolic labeling favored newly synthesized glycoproteins.27
Figure 1.
Principle of the boronic acid-based chemical enrichment method for comprehensive analysis of protein N-glycosylation. Reproduced with permission from ref 24. Copyright 2014 American Society for Biochemistry and Molecular Biology.
2.2. Capturing Low-Abundance Glycoproteins
2.2.1. Effect of Boronic Acid Derivatives on Glycopeptide Enrichment.
Although the boronic acid-based method can, in theory, universally cover glycopeptides with diverse glycans, the binding affinities between commonly used boronic acids, such as phenylboronic acid, and glycans are relatively weak. This renders the method inefficient to capture lowabundance glycopeptides/proteins, which are of great importance in biological systems. Therefore, enhancing the binding strength is critical for enriching low-abundance species.
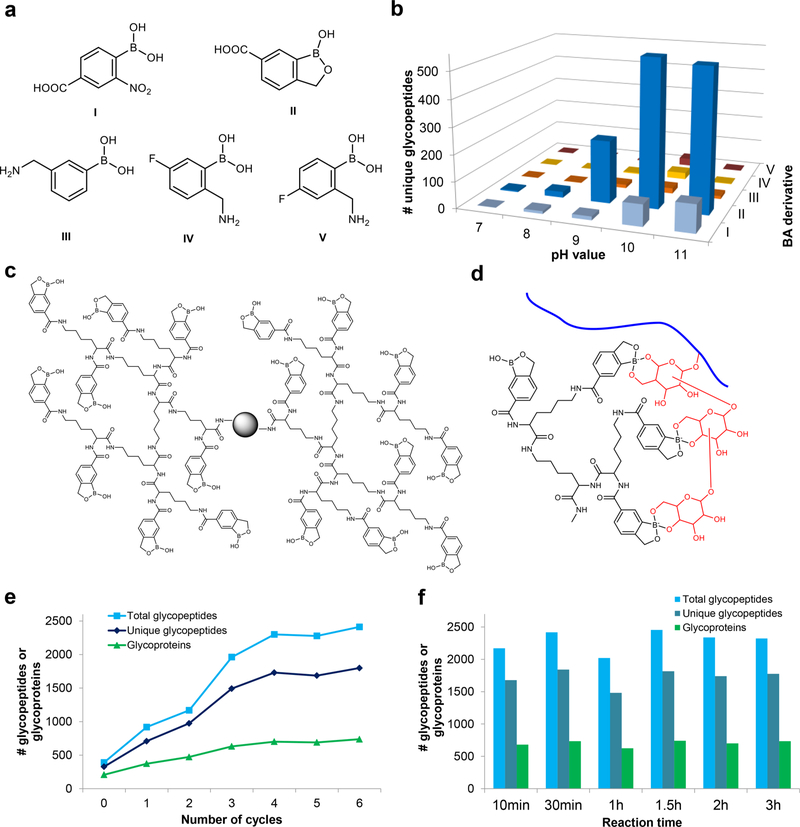
Different boronic acid derivatives have various binding affinities toward glycans.21,28,29 The performances of a group of boronic acid derivatives (Figure 2a) were evaluated for glycopeptide enrichment. Benzoboroxole was found to have dramatically better results than the other derivatives examined (Figure 2b).30 Benzoboroxole was reported to have strong interactions with sugars.28 Using a boronic acid derivative with high binding affinity can increase the coverage of glycoproteins in complex biological samples.
Figure 2.
(a) Structures of boronic acid derivatives tested for glycoproteomic analysis. (b) Numbers of unique N-glycopeptides identified with each derivative from the parallel experiments. (c) Structure of the dendrimer conjugated with benzoboroxole. (d) Example of the synergistic interactions between multiple benzoboroxole molecules in a dendrimer and several sugars within one glycan. (e) Effect of the number of synthesis cycles and corresponding dendrimer size on the enrichment of glycopeptides. (f) Effect of reaction time on identification of N-glycopeptides. Adapted with permission from ref 30. Copyright 2018 Nature.
2.2.2. Synergistic Interactions To Facilitate Glycopeptide Enrichment.
Further enhancement of the interactions between the boronic acid derivative and glycans can more effectively capture low-abundance glycoproteins. On the basis of the common features of a glycan containing multiple monosaccharides and one monosaccharide bearing several hydroxyl groups, the synergistic interactions between one glycan and multiple boronic acid derivative molecules surely facilitate glycopeptide enrichment. A dendrimer platform was employed because one dendrimer can bind many boronic acid derivative molecules and the dendrimer platform also provides the structural flexibility to synergistically interact with glycans. Dendrimers were previously used for binding of saccharide molecules,31 stereoselective recognition of monosaccharides,32 and selective enrichment of trace glycoproteins.33 We used a different dendrimer, and the enrichment was carried out at the glycopeptide level. We systematically optimized a boronic acidbased method by testing different boronic acid derivatives and using a dendrimer platform to benefit from the synergistic effect for glycoproteome analysis.30
The dendrimer-conjugated boronic acid derivative (DBA) (Figure 2c) enables synergistic interactions, as shown in Figure 2d.30 The dendrimer size was tested to maximize the glycopeptide enrichment (Figure 2e). In comparison with the same boronic acid magnetic beads without a dendrimer (cycle 0), many more unique N-glycopeptides were identified in human cells by DBA. The enrichment can be accomplished rapidly, and there is almost no glycopeptide degradation or other side reactions because prolonging the reaction time did not result in a lower number of glycopeptides (Figure 2f).
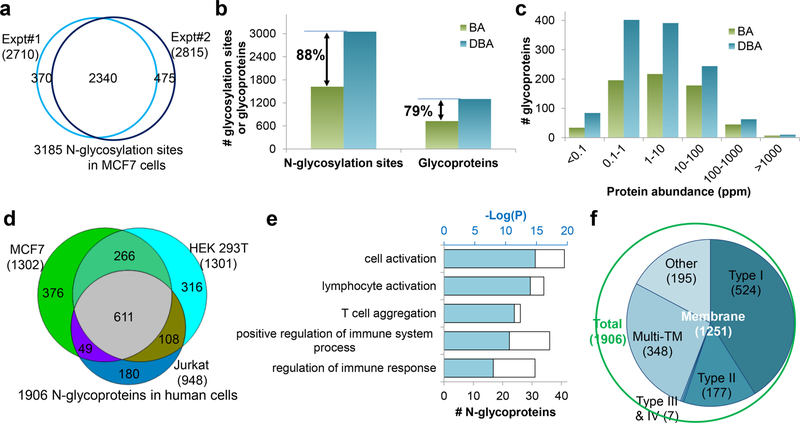
The DBA method was used to perform global analysis of glycoproteins in yeast cells and mouse brain tissue. For protein N-glycosylation, we identified over 1000 sites on 501 glycoproteins in yeast and 4195 sites on 1608 glycoproteins in mouse brain tissues. It was also employed for glycoprotein analysis in human cells (MCF7, HEK 293T, and Jurkat). Biologically duplicate experimental results for protein N-glycosylation in MCF7 cells are shown in Figure 3a, which are highly reproducible. Compared with the same derivative without the dendrimer (BA), the DBA method showed dramatic improvements in glycosylation site identification (Figure 3b) and glycoproteome coverage (Figure 3c), especially for glycoproteins with low abundance. From the three types of cells, 4691 sites were identified on 1906 N-glycopeptides (Figure 3d). The protein clustering results for 180 glycoproteins only in Jurkat cells showed that many of them were cell-typespecific (Figure 3e). Overall, most of the identified glycoproteins are membrane proteins (Figure 3f). We also applied this method to analyze protein O-glycosylation. In yeast, 234 O-mannosylated proteins were identified with glycan structure information. In human cells, we analyzed protein O-GlcNAcylation, which is normally more challenging to enrich, and over 200 O-GlcNAcylated proteins were identified.
Figure 3.
Comprehensive analysis of protein N-glycosylation in human cells using the DBA method. (a) Comparison of unique N-glycosylation sites identified in MCF7 cells in biologically duplicate experiments. (b) Comparison of glycosylation sites and glycoproteins identified with BA and DBA beads. (c) Abundance distributions of N-glycoproteins identified with the BA and DBA beads. (d) Overlap of N-glycoproteins in three types of cells. (e) Clustering results for 180 N-glycoproteins exclusively identified in Jurkat cells. (f) Distribution of membrane proteins among identified N-glycoproteins. Reproduced with permission from ref 30. Copyright 2018 Nature.
The DBA method can substantially enhance the interactions between the boronic acid and glycans, increasing the coverage of glycoproteins with low abundance. Besides, this method has several other advantages: First, it is fast and easy to operate. Second, it is robust, and the results are highly reproducible. Third, the enriched glycopeptides remain intact, allowing for site identification and glycan structure analysis. In addition, without sample restrictions, this method can readily be applied for glycoprotein analysis in different types of samples.
2.3. Generation of a Common Mass Tag for Global Analysis of N-Glycoproteins by MS
Different from many other types of protein modifications where the modified group has a fixed structure,34 glycans in glycoproteins have a wide variety of structures, resulting in a lack of a common mass tag for database searching and suboptimal MS fragmentation of glycopeptides. Therefore, generating a common small tag on glycosylation sites will be ideal for sitespecific identification of glycoproteins.
Enzymatic strategies are commonly used to generate mass tags on N-glycosylation sites.12,20 The enzyme peptide-N4-(N-acetyl-β-glucosaminyl) asparagine amidase F (PNGase F) is widely employed to remove N-glycans, which leaves a tag on the glycosylation sites for MS analysis. This reaction is frequently carried out in heavy-oxygen water (H218O) to distinguish the authentic N-glycosylation sites from the spontaneous deamidation sites.12 However, for glycans that cannot be cleaved with PNGase F, the corresponding glycosylation sites are lost. Besides PNGase F, endoglycosidases are another group of enzymes used to generate common tags on N-glycosylation sites. For instance, endoglycosidase H (Endo H) cleaves N-glycans and leaves the innermost GlcNAc residue as the tag. However, because of the inherent specificities of enzymes, no enzyme can generate a common tag for all N-glycopeptides with diverse glycans.
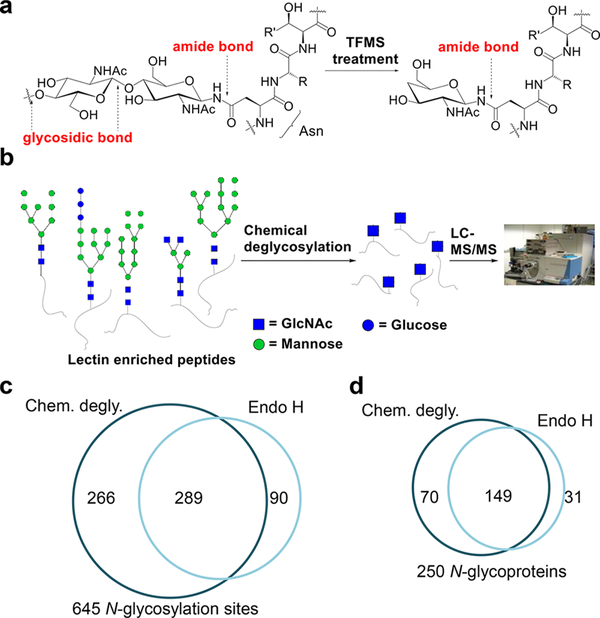
On the basis of the fact that all N-glycans have a common core structure and the first linkage to a peptide (an amide bond) is different from the rest of the linkages among sugars (glycosidic bonds), we first employed chemical deglycosylation to generate a common tag for global analysis of N-glycopeptides.35 Trifluoromethanesulfonic acid (TFMS) was used to remove all of the outer sugars by cleaving the glycosidic bonds, while the innermost GlcNAc remained (Figure 4a). The deglycosylated peptides were analyzed by MS (Figure 4b). Although this chemical deglycosylation generated the same tag as Endo H, it performed better on the identification of N-glycosylation sites (Figure 4c) and N-glycoproteins (Figure 4d).
Figure 4.
Principle of the chemical deglycosylation to break the glycosidic bonds but not the amide bond in an N-glycopeptide. (b) After the chemical deglycosylation, the innermost GlcNAc serves as a common tag for MS analysis. (c, d) Comparisons of the (c) glycosylation sites and (d) glycoproteins identified in yeast using the chemical deglycosylation and Endo H methods. Reproduced from ref 35. Copyright 2014 American Chemical Society.
3. TARGETING GLYCOPROTEINS LOCATED ONLY ON THE CELL SURFACE
Cell-surface proteins are extremely important for many cellular events, and most of them are glycosylated. They regulate nearly every extracellular activity, and about 70% of FDA-approved drugs target cell-surface proteins. As described above, global analysis of glycoproteins is extraordinarily challenging, and it is even more difficult to globally and specifically analyze glycoproteins located only on the cell surface. With the developments in chemical biology and MS-based proteomics in recent years, comprehensive analysis of cell-surface glycoproteins has become possible. Wollscheid et al.15 developed a beautiful MS-based method to analyze surface N-glycoproteins, termed cell-surface-capturing (CSC) technology, in which surface glycoproteins are oxidized and then captured through hydrazide chemistry. Boons and co-workers reported a chemical reporter strategy, named selective exoenzymatic labeling (SEEL), for cell-surface glycoconjugate analysis.36 Here we focus on the development of metabolic labeling and click chemistry-based methods for cell-surface glycoprotein analysis.
3.1. Integrating Click Chemistry and Enzymatic Reactions To Target Cell-Surface Glycoproteins
In recent years, unnatural sugars containing bio-orthogonal functional groups have been incorporated into glycoproteins,2,37–39 and in combination with bio-orthogonal chemistry,40,41 glycoproteins can be studied to obtain a better understanding of their functions in biological systems. The Bertozzi group has made significant contributions to this field and published multiple milestone papers about cell-surface glycoprotein engineering.2,38,42 Using MS-based proteomics, they performed cell-surface glycoproteomic analysis for prostate-cancer-derived PC-3 cells.39 The cells were treated with peracetylated N-azidoacetylgalactosamine (Ac4GalNAz), and surface-labeled glycoproteins were tagged with Phos-FLAG and then enriched for MS analysis.
Recently, we globally and site-specifically analyzed the surface glycoproteome by integrating metabolic labeling, copper-free click chemistry, enzymatic reaction, and MS-based proteomics.43 Similarly, a sugar analogue containing an azido group was used to label glycoproteins, including surface glycoproteins. Glycoproteins with the functional group on the cell surface were then tagged with dibenzocyclooctyne (DBCO)−sulfo−biotin via copper-free click chemistry under physiological conditions. After protein extraction and digestion, glycopeptides with the biotin tag were enriched with NeutrAvidin beads, and the enriched glycopeptides were deglycosylated with PNGase F in H218O. By this approach, 152 N-glycosylation sites on 110 proteins were identified in HEK293T cells. As expected, 95% of the identified glycoproteins were membrane proteins. Later we compared different sugar analogues, GalNAz, N-azidoacetylglucosamine (GlcNAz), and N-azidoacetylmannosamine (Man-NAz), and found that GalNAz labeling resulted in the greatest number of surface N-glycoproteins identified.44 A big advantage of copper-free click chemistry-based methods is that the reaction conditions are very mild and cell-surface glycoproteins are tagged under physiological conditions. This allows studies of surface glycoprotein dynamics as discussed below.
3.2. Global and Site-Specific Analysis of Sialoglycoproteins on the Cell Surface
Sialic acids presenting at the glycan termini carry negative charges, and sialoglycoproteins on the cell surface may dramatically impact cell properties and represent different cellular states. Metabolic labeling with sugar analogues is a powerful method to study sialoglycoproteins, and an azidecontaining ManNAc analogue, Ac4ManNAz, was employed to label sialic acid residues on the cell surface.45 Recently, Spiciarich et al.46 used Ac4ManNAz to label prostate cancer tissue slice cultures and performed glycoproteomic analysis. Yang et al.47 combined glycan metabolic labeling with MS to characterize metastasis-associated cell-surface sialoglycoproteins in PC3 (N2 (nonmetastatic) and ML2 (highly metastatic)) cells.
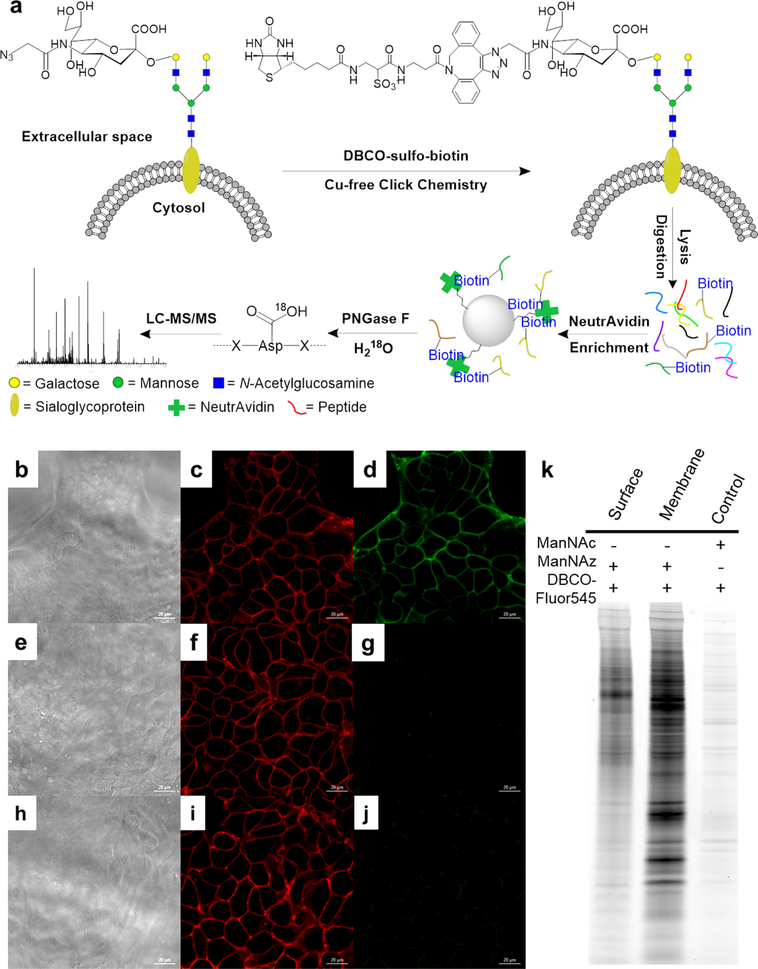
We performed global and site-specific analysis of N-sialoglycoproteins on the cell surface using an integrated method including metabolic labeling, click and enzymatic reactions, and MS-based proteomics (Figure 5a).48 After cells were labeled with Ac4ManNAz, copper-free click chemistry was performed to tag surface sialoglycoproteins with biotin for selective enrichment. The effectiveness of cell-surface sialoglycoprotein labeling and tagging was also tested by microscope imaging (Figure 5b−j) and gel electrophoresis (Figure 5k). The enrichment was performed at the peptide level, and the enriched glycopeptides were subjected to PNGase F treatment in H218O for MS analysis. A total of 395 unique N-sialoglycosylation sites on 274 surface proteins were identified in HEK 293T cells.
Figure 5.
(a) Principle of the site-specific identification of the surface N-sialoglycoproteome. (b−j) Microscope images of tagging sialoglycoprotein on the HEK 293T cell surface: (b, e, h) images of cells (scale bar is 20 μm); (c, f, i) fluorescence signals of labeled cells reacted with DBCO−Fluor545; (d) fluorescence signals of labeled cells bound to DBCO−sulfo−biotin, followed by streptavidin−FITC; (g) cells without the biotin tag treated with streptavidin−FITC show no green signal; (j) after surface sialoglycoproteins were tagged with DBCO−Fluor545, cells were further treated with DBCO−sulfo−biotin followed by streptavidin−FITC, but no green signals were detected. (k) Gel results for metabolic labeling and click chemistry. The control sample was from unlabeled cells. Reproduced with permission from ref 48. Copyright 2015 Royal Society of Chemistry.
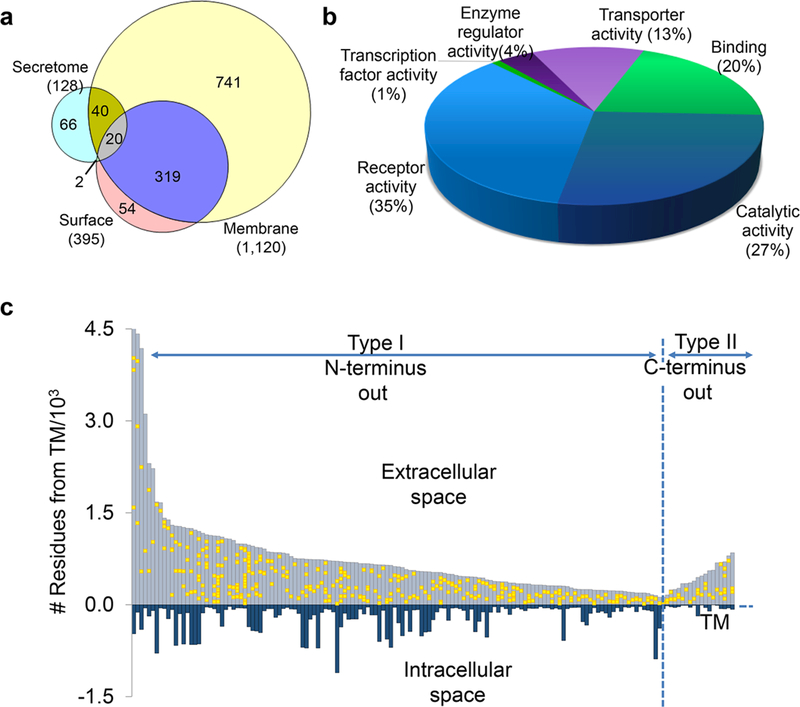
This method can also be used for sialoglycoprotein analysis in the secretome and whole-cell lysate. In the secretome, 128 sites were localized on 78 N-sialoglycoproteins, and 1120 N-sialoglycosylation sites on 482 proteins in the cellular membrane system were identified (Figure 6a). The protein clustering results (Figure 6b) show that the category with the most proteins is receptor activity (35%), followed by catalytic activity, binding, and transporter activity, which are well-known functions of surface glycoproteins. This method is site-specific, and Figure 6c illustrates the site location of types I and II transmembrane proteins. All of the glycosylation sites were located in the extracellular space, further demonstrating the effectiveness and specificity of this method. It was also applied to quantify surface N-sialoglycoproteins between cancer cells with distinct invasiveness, as discussed below. The click chemistrybased methods are effective for studying surface glycoproteins because the click reaction is relatively rapid and specific and occurs under mild conditions. Recently, we employed metabolic labeling and click chemistry to analyze protein O-GlcNAcylation, but unexpectedly, we found that protein S-GlcNAcylation extensively exists in human cells.49
Figure 6.
(a) Analysis of sialoglycoproteins in the secretome and whole-cell lysate. (b) Molecular function analysis of surface N-sialoglycoproteins. (c) Site locations of types I and II N-sialoglycoproteins based on the transmembrane domain. Reproduced with permission from ref 48. Copyright 2015 Royal Society of Chemistry.
Here surface glycoproteins were enriched at the peptide level, which is different from enrichment at the protein level. Protein-level enrichment often results in more nonspecifically bound proteins. In addition, the enzymatic deglycosylation reaction was integrated for site-specific analysis of cell-surface glycoproteins, which not only provides valuable information on site locations but also presents strong experimental evidence for glycopeptide identification.
4. SYSTEMATIC QUANTIFICATION OF GLYCOPROTEINS AND CELL-SURFACE GLYCOPROTEIN DYNAMICS
4.1. Quantification of Cell-Surface Glycoproteins
By combining the method discussed above with stable isotope labeling by amino acids in cell culture (SILAC), we systematically quantified surface glycoproteins. The experimental results demonstrated that many glycosylation sites on surface proteins were downregulated in Hep G2 cells treated with statin compared with untreated cells.44 Surface glycoprotein changes due to the statin treatment may result in cell property alteration, and quantification of surface glycoproteins aids in better understanding the pleiotropic effects of statin. Another example is that surface sialoglycoproteins in breast cancer cells with different invasiveness (MCF7 and MDA-MB-231) were quantified.48 Many sialoglycoproteins were upregulated on the cell surface in the highly invasive MDA-MB-231 cells. Protein clustering results demonstrated that upregulated surface sialoglycoproteins are related to cell adhesion, cell surface receptor linked signaling, and cell motion. Sialylation changes the protein properties, and correspondingly, surface sialoglycoproteins determine the cell-surface properties.
4.2. Systematic Quantification of Cell-Surface Glycoprotein Dynamics
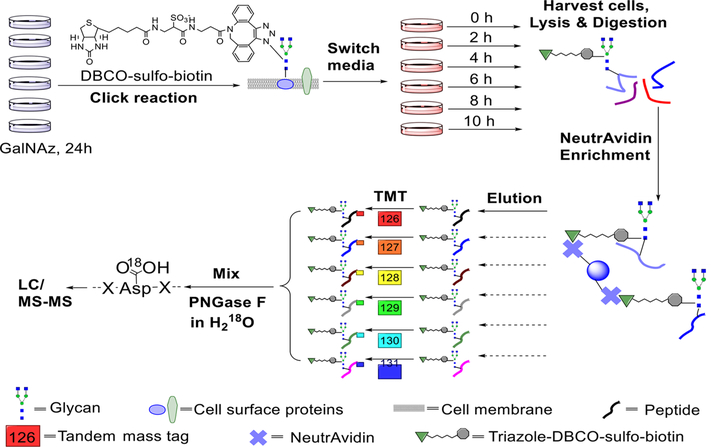
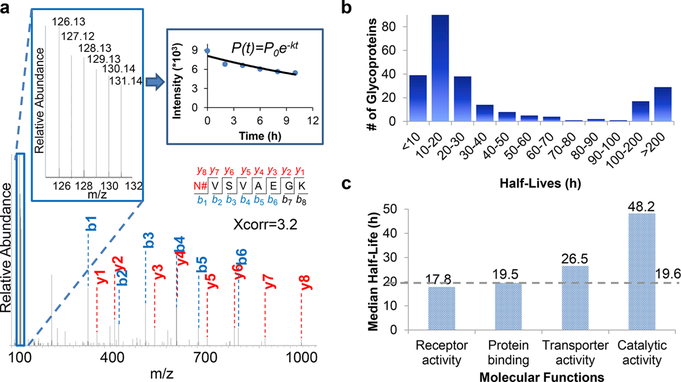
Surface glycoproteins are dynamic for cells to adapt to the everchanging extracellular environment. Besides the difficulties of surface glycoprotein identification described above, it is challenging to systematically quantify surface glycoprotein abundance changes. We recently studied surface glycoprotein dynamics using a method combining pulse-chase labeling, surface glycoprotein tagging, and selective enrichment with multiplexed proteomics50 (Figure 7). As shown in Figure 8a, the reporter ion intensities allow us to quantify the glycopeptide dynamics. The dynamics of 248 surface glycoproteins were quantified, and their half-lives were calculated (Figure 8b). Surface glycoproteins with catalytic activities were more stable than those with binding and receptor activities (Figure 8c). The half-lives of many important proteins, including cluster of differentiation (CD) proteins, were determined (some examples in Table 1). This method has several advantages. First, surface glycoproteins are selectively tagged and enriched. Second, site-specific information is obtained, and only the glycosylated forms of proteins are studied. Third, multiplexed proteomics enable the quantification of glycoproteins at several time points simultaneously, increasing the quantification accuracy. Furthermore, the high throughput allows us to study the surface glycoprotein dynamics systematically.
Figure 7.
Experimental procedure for studying the dynamics of surface glycoproteins and measuring their half-lives. Reproduced with permission from ref 50. Copyright 2017 Royal Society of Chemistry.
Figure 8.
(a) Example MSdata showing peptide identification and quantification. (b) Distribution of the half-lives of surface glycoproteins. (c) Median half-lives of glycoproteins with different molecular functions. Reproduced with permission from ref 50. Copyright 2017 Royal Society of Chemistry.
Table 1.
Half-Lives of Exemplary CD Proteins on the Cell Surface
| UniProt ID | gene symbol | CD name | Protein half-life (h) |
annotation | |
|---|---|---|---|---|---|
| this work50 | previous work (total protein)51 | ||||
| P02786 | TFRC | CD71 | 18.3 | 4.4 | transferrin receptor protein 1 |
| P05556 | ITGB1 | CD29 | 24.2 | integrin beta-1 | |
| P08069 | IGF1R | CD221 | 12.6 | insulin-like growth factor 1 receptor | |
| P08195 | SLC3A2 | CD98 | 27.2 | 10.1 | 4F2 cell-surface antigen heavy chain |
| P08962 | CD63 | CD63 | 24.2 | CD63 antigen | |
| P25445 | FAS | CD95 | 39.1 | tumor necrosis factor receptor superfamily member 6 | |
| P26006 | ITGA3 | CD49c | 37.3 | integrin alpha-3 | |
| P48960 | CD97 | CD97 | 13.2 | CD97 antigen | |
| P54709 | ATP1B3 | CD298 | 61.1 | sodium/potassium-transporting ATPase subunit beta-3 | |
| P78536 | ADAM17 | CD156b | 112.7 | disintegrin and metalloproteinase domain-containing protein 17 | |
4.3. Simultaneous Quantification of Glycoprotein Degradation and Synthesis Rates
Compared with protein dynamics, there are fewer reports about the study of glycoprotein turnover rates. Wang et al.8 metabolically labeled O-GlcNAc with 13C6-glucose and then analyzed protein O-GlcNAcylation turnover rates. We recently designed a method integrating isotopic labeling, chemical enrichment, and multiplexed proteomics to simultaneously quantify the degradation and synthesis rates for many glycoproteins in human cells.52 Most of the quantified proteins had higher synthesis rates than degradation rates because the cells were still growing throughout the time course, while a small group of proteins with lower synthesis rates mainly participated in adhesion, locomotion, localization, and signaling. This method can be widely applied for biochemical and biomedical research and provide insights into glycoprotein functions and the molecular mechanisms of biological events.
5. SPECIFIC IDENTIFICATION OF GLYCOPROTEINS WITH A PARTICULAR AND IMPORTANT GLYCAN
For glycoproteins, both the protein and glycan components contain a wealth of valuable information about the developmental and disease status of cells. Because of the heterogeneity of glycans, many studies have focused on site identification without glycan information or glycan analysis lacking protein information. The study of proteins containing a particular glycan remains largely unexplored. A tandem enzymatic strategy was developed to analyze glycoproteins with the Thomsen−Friedenreich (TF) antigen.53 Recently, another innovative method was designed to target glycoproteins with the sialylated TF antigen.54 More studies are needed to achieve global and site-specific analysis of glycoproteins with a particular and important glycan.
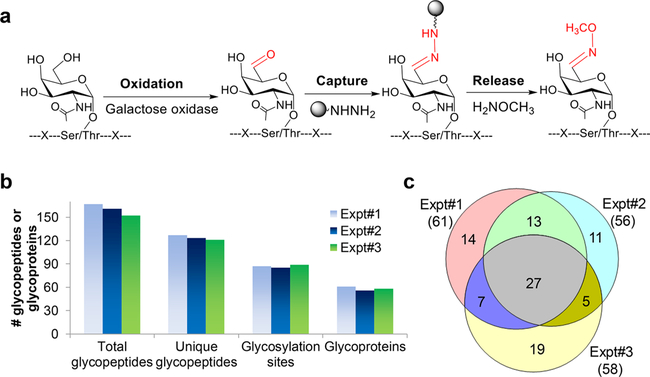
In cancer cells, some glycans (e.g., the Tn and TF antigens) are highly upregulated, but it remains largely unknown regarding glycoproteins with a particular glycan. We developed a method combining enzymatic and chemical reactions to enrich glycoproteins with the Tn antigen (Figure 9a). For synthetic glycopeptides with O-GalNAc (the Tn antigen) or O-GlcNAc, the results demonstrated that the method is highly selective for glycopeptides with O-GalNAc but not O-GlcNAc. The method was then applied to analyze glycoproteins with the Tn antigen in Jurkat cells, and the results from triplicate biological experiments were very consistent (Figure 9b). A total of 96 glycoproteins with the Tn antigen were identified (Figure 9c).10 Because there are no sample restrictions, this method can be applied to analyze clinical samples, leading to the identification of effective disease biomarkers.
Figure 9.
(a) Principle of enrichment of glycopeptides with the Tn antigen. (b) Comparison of experimental results from three biologically independent experiments. (c) Overlap of glycoproteins identified from the three experiments. Adapted with permission from ref 10. Copyright 2017 John Wiley and Sons.
6. CONCLUDING REMARKS
With the tremendous advancements in MS-based proteomics, considerable progress has been made in comprehensive analysis of proteins and protein modifications. However, it is still extraordinarily challenging to globally analyze protein glycosylation. The bottlenecks are that many glycoproteins have low abundance and glycan structures are highly heterogeneous. To overcome these difficulties, innovative methods have been developed for global analysis of glycoproteins.
In this Account, we have reviewed the existing methods for glycoprotein analysis and discussed the advantages and potential drawbacks of the widely used enrichment methods, such as lectin- and HILIC-based enrichment methods. Boronic acid-based methods are highly promising because of the formation of reversible covalent bonds between boronic acids and glycans, but the weak interactions sacrifice low-abundance glycoproteins containing more important biological information. To capture the low-abundance glycopeptides, the binding between boronic acids and glycans was strengthened by using benzoboroxole and conjugating it to a dendrimer that enables synergistic interactions. On the basis of the same core structure of Nglycans, we first employed chemical deglycosylation to remove N-glycans, leaving the innermost GlcNAc as a common tag for MS analysis. Because of the importance of surface glycoproteins in regulating extracellular events, we developed enzymatic and chemical methods to qualitatively and quantitatively study the cell-surface glycoproteome. Furthermore, glycoproteins with an important and specific glycan (the Tn antigen) were selectively enriched for MS analysis. With these and other existing methods as well as further developments, global analysis of protein glycosylation will be achievable, including intact glycopeptide analysis with glycosylation site and glycan structure information. Global analysis of protein glycosylation will lead to a better understanding of glycoprotein functions, cellular activities, and the molecular mechanisms of human diseases. Eventually we will be able to identify the glycoproteins and/or enzymes responsible for protein glycosylation as effective biomarkers for disease detection and drug targets for disease treatment.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (CAREER Award, CHE-1454501) and the National Institutes of Health (R01GM118803).
Biographies
Haopeng Xiao received his B.S. and B.A. from Peking University and his Ph.D. in Analytical Chemistry from Georgia Tech under the supervision of Dr. Ronghu Wu. His doctoral studies focused on developing novel MS methods for glycoproteome analysis. He is currently a postdoctoral fellow at Dana-Farber Cancer Institute and Harvard Medical School.
Suttipong Suttapitugsakul received his B.S. from Rensselaer Polytechnic Institute and came to Georgia Tech for his graduate studies in Dr. Wu’s lab. His research is mainly about the investigation of cell-surface glycoproteins.
Fangxu Sun received his B.S. from East China University of Science and Technology and his M.S. from Shanghai Institute of Organic Chemistry before joining Dr. Wu’s lab. He has developed new methods to analyze glycoproteins.
Ronghu Wu received his M.Sc. and Ph.D. from the University of Science and Technology of China. He is an Associate Professor at Georgia Tech, where he works on global analysis of protein modifications, especially glycosylation.
Footnotes
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Essentials of Glycobiology, 2nd ed.; Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2008. [PubMed] [Google Scholar]
- (2).Mahal LK; Yarema KJ; Bertozzi CR Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 1997, 276, 1125–1128. [DOI] [PubMed] [Google Scholar]
- (3).Ju TZ; Otto VI; Cummings RD The Tn antigen-structural simplicity and biological complexity. Angew. Chem., Int. Ed 2011, 50, 1770–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gilgunn S; Conroy PJ; Saldova R; Rudd PM; O’Kennedy RJ Aberrant PSA glycosylation-a sweet predictor of prostate cancer. Nat. Rev. Urol 2013, 10, 99–107. [DOI] [PubMed] [Google Scholar]
- (5).Kailemia MJ; Park D; Lebrilla CB Glycans and glycoproteins as specific biomarkers for cancer. Anal. Bioanal. Chem 2017, 409, 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rexach JE; Rogers CJ; Yu SH; Tao JF; Sun YE; Hsieh-Wilson LC Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol 2010, 6, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Segu ZM; Hussein A; Novotny MV; Mechref Y Assigning N-glycosylation sites of glycoproteins using LC/MSMS in conjunction with Endo-M/exoglycosidase mixture. J. Proteome Res 2010, 9, 3598–3607. [DOI] [PubMed] [Google Scholar]
- (8).Wang XS; Yuan ZF; Fan J; Karch KR; Ball LE; Denu JM; Garcia BA A novel quantitative mass spectrometry platform for determining protein O-GlcNAcylation dynamics. Mol. Cell. Proteomics 2016, 15, 2462–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Yang Y; Liu F; Franc V; Halim LA; Schellekens H; Heck AJR Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat. Commun 2016, 7, 13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zheng JN; Xiao HP; Wu RH Specific identification of glycoproteins bearing the Tn antigen in human cells. Angew. Chem., Int. Ed 2017, 56, 7107–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Steentoft C; Vakhrushev SY; Vester-Christensen MB; Schjoldager K; Kong Y; Bennett EP; Mandel U; Wandall H; Levery SB; Clausen H Mining the O-glycoproteome using zincfinger nuclease-glycoengineered SimpleCell lines. Nat. Methods 2011, 8, 977–982. [DOI] [PubMed] [Google Scholar]
- (12).Kaji H; Saito H; Yamauchi Y; Shinkawa T; Taoka M; Hirabayashi J; Kasai K; Takahashi N; Isobe T Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify Nlinked glycoproteins. Nat. Biotechnol 2003, 21, 667–672. [DOI] [PubMed] [Google Scholar]
- (13).Zhang H; Li XJ; Martin DB; Aebersold R Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 2003, 21, 660–666. [DOI] [PubMed] [Google Scholar]
- (14).Nilsson J; Rüetschi U; Halim A; Hesse C; Carlsohn E; Brinkmalm G; Larson G Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 2009, 6, 809–811. [DOI] [PubMed] [Google Scholar]
- (15).Wollscheid B; Bausch-Fluck D; Henderson C; O’Brien R; Bibel M; Schiess R; Aebersold R; Watts JD Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009, 27, 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zielinska DF; Gnad F; Wisniewski JR; Mann M Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141, 897–907. [DOI] [PubMed] [Google Scholar]
- (17).Woo CM; Iavarone AT; Spiciarich DR; Palaniappan KK; Bertozzi CR Isotope-targeted glycoproteomics (IsoTaG): a massindependent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 2015, 12, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sun SS; Shah P; Eshghi ST; Yang WM; Trikannad N; Yang S; Chen LJ; Aiyetan P; Hoti N; Zhang Z; Chan DW; Zhang H Comprehensive analysis of protein glycosylation by solidphase extraction of N-linked glycans and glycosite-containing peptides. Nat. Biotechnol 2016, 34, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wohlgemuth J; Karas M; Eichhorn T; Hendriks R; Andrecht S Quantitative site-specific analysis of protein glycosylation by LC-MS using different glycopeptide-enrichment strategies. Anal. Biochem 2009, 395, 178–188. [DOI] [PubMed] [Google Scholar]
- (20).Hagglund P; Bunkenborg J; Elortza F; Jensen ON; Roepstorff P A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res 2004, 3, 556–566. [DOI] [PubMed] [Google Scholar]
- (21).Arnaud J; Audfray A; Imberty A Binding sugars: from natural lectins to synthetic receptors and engineered neolectins. Chem. Soc. Rev 2013, 42, 4798–4813. [DOI] [PubMed] [Google Scholar]
- (22).Wang W; Gao XM; Wang BH Boronic acid-based sensors. Curr. Org. Chem 2002, 6, 1285–1317. [Google Scholar]
- (23).Zhang LJ; Xu YW; Yao HL; Xie LQ; Yao J; Lu HJ; Yang PY Boronic acid functionalized core-satellite composite nanoparticles for advanced enrichment of glycopeptides and glycoproteins. Chem. - Eur. J 2009, 15, 10158–10166. [DOI] [PubMed] [Google Scholar]
- (24).Chen WX; Smeekens JM; Wu RH A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS). Mol. Cell. Proteomics 2014, 13, 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Xiao HP; Smeekens JM; Wu RH Quantification of tunicamycin-induced protein expression and N-glycosylation changes in yeast. Analyst 2016, 141, 3737–3745. [DOI] [PubMed] [Google Scholar]
- (26).Smeekens JM; Xiao HP; Wu RH Global analysis of secreted proteins and glycoproteins in Saccharomyces Cerevisiae. J. Proteome Res. 2017, 16, 1039–1049. [DOI] [PubMed] [Google Scholar]
- (27).Xiao HP; Hwang JE; Wu RH Mass spectrometric analysis of the N-glycoproteome in statin-treated liver cells with two lectinindependent chemical enrichment methods. Int. J. Mass Spectrom 2018, 429, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dowlut M; Hall DG An improved class of sugar-binding boronic acids, soluble and capable of complexing glycosides in neutral water. J. Am. Chem. Soc 2006, 128, 4226–4227. [DOI] [PubMed] [Google Scholar]
- (29).Adamczyk-Wozniak A; Cyranski MK; Zubrowska A; Sporzynski A Benzoxaboroles - old compounds with new applications. J. Organomet. Chem 2009, 694, 3533–3541. [Google Scholar]
- (30).Xiao HP; Chen WX; Smeekens JM; Wu RH An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins. Nat. Commun 2018, 9, 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).James TD; Shinmori H; Takeuchi M; Shinkai S A saccharide “sponge”. Synthesis and properties of a dendritic boronic acid. Chem. Commun 1996, 705–706. [Google Scholar]
- (32).Smith DK; Diederich F Dendritic hydrogen bonding receptors: enantiomerically pure dendroclefts for the selective recognition of monosaccharides. Chem. Commun 1998, 2501–2502. [Google Scholar]
- (33).Wang HY; Bie ZJ; Lu CC; Liu Z Magnetic nanoparticles with dendrimer-assisted boronate avidity for the selective enrichment of trace glycoproteins. Chem. Sci. 2013, 4, 4298–4303. [Google Scholar]
- (34).Wu RH; Haas W; Dephoure N; Huttlin EL; Zhai B; Sowa ME; Gygi SP A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 2011, 8, 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chen WX; Smeekens JM; Wu RH Comprehensive analysis of protein N-glycosylation sites by combining chemical deglycosylation with LC-MS. J. Proteome Res 2014, 13, 1466–1473. [DOI] [PubMed] [Google Scholar]
- (36).Sun TT; Yu SH; Zhao P; Meng L; Moremen KW; Wells L; Steet R; Boons GJ One-step selective exoenzymatic labeling (SEEL) strategy for the biotinylation and identification of glycoproteins of living cells. J. Am. Chem. Soc. 2016, 138, 11575–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sprung R; Nandi A; Chen Y; Kim SC; Barma D; Falck JR; Zhao YM Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res 2005, 4, 950–957. [DOI] [PubMed] [Google Scholar]
- (38).Prescher JA; Dube DH; Bertozzi CR Chemical remodelling of cell surfaces in living animals. Nature 2004, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- (39).Hubbard SC; Boyce M; McVaugh CT; Peehl DM; Bertozzi CR Cell surface glycoproteomic analysis of prostate cancerderived PC-3 cells. Bioorg. Med. Chem. Lett 2011, 21, 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hong V; Presolski SI; Ma C; Finn MG Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem., Int. Ed 2009, 48, 9879–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lutz JF Copper-free azide-alkyne cycloadditions: New insights and perspectives. Angew. Chem., Int. Ed 2008, 47, 2182–2184. [DOI] [PubMed] [Google Scholar]
- (42).Bertozzi CR; Kiessling LL Chemical glycobiology. Science 2001, 291, 2357–2364. [DOI] [PubMed] [Google Scholar]
- (43).Smeekens JM; Chen WX; Wu RH Mass spectrometric analysis of the cell surface N-glycoproteome by combining metabolic labeling and click chemistry. J. Am. Soc. Mass Spectrom 2015, 26, 604–614. [DOI] [PubMed] [Google Scholar]
- (44).Xiao HP; Tang GX; Wu RH Site-specific quantification of surface N-glycoproteins in statin-treated liver cells. Anal. Chem 2016, 88, 3324–3332. [DOI] [PubMed] [Google Scholar]
- (45).Saxon E; Bertozzi CR Cell surface engineering by a modified Staudinger reaction. Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- (46).Spiciarich DR; Nolley R; Maund SL; Purcell SC; Herschel J; Iavarone AT; Peehl DM; Bertozzi CR Bioorthogonal labeling of human prostate cancer tissue slice cultures for glycoproteomics. Angew. Chem., Int. Ed 2017, 56, 8992–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yang LF; Nyalwidhe JO; Guo SQ; Drake RR; Semmes OJ Targeted identification of metastasis-associated cell-surface sialoglycoproteins in prostate cancer. Mol. Cell. Proteomics 2011, 10, M110.007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Chen WX; Smeekens JM; Wu RH Systematic and sitespecific analysis of N-sialoglycosylated proteins on the cell surface by integrating click chemistry and MS-based proteomics. Chem. Sci 2015, 6, 4681–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Xiao HP; Wu RH Global and site-specific analysis revealing unexpected and extensive protein S-GlcNAcylation in human cells. Anal. Chem 2017, 89, 3656–3663. [DOI] [PubMed] [Google Scholar]
- (50).Xiao HP; Wu RH Quantitative investigation of human cell surface N-glycoprotein dynamics. Chem. Sci. 2017, 8, 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chen WX; Smeekens JM; Wu RH Systematic study of the dynamics and half-lives of newly synthesized proteins in human cells. Chem. Sci 2016, 7, 1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Xiao HP; Wu RH Simultaneous quantitation of glycoprotein degradation and synthesis rates by integrating isotope labeling, chemical enrichment, and multiplexed proteomics. Anal. Chem 2017, 89, 10361–10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Li Q; Li ZH; Duan XT; Yi W A tandem enzymatic approach for detecting and imaging tumor-associated Thomsen-Friedenreich antigen disaccharide. J. Am. Chem. Soc 2014, 136, 12536–12539. [DOI] [PubMed] [Google Scholar]
- (54).Wen L; Liu D; Zheng Y; Huang K; Cao X; Song J; Wang PG A one-step chemoenzymatic labeling strategy for probing sialylated Thomsen−Friedenreich antigen. ACS Cent. Sci 2018, 4, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]