Abstract
Data mapping plays an important role in data integration and exchanges among institutions and organizations with different data standards. However, traditional rule-based approaches and machine learning methods fail to achieve satisfactory results for the data mapping problem. In this paper, we propose a novel and sophisticated deep learning framework for data mapping called mixture feature embedding convolutional neural network (MfeCNN). The MfeCNN model converts the data mapping task to a multiple classification problem. In the model, we incorporated multimodal learning and multiview embedding into a CNN for mixture feature tensor generation and classification prediction. Multimodal features were extracted from various linguistic spaces with a medical natural language processing package. Then, powerful feature embeddings were learned by using the CNN. As many as 10 classes could be simultaneously classified by a softmax prediction layer based on multiview embedding. MfeCNN achieved the best results on unbalanced data (average F1 score, 82.4%) among traditional state-of-the-art machine learning models and CNN without mixture feature embedding. Our model also outperformed a very deep CNN with 29 layers, which took free texts as inputs. The combination of mixture feature embedding and a deep neural network can achieve high accuracy for data mapping and multiple classification.
Keywords: Data mapping, convolutional neural network, mixture feature embedding, multimodal, multiview, deep learning
I. Introduction
WHEN data is exchanged among institutions with different data standards, data mapping is often required to convert the various data formats to a common standard. In health care, health-related institutions and organizations including hospitals, pharmaceutical factories, medical insurance companies, and medical device industries have historically used internal systems for document composition. The World Health Organization, National Institutes of Health, Health Level Seven International (HL7), and other world-class institutions have made great progress in developing international standards for electronic health records that supports clinical practice and management as well as delivery and evaluation of health services. Most health institutions have adopted some of these international standards. However, inconsistencies remain because of diverse interpretations of the same standard or missing data and errors made by medical staffs who handle the data. Most documents have large volumes of free texts, which are unstructured or semi-structured. Consequently, automatic data mapping of one format to another has become an important research topic among the communities of natural language processing (NLP) and data science.
Data transformation and mapping [1] have been becoming important components of information integration. With the increasement of the requirement for relational database integration, research has concentrated on the problem of schema mapping [2], [3], which refers to automatic detection of the mapping relationship between source and target data. Schema mapping methods were classified by Rahm et al. [4] as schema-only based, instance/content-based, and combination approaches. Schema-only based methods uncover a mapping relationship with the information in the schema metadata [5], whereas instance-based mapping discovers the relationship through real instances of data, such as value patterns, word frequencies, and ranges [6]–[10]. Combination approaches [11] handle the data mapping problem by using both schema and real instances.
Machine learning and sophisticated statistical techniques have recently been used by researchers to determine schema elements mapping in the instance level. Doan et al. [12] showed that new mappings can be learned from existing mappings between source and target schema. Researchers have used machine learning algorithms to train models with known mappings, and to apply the learned models to the new schema elements for mapping them to the target elements [13]. In addition, the models catch probabilistic knowledge in the training examples given by domain experts. The models trained in a domain can be applied to the same domain for new data mappings [14]. This work formed the basis of our approach, although we were particularly interested in integrating clinical data represented by different standards and associating these data with a unified data model, with the ultimate goal of seamless exchange of clinical data.
For the data mapping task, we propose a sophisticated machine learning model calledmixture feature embedding convolutional neural network (MfeCNN), which expands on our previous work [15]. The innovation of our approach lies in applying the deep learning method to the data mapping problem. Our semi-structured data, HL7 messages from health providers, use a non-XML encoding syntax based on segment format, which is supported by major medical information system vendors in the United States. The standards of HL7 v2 allow some custom fields, and fields can contain free-text content. However, the data set we used was unbalanced, and 1 target category was dominant. These data characteristics led to the complex mapping problem and our work to solve the data mapping problem by training an advanced model with many NLP features.
We basically converted the data mapping task to a classification task and made the best of extracted features from given texts as inputs. In addition to free texts as one type of features, we retrieved medical terms from the HL7 messages as language-relevant features, including concepts and syntax. Traditional machine learning models, such as support vector machine (SVM) or conditional random field, just treat language-relevant features as uniform features without distinguishing their semantic categories. All of those features are, in fact, from different semantic spaces and can be regarded as from different modals and thus should be considered or viewed differently. Therefore, it seemed reasonable to construct a multimodal and multiview model for the mapping task.
Previous relevant studies further aided us to develop a deep neural network model with both multimodal and multiview characteristics. Deep neural networks have been proposed and used by Ngiam et al. [18] to learn features over multiple modalities. Their deep neural network was based on sparse-restricted Boltzmann machines and demonstrated the capacity of cross-modal feature learning, where better features for 1 modality, such as video, can be learned for other modalities when multiple modalities (e.g., audio and video) are needed at the feature-learning stage. This work motivated us to use diverse modals to learn rich features from health data sets. From those data sets, we could extract multiple modalities, including words, syntax, semantics of sentences, and terminology codes. Twin-view (TV) embedding for CNN, another area of research [19], was also important in developing our model. According to the model, variable X1 may have TV-embedding for any X2 if a function g1 exists such that P(X2|X1) = g1(f1(X1), X2), where (X1, X2) ∈ X1 × X2. The TV-embedding can be expressed as a function, f1. This model makes it possible for current data to find TV-embedding from unlabeled data and accordingly enhance the data representation. Further, the learned TV-embedding can be integrated into supervised CNN with a compound sigmoid function as . If there are multiple TV-embeddings, [summationtext] a summation can be added to the former equation as , which can be regarded as multiview embedding.
In this study, first, we extracted many relevant features using third-party tools as multimodal inputs, including bag-of-words (BOW), part of speech (POS), concepts, syntax, and semantic roles [16], [17]. Second, feature embedding representations were learned with a CNN model to generate feature tensors. Third, these feature tensors were fed into a multiview-based CNN model to predict the data mappings. The MfeCNN model was evaluated for data mapping from the HL7 message to CommonSIF (Common Standard Input Format) and compared with baseline classification models based on SVM and other deep learning models, such as CNN. The results showed that our model yields better performance than baseline models and indicates that our approach is a promising way to resolve the automatic data mapping problem. Although we tested our model in the health care field, the methodology is generic enough to handle similar tasks in other fields.
II. Methods
We propose a deep learning method, MfeCNN, to solve the challenging problem of mapping clinical data. It is a multimodal model that employs multiview embedding for feature integration. An essential contribution of this model comes from how we integrate features with multiview embedding. Features in MfeCNN are from totally different semantic spaces or modalities and include words, concepts, and syntax. Different integration functions for multiview embedding are deployed as sigmoid functions in MfeCNN. Word embedding [20]–[22] can be trained with recurrent neural networks (RNN) and long short-term memory networks (LSTMs) to reflect the sequential relationships among words. However, those models allow only for strictly sequential information propositions. In human languages, the order and dependency between words and phrases are often important. Therefore, a tree-LSTM [23] was proposed as a generalization of LSTMs to tree-structured network topologies to address the issue of dependencies between words and phrases and to provide better semantic embedding of words. We used tree-LSTM to generate compound-feature embedding to enrich feature modalities for mixture feature embedding (Mfe).
Thus, in the following subsections, we will sequentially describe in details the workflow of data mapping, the MfeCNN and CNN models, the mixture feature embedding method, and the tree-LSTM for enriched semantic features.
A. Mapping Workflow
Figure 1 depicts the end-to-end data mapping workflow of our MfeCNN model. The workflow contains 2 pipelines: a training pipeline and an application pipeline. The training pipeline first takes the training set, including the message and target data and their mapping relationships, and then extracts features to train the MfeCNN model for data mapping. In the application pipeline, new messages are mapped through the same pipeline. These messages are classified by the trained MfeCNN model, and each field of message contents can be mapped to a schema element of target data.
Fig. 1.
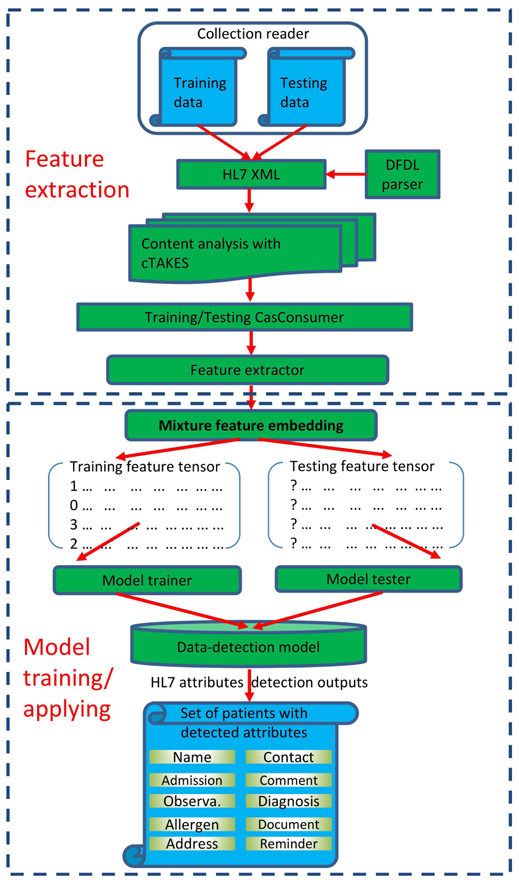
Data Mapping Workflow With MfeCNN. cTAKES indicates clinical Text Analysis and Knowledge Extraction System; DFDL, Data Format Description Language; HL7, Health Level Seven International.
Both training and testing pipelines consist of 2 stages (a feature extraction stage and a model training/application stage). In the feature extraction stage, source data in HL7 message format is preprocessed by a DFDL (Data Format Description Language) parser (IBM). The parsed data is then converted to a mediated format, HL7 XML. The mediated format helps delimitate HL7 contents into different segments, fields, and subfields, which makes it possible to analyze each segment of the contents. We then employed a third-party tool called cTAKES (clinical Text Analysis and Knowledge Extraction System) [24], [25] to preprocess and annotate each field of message contents parsed by DFDL and to extract multimodal features using the CasConsumer module in the cTAKES tool. In the model training stage, these retrieved multimodal features were used to perform mixture feature embedding to train the MfeCNN model against the message and target data to establish the mappings. After the MfeCNN model was trained and validated, test data was mapped into target attributes by using a similar pipeline that included feature extraction and model application, as shown in Figure 1.
B. Mixture Feature Embedding Convolutional Neural Network
Figure 2 shows the structure of our MfeCNN model, which includes 2 main components: mixture feature embedding and deep network learning. Feature extraction yields multimodal features including the BOW modal, POS modal, concept modal, and syntax modal as well as a semantic role modal from the tree-LSTM method. We use these features to build a feature matrix for each sentence and feed them into the Mfe layer, which outputs feature tensors after feature learning. The Mfe layer consists of a shared representation model for multimodalities and CNN, together with a max-pooling layer to learn feature embedding presentations as tensors. The deep network in the last layer takes the mixture feature embedding as inputs to perform model learning and make mapping predictions. The deep network layer is a CNN with a convolution layer, a max-pooling layer, and a modified, fully connected softmax layer based on multiview embedding for data mapping prediction.
Fig. 2.
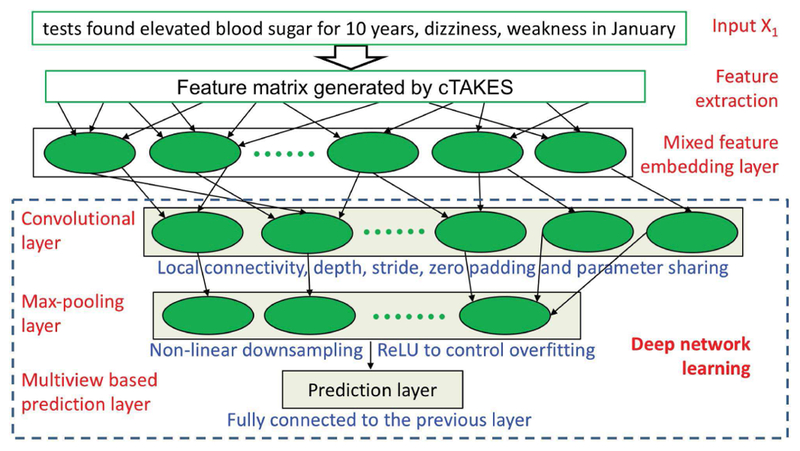
Architecture of MfeCNN Model for Data Mapping. cTAKES indicates clinical Text Analysis and Knowledge Extraction System; ReLU, rectified linear unit.
Our main contribution and innovation in the network architecture of our MfeCNN model are the inclusion of the mixture feature embedding layer and the multiview-based prediction layer, which are described in more detail in the following subsections. The mixture feature embedding layer enables the neural network model to incorporate rich external data resources and generate more expressive representations of features. The prediction layer with multiview embedding can incorporate multimodal feature embedding in a multiview style and has the potential to enhance the prediction performance of data mapping. Our experimental results showed the advantages of this novel model.
C. Convolutional Neural Network
We use CNN in 2 places for our model training. First, a regular CNN model is used for mixture feature embedding to generate embedding representation as feature tensors, as shown in Figure 3. Then, a multiview-based CNN model is used in the network model learning to improve the performance of data mapping prediction (Figure 2).
Fig. 3.

Details of the Mixture Feature Embedding Layer. BOW indicates bag-of-words; POS, parts of speech; cTAKES, clinical Text Analysis and Knowledge Extraction System.
Both the regular and modified CNN models have a triple-layer architecture: a convolutional layer, a max-pooling layer, and a fully connected softmax prediction layer. The main difference between the 2 CNN models is in the prediction layer. The multiview-based CNN model has a prediction layer based on multiview embedding. Specifically, a softmax classifier (based on multiview embedding) was constructed to predict the class labels {yi}, given the inputs {xi} from the mixture feature embedding layer as follows:
| (1) |
where y refers to the final label to predict; t refers to each sample instance;.and is the transformed feature tensor vector from the mixture feature embedding layer as the input of the prediction layer; v and w are the weight vectors for hidden units z and multiview embedding ul(xl), respectively.
D. Mixture Feature Embedding
Mixture feature embedding is the combination of multi-modal learning and multiview embedding (Figure 3). Given a free text input, x1, multimodal features are generated by the cTAKES tool. The extracted semantic modals include the POS modal, concept modal, and syntax modal. The POS modal captures the property of each word in input sentences. The concept modal provides knowledge in specific domains, such as medicine, and serves as a good discriminator of targets. The syntax modal conveys important dependent relations between words and the composition of a sentence, which usually includes phrases. The syntax modal also indicates the structural nature of a sentence. Because of the importance of the syntax modal for human languages, a semantic-role modal is generated by tree-LSTM, described below, to provide rich features for better feature embedding.
With these generated feature modals, a shared feature representation layer is used to learn cross-modal features. The cross-modal features are then fed into a basic CNN model that includes the convolutional layer, max-pooling layer, and prediction layer (Figure 3) to obtain embedded feature representations as feature tensors for the network model learning. During the model learning, the Mfe stretches itself to transform multimodal learning into a multiview embedding problem.
| (2) |
By following the idea of TV -embedding described in equation 2 [19], Mfe attempts to learn multiple TV embed dings (multiview embedding) for input feature tensors Xl from the mixture feature embedding layer. In addition, those multiview embeddings are obtained from different modals or various syntactic/semantic spaces. They differ from original TV embeddings, which are all taken from word levels. For multiview feature embedding from multiple syntactic/semantic spaces, equation 2 is expanded:
| (3) |
E. Tree-LSTM for Enriched Semantic Features
Tree-LSTM [23] is used to generate complex semantic-role features for mixture feature embeddings to enrich feature modalities. The tree-LSTM extends the LSTMs to tree-structured network topologies and can reflect the important order and dependencies between words and phrases and provide better semantic embeddings. The complex feature embedding from tree-LSTM is concatenated into Mfe with some weights. The tree-LSTM has 2 different implementing versions, the child-sum tree-LSTM and N-ary tree-LSTM [23]. Both variants have rich network topologies and can incorporate information from multiple child units. N-ary tree-LSTM takes the order and the importance of the children into consideration. We used N-ary tree-LSTM to generate extra feature embeddings because the model can catch the subtle importance of each word feature, which is important for training the classification model.
A constituent parser is used to parse a sentence into binary constituents. For example, a sentence with subject, predicate, and object may be parsed as a noun phrase (NP), the subject (NP), and a verbal phrase (VP). The VP is then parsed into a verb (V) and another NP (object). Given a constituent tree, let C(j) denote the set of children of node j. The constituent tree has at most N branching factors and for each child k, a separate parameter matrix is introduced to allow the N-ary tree-LSTM model to learn more fine-grained conditioning on the state of a unit’s children than both child-sum tree-LSTM and the flat-LSTM. A sentence like the one above will assign the V or VP the highest weights and the subject NP and the object NP lower weights.
III. Evaluations
In this section, we evaluated the performance of our MfeCNN model for data mapping from an HL7 message to CommonSIF by comparing the MfeCNN model with traditional state-of-the-art machine learning models and CNN models with BOW features and extracted features from cTAKES as inputs. Firstly, we introduced the mapping data sets used in the model evaluation. Then, we described the settings of evaluation experiments for each model in details. Finally, we showed the evaluation metrics used for comparison.
A. Data for Evaluation
In our evaluation experiments, we chose the HL7 message v2 [26] as the standard for input data. We collected 3 data sets as show in Table 1 to compose a HL7 message data set for evaluation: (1) HL7 message sample data sets from a health care solution provider, which includes hospital charge and patient discharge, medical treatment, and laboratory tests; (2) de-identified clinical document architecture (CDA) sample data sets on patient health records obtained from a hospital; (3) I2B2 data sets [27] containing public clinical notes used for NLP research. All data sets were converted to consistent HL7 v2 messages in advance so that we could process data sets with different raw formats by using the same pipeline, as shown in Figure 1.
TABLE I.
Data With Message and Map Numbers.
| Data | Message | Mapping |
|---|---|---|
| Customer HL7 | 2,000 | 23,251 |
| Customer CDA | 3,000 | 47,503 |
| I2B2 | 20,000 | 65,467 |
| Total | 25,000 | 136,221 |
Abbreviations: CDA, clinical document architecture; HL7, Health Level Seven International.
We totally collected 25,000 HL7 documents, each of which included only 1 HL7 message. Domain experts manually annotated all the fields of each HL7 message by using CommonSIF schema elements as target data (Annotated target data are considered the criterion standard for this type of work). Table 1 shows our data sets in the HL7 v2 format with annotated mappings. The whole HL7 data was divided into training (50%), validation (20%), and testing (30%) sets, respectively. Total 40,866 mapping instances were used to evaluate our models. All instances were mapped to 10 classes, each of which represents one schema element of target data (See Table 2).
TABLE II.
Ten Schema Elements in Target Data.
| Class ID | Class Name |
|---|---|
| 0 | PID.5.PatientName |
| 1 | NK1.7.ContactRole |
| 2 | PV2.3.AdmitReason |
| 3 | NTE.3.Comment |
| 4 | OBX.5.Observation Value |
| 5 | DG1.4.DiagnosisDescription |
| 6 | AL1.3.AllergenCode MnemonicDescription |
| 7 | TXA.2.DocumentType |
| 8 | PID.11.PatientAddress |
| 9 | Remainder |
Abbreviation: ID, identification.
B. Evaluation Designs
To evaluate the effectiveness of mixture feature embedding, 2 kinds of baselines were trained and tested for model comparison, as shown in Table 3. Because our model is based on CNN, the first baseline is a basic CNN model with only BOW features as input data, which is referred to as “basic CNN” for convenience. For the second baseline, we trained and tested both SVM and CNN models for data mapping by using extracted features from cTAKES as inputs.
TABLE III.
Baseline Models and MfeCNN.
| Model | Input |
|---|---|
| CNN | Bag-of-words |
| SVM | Features extracted by cTAKES |
| CNN | Features extracted by cTAKES |
| MfeCNN | Features extracted by cTAKES |
Abbreviations: CNN, convolutional neural network; cTAKES, clinical Text Analysis and Knowledge Extraction System; MfeCNN, mixture feature embedding convolutional neural network; SVM, support vector machine.
All experiments were performed on a Linux operating system with 24 Intel Xeon 2.4 GHz central processing unit (CPU) cores, 2 Nvidia Grid K2 graphics processing units (GPUs), and 64 GB RAM. For the SVM model, we used a LIBSVM 28library for the linear classification model. We implemented our deep learning models by using TensorFlow [29], a well-known, open-source software library for deep learning from Google LLC. Namely, both CNN and MfeCNN were implemented with TensorFlow with the same network configuration as that described in the Methods section.
C. Evaluation Metrics
We deployed precision (Pre%), recall (Rec%), F1 score (F1%), and standard support as evaluation metrics, which are defined as,
| (4) |
| (5) |
| (6) |
| (7) |
IV. Results and Discussion
Table 4 shows the baseline results of our data mapping prediction on test data conducted by the basic CNN model with only BOW features as inputs, i.e., the precision, recall, F1 score, and support of each target category and their overall micro and macro means. The range of F1 scores was 26.2% to 60.7%. The micro and macro means of the F1 scores were 50.6% and 47.8%, respectively.
TABLE IV.
Results of Basic CNN With Only BOW Features as Inputs.
| Class ID | Pre% | Rec% | F1% | Support |
|---|---|---|---|---|
| 0 | 68.9 | 50.9 | 58.6 | 15/31 |
| 1 | 40.7 | 49.5 | 44.6 | 26/54 |
| 2 | 58.7 | 56.4 | 57.5 | 93/166 |
| 3 | 28.2 | 44.2 | 34.4 | 340/771 |
| 4 | 71.3 | 31.6 | 43.7 | 262/832 |
| 5 | 47.4 | 37.1 | 41.7 | 439/1,183 |
| 6 | 43.4 | 52.3 | 47.5 | 644/1,232 |
| 7 | 53.9 | 44.4 | 48.7 | 1,768/3,981 |
| 8 | 20.9 | 35.2 | 26.2 | 1,687/4,797 |
| 9 | 67.2 | 55.4 | 60.7 | 15,412/27,819 |
| Micro mean | 50.6 | 50.6 | 50.6 | 20,686/40,866 |
| Macro mean | 50.1 | 45.7 | 47.8 | … |
Abbreviations: CNN, convolutional neural network; BOW, bag of words; ID, identification; Pre%, precision; Rec%, recall; F1%, F1 score.
Tables 5, 6, and 7 show the evaluation results of 3 models with features extracted by cTAKES as inputs. The F1 score ranges of SVM, CNN, and MfeCNN were 12.5% to 100%, 35.6% to 97.2%, and 38.2% to 100%, respectively. The macro means of SVM, CNN, and MfeCNN were 59.7%, 65.5%, and 82.4%, respectively. Compared with the basic CNN with only BOW as inputs, the macro means of the F1 score of all 3 models (SVM, CNN, and MfeCNN) increased by 11.9%, 17.7%, and 34.6%. The results showed that the 3 approaches are much better than the basic CNN baseline, even though the baseline uses CNN, which is an advanced deep learning framework. This finding suggests that enriched features are very important to the performance of machine learning models.
TABLE V.
Results of SVM With Features Extracted by cTAKES.
| Class ID | Pre% | Rec% | F1% | Support |
|---|---|---|---|---|
| 0 | 100 | 100 | 100 | 31/31 |
| 1 | 100 | 22.1 | 36.2 | 11/54 |
| 2 | 41.8 | 13.0 | 19.8 | 21/166 |
| 3 | 45.2 | 66.7 | 53.9 | 514/771 |
| 4 | 64.5 | 14.4 | 23.5 | 119/832 |
| 5 | 72.3 | 8.2 | 14.8 | 97/1,183 |
| 6 | 63.2 | 89.8 | 74.2 | 1,106/1,232 |
| 7 | 96.4 | 99.2 | 97.8 | 3,947/3,981 |
| 8 | 7.8 | 31.4 | 12.5 | 1,504/4,797 |
| 9 | 87.1 | 88.4 | 87.7 | 24,584/27,819 |
| Micro Mean | 78.1 | 78.1 | 78.1 | 31,934/40,866 |
| Macro Mean | 67.8 | 53.3 | 59.7 | … |
Abbreviations: SVM, support vector machine; cTAKES, clinical Text Analysis and Knowledge Extraction System; ID, identification; Pre%, precision; Rec%, recall; F1%, F1 score.
TABLE VI.
Results of CNN With Features Extracted by cTAKES.
| Class ID | Pre% | Rec% | f1% | Support |
|---|---|---|---|---|
| 0 | 95.9 | 49.7 | 65.4 | 15/31 |
| 1 | 59.9 | 74.5 | 66.4 | 40/54 |
| 2 | 72.2 | 24.2 | 36.2 | 40/166 |
| 3 | 29.0 | 84.9 | 43.3 | 654/771 |
| 4 | 99.7 | 32.2 | 48.6 | 267/832 |
| 5 | 52.1 | 47.2 | 49.5 | 558/1,183 |
| 6 | 66.8 | 64.6 | 65.7 | 795/1,232 |
| 7 | 95.2 | 96.1 | 95.6 | 3,824/3,981 |
| 8 | 27.9 | 49.0 | 35.6 | 2,351/4,797 |
| 9 | 98.2 | 96.2 | 97.2 | 26,753/27,819 |
| Micro Mean | 86.4 | 86.4 | 86.4 | 35,297/40,866 |
| Macro mean | 69.7 | 61.8 | 65.5 | … |
Abbreviations: CNN, convolutional neural network; cTAKES, clinical Text Analysis and Knowledge Extraction System; ID, identification; Pre%, precision; Rec%, recall; F1%, F1 score.
TABLE VII.
Results of MfeCNN With Features Extracted by cTAKES.
| Class ID | Pre% | Rec% | F1% | Support |
|---|---|---|---|---|
| 0 | 100 | 100 | 100 | 31/31 |
| 1 | 100 | 100 | 100 | 54/54 |
| 2 | 100 | 39.8 | 57.0 | 66/166 |
| 3 | 95.4 | 66.7 | 78.5 | 514/771 |
| 4 | 100.0 | 37.6 | 54.6 | 312/832 |
| 5 | 71.9 | 58.6 | 64.6 | 693/1,183 |
| 6 | 97.8 | 69.0 | 80.9 | 849/1,232 |
| 7 | 100 | 100 | 100 | 3,981/3,981 |
| 8 | 23.6 | 100 | 38.2 | 4,797/4,797 |
| 9 | 96.6 | 98.3 | 97.4 | 27,340/27,819 |
| Micro Mean | 94.5 | 94.5 | 94.5 | 38,637/40,866 |
| Macro Mean | 88.5 | 77.0 | 82.4 | … |
Abbreviations: cTAKES, clinical Text Analysis and Knowledge Extraction System; ID, identification; MfeCNN, mixture feature embedding convolutional neural network; Pre%, precision; Rec%, recall; F1%, F1 score.
When the same input features from cTAKES were used, SVM had lower overall recalls and F1 scores than those of deep learning models, (CNN and MfeCNN). The possible reason for this difference is that our SVM model is linear and simply transforms the input features to a higher dimensional space in order to reveal the differences of target classes.
However, the deep architecture of the deep learning model has multiple layers, which combine and nonlinearly transform features from layer to layer. This deep and nonlinear architecture could help to achieve better classification results.
The macro means of F1 scores of the 3 deep learning models (basic CNN, CNN and MfeCNN) reported herein were 47.8%, 65.5%, and 82.4%, respectively. Among these deep learning models, MfeCNN achieved the best overall performance for data mapping. The macro mean F1 score for MfeCNN was as high as 82.4%. With features generated by cTAKES, the overall F1 score of CNN increased by 17.7%. Compared with CNN, MfeCNN had much better results for most classes. With MfeCNN, we achieved a 16.9% improvement over the CNN results. The accuracy (micro mean) of our MfeCNN model for data mapping with ten classes is 94.5%. Lecun et al. [30] recently developed a very deep CNN model with 29 network layers to perform topic classification of 10 topics for free texts without preprocessing these texts as inputs, and the prediction accuracy was 73.4%.
As shown in Table 7, our data is very unbalanced. The differences between F1 micro and macro means of the 3 models (SVM, CNN, and MfeCNN) are 18.4%, 20.9% and 12.1%, respectively. It indicates that the issue of data imbalance can be somewhat improved by our MfeCNN model with mixture feature embedding.
The study results validate that our MfeCNN model can indeed map customer HL7 messages to canonical data types effectively and can handle data imbalance to some degree. When the results were compared with those of traditional SVM model and CNN models, the combination of mixture feature embedding and CNN in MfeCNN allowed for the development of a sophisticated deep learning model, which achieved excellent mapping accuracy.
V. Conclusion
We implemented a novel and sophisticated deep learning framework, MfeCNN, for clinical data mapping. With this framework, we successfully converted the data mapping task to a multi-label classification problem. Our model innovatively incorporates multimodal learning and multiview embedding into CNN for mixture feature tensor generation and classification prediction. cTAKES, an open-source tool, was used to perform deep language analysis of unstructured free texts to extract rich linguistic features. To make complete use of diverse features in the multimodal semantic spaces, we used mixture feature embedding to digest features from different semantic spaces, which was done with multimodal and multi-view approaches. These approaches are quite different from those used previously for feature embedding, which have usually focused on word spaces. In contrast, we combined the feature embedding of the semantic space, syntax space, domain space (medical concepts), and word space. Our experimental results showed that our approach achieved satisfactory results. The combination of mixture feature embedding and a deep neural network was important for achieving a high accuracy rate. Although we tested our model for problems with clinical data, our MfeCNN can also be used for data mapping and multi-label classification in fields other than health care.
Acknowledgments
Funding for this study was provided by the Mayo Clinic Center for Clinical and Translational Science (UL1TR002377) from the National Institutes of Health/National Center for Advancing Translational Sciences (MH and LY) and the Career Development Award (5K01LM012102) from the National Library of Medicine (LY).
Biographies

Dingcheng Li received the Ph.D. degree in Computational Linguistics in 2011, the M.Sc. degree in Computer Science in 2011, and the M.A. degree in Lunguistics in 2007 from University of Minnesota-Twin Cities, Minneapolis, MN, USA.
From Dec. 2016 to present, he has been a Research Scientist at the Big Data Lab in Baudu USA in Sunnyvale, CA. From May 2016 to Dec. 2016, he was a Senior Software Engineer in IBM TJ Watson Health Cloud in Rochester, MN, USA. From May 2011 to Apr. 2016, he was a Researcher and Programmer in Mayo Clinic in Rochester, MN, USA. In 2010, he was a Research Intern in Siemens. His research intersests include Large Scale Real-time Cognitive Computing, Intelligent Text Mining and Natural Language Processing, and Data Analysis and Data Semantics Discoveries.
Dr. Li was the recipient of NIH Pathway to Independence Award (K99/R00) in 2015. He is now a member of AMIA, ACL and ACM.

Ming Huang was born in Jiangxi province, China in 1983. He received his dual B.Sc. degrees in Computer Science and Applied Chemistry from Beijing Normal University, Beijing, China in 2004 and the Ph.D. degree in Scientific Computation from the University of Minnesota-Twin Cities, Minneapolis, MN, USA in 2014.
From 2014 to 2017, he worked as a Postdoctoral Associate in the Institute of Quantitative Biomedicine, Rutgers University. Since 2017, he has been a Research Associate with the Division of Biomedical Statistics and Informatics, Health Sciences Department in Mayo Clinic. He has published 13 peer-reviewed articles. His research interests include semiempirical quantum mechanical methods, ribozymes, biocatalysis, machine learning, and medical data mining.
Dr. Huang was a recipient of Biomedical Informatics and Computational Biology Training Fellowship sponsored by UMN, Mayo Clinic and IBM from 2008 to 2010, and the American Chemistry Society Conference Travel Award in 2015.

Xiaodi Li was born in Chongqing, China in 1996. She will receive the B.Eng. degree in Mechatronics Engineering from Donhua University, Shanghai, China in July 2018 and will start her Ph.D. study in the University of Texas at Dallas in August 2018.
From July 2015 to August 2015, she was a Software Test Engineer intern in Tektronix Company in Shanghai, China. From 2016 Summer to August 2017, she did research in WSN Lab in Sichuan University. From Sept. 2017 to Feb. 2018, she worked on research projects in the Key Lab of Cloud Computing and Intelligent Technology in Southwest Jiaotong University. Her research interests include Machine Learning, Natural Language Processing, Image Processing and Data Mining.

Yaoping Ruan received the Ph.D. degree in Computer Science from Princeton University, Princeton, NJ, USA in 2005, M.Sc. degree in Computer Science and Technology from University of Science and Technology of China, Hefei, Anhui, China in 2000, and B.Sc. degree in Precision Instruments from Tsinghua University, Beijing, China in 1997.
From Jun. 2016 to present, he is a RSM & Senior Manager in IBM Watson Health in Yorktown Heights, NY, USA. From May 2015 to Jun. 2016, he was a RSM & Development Manager in IBM Watson Health. From Aug. 2013 to May 2015, he was a Research Manager in IBM Research. From 2005 to Aug. 2013, he was a Researcher (Research Staff Member) in IBM Research. From Sep. 2001 to Sep. 2005, he was a Research Assistant in Princeton University. His research focuses on cloud-scale data management for data lake, data reservoir, and data curation; cloud platform performance optimization; cloud platform operational analysis with logging, monitoring, and alerting; networking infrastructure for data-centric service placement; dimensional-model-based and unstructured data analytical platform.
Dr. Ruan has been recognized with one IBM Corporate Award (highest technical award), multiple IBM Outstanding Technical Achievement Awards, IBM Research Accomplishments, and IBM Invention Achievement Awards.

Lixia Yao was born in China in 1980. She received a B.Eng. degree in Chemical Engineering from Dalian University of Technology, China in 2002, a M.Sc. degree in Computational Science from the National University of Singapore, Singapore in 2004, and a Ph.D. degree in Biomedical Informatics from Columbia University, NY, USA in 2010.
She then worked as a Principal Investigator at GlaxoSmithKline Pharmaceuticals from 2010 to 2013 and as an assistant professor at the University of North Carolina at Charlotte from 2013 to 2016. She becomes an Assistant Professor at Mayo Clinic since 2016. Her research focuses on mining, integrating, and transforming unstructured, incomplete, and noisy data (i.e., electronic health records and claims databases, literature, patents, and social media) into meaningful biomedical knowledge and informatics applications. She has published 24 peer-reviewed articles.
Dr. Yao is the recipient of Career Development Award in Biomedical Informatics from the National Library of Medicine for 2016–2019.
Contributor Information
Dingcheng Li, Big Data Lab, Baidu, Sunnyvale, CA, 94085 USA (lidingcheng@baidu.com)..
Ming Huang, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, 55906 USA (huang.ming@mayo.edu)..
Xiaodi Li, Department of Mechatronics Engineering, Donhua University, Shanghai, 200336 China (xiaodi_li327@qq.com)..
Yaoping Ruan, Watson Health Cloud, IBM, Yorktown Heights, NY, 10598 USA (yaoping.ruan@us.ibm.com).
Lixia Yao, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, 55906 USA (yao.lixia@mayo.edu)..
References
- [1].Liu S, Wang Y, Hong N, Shen F, Wu S, Hersh W, and Liu H, “On mapping textual queries to a common data model,” in 2017 IEEE International Conference on Healthcare Informatics (ICHI), IEEE, 2017, pp. 21–25. [Google Scholar]
- [2].Geller J, He Z, Perl Y, Morrey CP, and Xu J, “Rule-based support system for multiple umls semantic type assignments,” Journal of Biomedical Informatics, vol. 46, no. 1, pp. 97–110, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gu H, Chen Y, He Z, Halper M, and Chen L, “Quality assurance of umls semantic type assignments using snomed ct hierarchies,” Methods of Information in Medicine, vol. 55, no. 2, pp. 158–65, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rahm E and Bernstein PA, “A survey of approaches to automatic schema matching,” the VLDB Journal, vol. 10, no. 4, pp. 334–350, 2001. [Google Scholar]
- [5].Eisenstein J, O’Connor B, Smith NA, and Xing EP, “A latent variable model for geographic lexical variation,” in Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing. Stroudsburg, PA, USA: Association for Computational Linguistics, 2010. [Google Scholar]
- [6].Berlin J and Motro A, “Autoplex: Automated discovery of content for virtual databases,” in International Conference on Cooperative Information Systems. Springer, 2001, pp. 108–122. [Google Scholar]
- [7].Bilke and Naumann F, “Schema matching using duplicates,” in ICDE 2005 Proceedings of 21st International Conference on Data Engineering, IEEE, 2005, pp. 69–80. [Google Scholar]
- [8].Yan LL, Miller RJ, Haas LM, and Fagin R, “Data-driven understanding and refinement of schema mappings,” in ACM SIGMOD Record, vol. 30, no. 2 ACM, 2001, pp. 485–496. [Google Scholar]
- [9].Herna’ndez MA, Miller RJ, and Haas LM, “Clio: Asemi-automatic tool for schema mapping,” ACM SIGMOD Record, vol. 30, no. 2, p. 607, 2001. [Google Scholar]
- [10].Bernstein PA, Melnik S, and Mork P, “Interactive schema translation with instance-level mappings,” in Proceedings of the 31st international conference on very large data bases. VLDB Endowment, 2005, pp. 1283–1286. [Google Scholar]
- [11].Drumm, Schmitt M, Do H-H, and Rahm E, “Quickmig: automatic schema matching for data migration projects,” in Proceedings of the sixteenth ACM conference on Conference on information and knowledge management. ACM, 2007, pp. 107–116. [Google Scholar]
- [12].Doan, Domingos P, and Halevy AY, “Reconciling schemas of disparate data sources: A machine-learning approach,” in ACM Sigmod Record, vol. 30, no. 2 ACM, 2001, pp. 509–520. [Google Scholar]
- [13].Alexe, Ten Cate B, Kolaitis PG, and Tan W-C, “Designing and refining schema mappings via data examples,” in Proceedings of the 2011 ACM SIGMOD International Conference on Management of data. ACM, 2011, pp. 133–144. [Google Scholar]
- [14].Madhavan J, Bernstein PA, Doan A, and Halevy A, “Corpus-based schema matching,” in ICDE 2005 Proceedings of 21st International Conference on Data Engineering,IEEE, 2005, pp. 57–68. [Google Scholar]
- [15].Li, Liu P, Huang M, Gu Y, Zhang Y, Li X, Dean D, Liu X, Xu J, Lei H et al. , “Mapping client messages to a unified data model with mixture feature embedding convolutional neural network,” in 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, 2017, pp. 386–391. [Google Scholar]
- [16].Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, and Kuksa P, “Natural language processing (almost) from scratch,” Journal of Machine Learning Research, vol. 12, pp. 2493–2537, 2011. [Google Scholar]
- [17].Miwa M and Bansal M, “End-to-end relation extraction using lstms on sequences and tree structures,” arXiv preprint arXiv:1601.00770, 2016. [Google Scholar]
- [18].Ngiam J, Khosla A, Kim M, Nam J, Lee H, and Ng AY, “Multimodal deep learning,” in Proceedings of the 28th international conference on machine learning (ICML-11), 2011, pp. 689–696. [Google Scholar]
- [19].Johnson R and Zhang T, “Semi-supervised convolutional neural networks for text categorization via region embedding,” in Advances in neural information processing systems, 2015, pp. 919–927. [PMC free article] [PubMed] [Google Scholar]
- [20].Mikolov T, Sutskever I, Chen K, Corrado GS, and Dean J, “Distributed representations of words and phrases and their compositionality,” in Advances in neural information processing systems, 2013, pp. 3111–3119. [Google Scholar]
- [21].Liu S, Shen F, Chaudhary V, and Liu H, “Mayonlp at semeval 2017 task 10: Word embedding distance pattern for keyphrase classification in scientific publications,” in Proceedings of the 11th International Workshop on Semantic Evaluation (SemEval-2017), 2017, pp. 956–960. [Google Scholar]
- [22].Wang Y, Liu S, Afzal N, Rastegar-Mojarad M, Wang L, Shen F, and Liu H, “A comparison of word embeddings for the biomedical natural language processing,” arXiv preprint arXiv:1802.00400, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tai KS, Socher R, and Manning CD, “Improved semantic representations from tree-structured long short-term memory networks,” arXiv preprint arXiv:1503.00075, 2015. [Google Scholar]
- [24].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, and Chute CG, “Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications,” Journal of the American Medical Informatics Association, vol. 17, no. 5, pp. 507–513, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, Liu S, Zeng Y, Mehrabi S, Sohn S et al. , “Clinical information extraction applications: A literature review,” Journal of biomedical informatics, vol. 77, pp. 34–49, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Benson T and Grieve G, “HL7 version 2,” in Principles of Health Interoperability. Springer, 2016, pp. 223–242. [Google Scholar]
- [27].Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, and Kohane I, “Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2),” Journal of the American Medical Informatics Association, vol. 17, no. 2, pp. 124–130, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chang C-C and Lin C-J, “Libsvm: a library for support vector machines,” ACM Transactions on Intelligent Systems and Technology (TIST), vol. 2, no. 3, p. 27, 2011. [Google Scholar]
- [29].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M et al. , “Tensorflow: Large-scale machine learning on heterogeneous distributed systems,” arXiv preprint arXiv:1603.04467, 2016. [Google Scholar]
- [30].Conneau, Schwenk H, Barrault L, and Lecun Y, “Very deep convolutional networks for text classification,” arXiv preprint arXiv:1606.01781, 2016. [Google Scholar]


