Abstract
Anaplastic thyroid carcinomas (ATCs) have a high prevalence of BRAF and TP53 mutations. A trial of vemurafenib in nonmelanoma BRAFV600E-mutant cancers showed significant, although short-lived, responses in ATCs, indicating that these virulent tumors remain addicted to BRAF despite their high mutation burden. To explore the mechanisms mediating acquired resistance to BRAF blockade, we generated mice with thyroid-specific deletion of p53 and dox-dependent expression of BRAFV600E, 50% of which developed ATCs after dox treatment. Upon dox withdrawal there was complete regression in all mice, although recurrences were later detected in 85% of animals. The relapsed tumors had elevated MAPK transcriptional output, and retained responses to the MEK/RAF inhibitor CH5126766 in vivo and in vitro. Whole-exome sequencing identified recurrent focal amplifications of chromosome 6, with a minimal region of overlap that included Met. Met-amplified recurrences overexpressed the receptor as well as its ligand Hgf. Growth, signaling, and viability of Met-amplified tumor cells were suppressed in vitro and in vivo by the Met kinase inhibitors PF-04217903 and crizotinib, whereas primary ATCs and Met-diploid relapses were resistant. Hence, recurrences are the rule after BRAF suppression in murine ATCs, most commonly due to activation of HGF/MET signaling, which generates exquisite dependency to MET kinase inhibitors.
Keywords: Oncology
Keywords: Mouse models, Oncogenes, P53
Introduction
Anaplastic thyroid cancers (ATCs) are extraordinarily virulent tumors that affect mostly elderly patients and have a median survival of 6 months. Cytotoxic chemotherapy with antimicrotubule agents and/or anthracyclines in combination with surgery and radiation therapy may extend survival to a modest extent when the ATC is confined to the neck, but has minimal activity in patients with metastatic disease (1). Most ATCs are thought to arise from preexisting well-differentiated tumors as a result of acquiring additional alterations (2–4). We recently reported the massively parallel exon sequencing of a 341 cancer gene panel in 84 poorly differentiated thyroid cancers (PDTCs) and 33 ATCs (5). Consistent with other reports, BRAFV600E was the most common oncogenic driver, with most of the BRAFV600E-mutant ATCs also having mutations in TRP53 and/or TERT (2, 5, 6).
Until recently, the general presumption was that ATCs were unlikely to respond to monotherapy with compounds selectively targeting the oncogenic drivers of the disease, primarily because of their high mitotic index and large burden of mutations. However, several isolated case reports in the literature reported significant responses of BRAF-mutant ATCs to RAF kinase inhibitors (7–10). A prospective trial of vemurafenib in patients with nonmelanoma BRAFV600E-mutant cancers included 7 patients with ATC, 3 of whom had major responses (11). The overall response rate in ATCs is lower than that of BRAFV600E-positive metastatic melanomas, which is likely due in part to feedback-induced activation of RTKs that overcomes inhibition of the MAPK pathway (12). Similar to what is observed in BRAFV600E melanomas, all but one patient with ATC progressed within 13 months. The evidence that overcoming adaptive resistance to RAF kinase inhibitors in ATCs is clinically relevant is further cemented by the results of a clinical trial with a combination of the RAF inhibitor dabrafenib with the MEK inhibitor trametinib, in which an overall response rate of 69% (11 of 16) was achieved (13).
Multiple mechanisms of acquired resistance to BRAF inhibitors have been identified in patients with melanoma, including overexpression of CRAF or COT1 (14), BRAFV600E amplification (15–19), activating mutations in NRAS, KRAS, MEK1, or AKT1 (15–22), BRAFV600E alternative splicing (15–19), activation of phosphatidylinositol-3-OH kinase (15, 19, 23), and increased expression of receptor tyrosine kinases (15–17). There is currently no information on the mechanisms of acquired resistance of BRAF-mutant ATC to RAF kinase inhibition. Here we explore the responses to downregulation of oncogenic BRAF in a mouse model of BRAFV600E-driven ATC. Genetic inhibition of BRAFV600E induced consistent and profound tumor regressions, although nearly all mice developed recurrences within 1 year. Analysis of the recurrent tumors indicated that reactivation of the MAPK pathway was universally associated with tumor regrowth, primarily via concomitant Met amplification and cell-autonomous Hgf overexpression.
Results
Development of mouse anaplastic thyroid cancer by inducible expression of BRAFV600E and loss of p53.
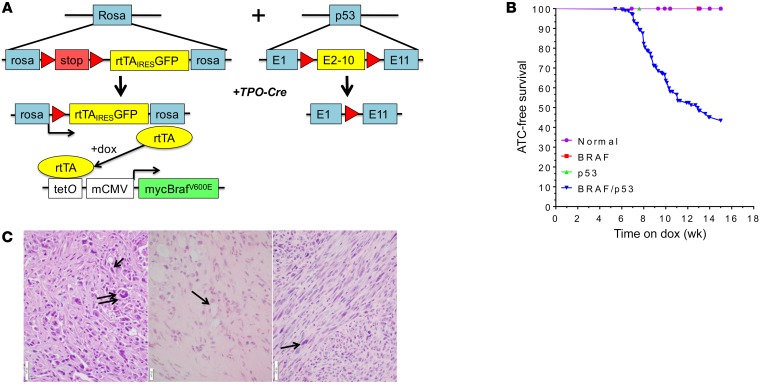
We generated mice with thyroid-specific loss of p53 and doxycycline-inducible (dox-inducible) expression of BRAFV600E (Figure 1A). The dox-dependent transactivator rtTA gene is present as a latent allele within the Rosa locus, requiring excision of a Lox-stop-Lox (LSL) cassette by Cre recombinase for its expression (24, 25). Dox-treated Tpo-Cre/LSL-rtTA_GFP/tetO-BRAFV600E mice have an approximately 2-fold increase in Braf mRNA, with a BrafWT/BRAFV600E ratio of approximately 1:1 (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI120966DS1). Despite receiving dox for 6 months, these mice did not develop thyroid cancer (data not shown). However, when crossed with Trp53fl/fl mice (26) to generate the quadruple transgenic line Tpo-Cre/LSL-rtTA_GFP/tetO-BRAFV600E/Trp53fl/fl (BRAF/p53), approximately 50% of compound animals developed large thyroid cancers after a mean of 9 weeks on dox (Figure 1B). Tumors were only found in quadruple transgenic mice treated with dox, and in the vast majority of those mice, tumors were unilateral. Histology of 15 BRAF/p53 tumors showed them all to be ATC, as evidenced by spindle-shaped cells, giant cells, high mitotic rate, necrosis, and extrathyroidal invasion (Figure 1C and Supplemental Figure 2A). Examination of the lungs from 10 animals with ATC found metastatic foci in 5. In addition, 5 mice had metastases to either lymph nodes (n = 3) or skull (n = 2) (Supplemental Figure 3). The ATCs showed strong pERK staining (Supplemental Figure 2B) and, consistent with human ATCs (27, 28), they were heavily infiltrated with macrophages, marked by Iba1 immunostaining (Supplemental Figure 2B).
Figure 1. BRAFV600E-driven mouse model of ATC.
(A) Genetic schema of mouse model of dox-inducible BRAFV600E-driven ATCs. Left: LSL-rtTA-ires-GFP construct targeted to the Rosa allele, which only expresses rtTA after Cre excision. Expression of rtTA transduces expression of a tetO-driven myc-tagged BRAFV600E in the presence of dox. Right: Exons 2–10 of Trp53 are flanked by loxP sites (red arrows) in Trp53fl mice. Thyroid-specific inactivation of Trp53 and expression of rtTA is achieved by crosses with TPO-Cre mice. (B) ATC-free survival of TPO-Cre negative (normal), TPO-Cre/LSL-rtTA-GFP/tetO-BRAFV600E (BRAF), TPO-Cre/Trp53fl/fl (p53), and BRAF/p53 mice after switching to dox-impregnated chow. Each cohort consisted of at least 9 mice. (C) Left: H&E staining of thyroid tumor sections from TPO-Cre/LSL-rtTA-GFP/tetO-BRAFV600E/Trp53fl/fl (BRAF/p53) mouse on dox showing an ATC with giant cells (double arrow) and spindle-shaped cells (single arrow). Scale bar, 50 μm. Middle: Representative H&E staining of ATC remnant 3 weeks after dox withdrawal showing marked fibrosis with small foci of thyroid cells with benign morphology (arrow). Scale bar, 20 μm. Right: ATCs that recurred after genetic inhibition of BRAFV600E (arrow: spindle-shaped tumor cell). Scale bar, 50 μm.
Transition from PTC to ATC is associated with increased MAPK output.
To investigate possible differences in the intensity of MAPK pathway activation between BrafV600E-driven PTCs and ATCs, we calculated a MAPK score by adapting previously reported approaches that integrate the magnitude of expression changes in a number of transcripts known to be regulated by MAPK signaling (refs. 29, 30, and Supplemental Table 1). We found a highly significant increase in MAPK transcriptional output between PTCs from TPO-Cre/LSL-BRAFV600E (31) and the BRAF/p53 ATCs. We corroborated this by comparing PTCs to ATCs in previously published BrafV600E-driven mouse models (32), as well as in human ATCs (refs. 5, 33, and Supplemental Figure 1B). The higher MAPK output in the current BRAF/p53 ATC model was associated with increased expression of transgenic BRAF, likely resulting from higher levels of rtTA (Supplemental Figure 1, C and D). However, this was not the case in ATCs driven by postnatal, tamoxifen-induced, Cre-dependent knockin of BrafV600E in the context of Tp53 loss, where Braf levels were comparable to those of PTCs developing in mice with intact Trp53 alleles (Supplemental Figure 1E). Interestingly, expression of a number of genes encoding effectors that downregulate MAPK signaling, including dual-specificity phosphatases (DUSPs) and the Sprouty-related proteins SPRED1 and 2, was lower in BrafV600E murine and human ATCs than in Braf-mutant PTCs (Supplemental Table 2).
BRAF/p53 ATCs regress upon switching off BRAF expression, but eventually recur in a BRAF-independent manner.
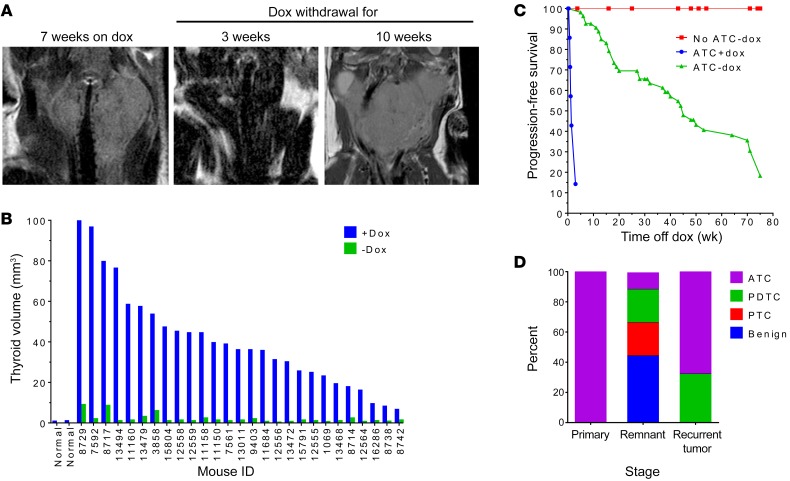
To investigate if the ATCs were addicted to oncogenic BRAF, animals confirmed by MRI to have tumors were placed on a dox-free diet, and imaged 3–4 weeks later (Figure 2, A and B). Approximately 20% of the mice died in the first week after dox withdrawal (67% died within 1–4 days, and 33% died on days 5–7). MRIs of most of these mice showed marked tracheal compression at the time that dox was removed from the diet, and death was attributed to asphyxia due to the large tumor burden at the time of diagnosis. All others had profound tumor regression by 3 weeks (Figure 2, A and B), and markedly increased progression-free and overall survival as compared with mice continued on dox, none of which survived longer than 3 weeks (Figure 2C). The response to dox withdrawal was also seen in lymph node and skull metastases (Supplemental Figure 3). Histological examination of the residual lesions revealed debris, fibrosis, and small nests of tumor cells (Figure 1C and Supplemental Figure 4A). The majority of the remaining thyroid follicular cells had a benign appearance. However, a few tumor cells with PTC-, PDTC-, or ATC-like features were observed in 5 of 9 tumor remnants (Figure 2D and Supplemental Figure 4A). IHC for myc-tagged BRAFV600E and pERK demonstrated strong expression of the oncoprotein and activation of the MAPK pathway in the primary ATCs, which was absent in the tumor remnants (Supplemental Figure 4B).
Figure 2. BRAFV600E-driven mouse ATCs are addicted to the oncoprotein, but frequently recur.
(A) MRI images of a representative BRAF/p53 mouse showing an ATC (left), then regression 3 weeks after dox withdrawal (middle), and a recurrence 7 weeks later (right). (B) Tumor volume before and 3–4 weeks after withdrawal of dox (n = 27). Volume of 2 normal thyroid glands is shown for comparison. (C) Kaplan-Meier curve showing progression-free survival of BRAF/p53 mice harboring an ATC and continued on dox (ATC+dox) versus after dox withdrawal (ATC-dox). Control BRAF/p53 mice without MRI evidence of ATC at the time of dox withdrawal (no ATC-dox) are shown in red. Each cohort consisted of at least 13 mice. (D) Histological diagnoses of 15 primary tumors, 9 tumor remnants after dox withdrawal, and 22 recurrent tumors.
Over an 18-month period of dox withdrawal, 87% of mice off dox experienced a recurrence, with a median progression-free survival of 45 weeks (95% CI: 36.6–71). Recurrences occurred in the thyroid bed in 84% (32 of 38) of mice, consistently within the lobe where the original ATC was detected. Relapses were only present at distant sites in 16% (6 of 38) of mice, including lung, lymph node, and skull. Histology of relapses showed greater heterogeneity than in the primary tumors (Figure 2D and Supplemental Figure 5). The majority resembled primary ATCs (Figure 1C, Figure 2D, and Supplemental Figure 5). However, others were classified as PDTCs, ATCs with osteosarcoma transformation, or mucin-producing adenocarcinomas (Supplemental Figure 5). A cohort of mice without detectable tumors after 1 year off dox was placed back on a dox-containing diet. Tumors regrew within 2 weeks in 8 of 9 mice (Supplemental Figure 6), and the remaining one became ill after 6 weeks as a result of a large lung metastasis. One mouse with a dox-induced recurrence was put back on the dox-free diet and the tumor remained stable at 4 weeks, ultimately progressing 4 weeks later (Supplemental Figure 6).
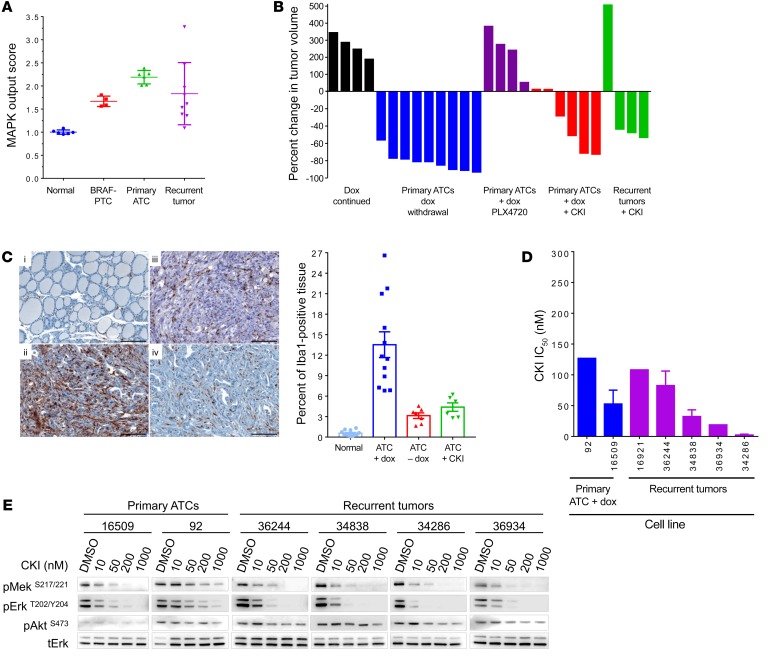
To explore the mechanisms driving tumor recurrence, we first examined whether BRAFV600E was expressed despite withdrawal of dox, as dox-independent expression of rtTA-driven oncoproteins has been found to be a common mechanism for tumor regrowth in similarly designed mouse models. This is often associated with acquired mutations in the rtTA transgene, which allow it to transactivate the tetO promoter in a dox-independent manner (34–36). Only 3 of 20 relapses expressed the myc-tagged BRAFV600E mRNA (Supplemental Figure 7A), although none of these developed mutations in rtTA. Primary ATCs clustered together by gene expression profiling, whereas ATC recurrences clustered apart from the primary tumors and from each other, consistent with their heterogeneous histological characteristics (Supplemental Figure 7B). As the primary ATCs were driven by an oncoprotein that signals exclusively through MAPK, we next explored whether the recurrent tumors had a high MAPK transcriptional output in the absence of BRAFV600E expression). The ATC recurrences were more variable in this respect than the primary tumors, but they consistently showed an increase in the MAPK output score compared with normal thyroid (Figure 3A).
Figure 3. Reactivation of the MAPK pathway drives tumor recurrences.
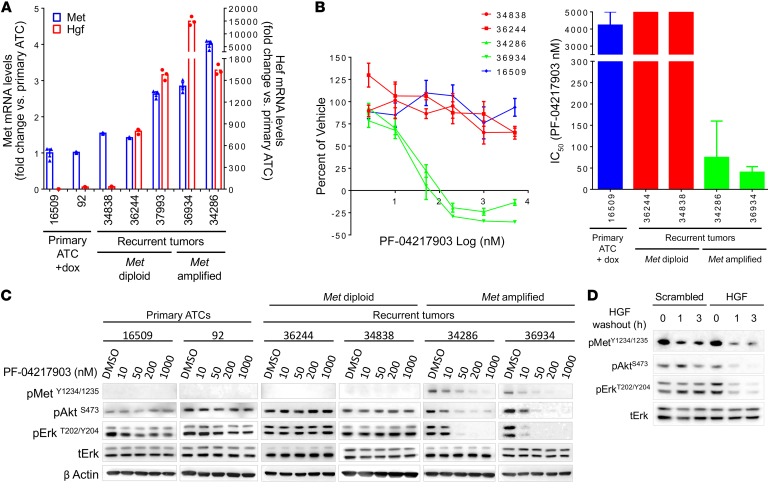
(A) MAPK output score for normal mouse thyroid, BRAF-PTCs (from TPO-Cre/LSL-BrafV600E mice) (31), BRAF/p53 ATCs, and recurrent tumors was calculated from expression array data of flash-frozen tissue from recurrent tumors using MAPK transcriptional targets (n = 4–7) (29). (B) Response of primary or recurrent tumors to dox withdrawal or to treatment with PLX4720 (administered in drug-impregnated chow) or CKI (1.5 mg/kg/d). Tumor volume was measured 11 days after starting the intervention (n = 4–9). (C) Iba1 IHC (left) in (i) normal thyroid, (ii) untreated primary ATCs, (iii) primary ATCs 11 days after withdrawal of dox, or (iv) starting CKI. Bars (right) represent average percent of tumor positive for Iba1 from at least 6 animals. (D) IC50 of the MEK inhibitor CKI in cell lines derived from primary or recurrent tumors. Each dose was run in triplicate. Bars represent the IC50 values for cell lines 16509 (n = 4), 36244 (n = 3), and 34286 (n = 2). IC50 for cell lines 92, 16921, and 36934 was calculated from a single experiment. (E) Dose response of CKI on MEK and ERK phosphorylation in the cell lines listed in D. Cells from the primary ATCs were grown on dox, whereas recurrent tumors were off dox. Cells were treated for 1 hour with the indicated doses of CKI, collected, and protein was isolated for Western blotting with antibodies to pERKT202/Y204, tERK, pAKTS473, and pMEKS217/221.
Primary ATCs were refractory to the RAF kinase inhibitor PLX4720 (Figure 3B), consistent with the previously reported adaptive resistance to these compounds in BRAFV600E human ATC cell lines (12). By contrast, CH5126766 (CKI), an allosteric inhibitor of MEK that locks this kinase in an inactive complex with RAF proteins and blunts feedback-induced reactivation of MAPK (37), evoked major responses in the primary tumors as well as in 3 of 4 ATC recurrences (Figure 3B). The marked reduction of primary ATC volume after 11 days of suppressing BrafV600E expression was associated with a decrease in tumor-associated macrophages, as demonstrated by Iba1 IHC. This effect was also observed in mice treated with CKI (Figure 3C). CKI also blocked growth and MAPK signaling in cell lines derived from primary and relapsed ATCs (Figure 3, D and E).
We next investigated potential differences in gene expression pathways or programs between the primary ATCs and the recurrent tumors. Gene set enrichment analysis (GSEA) of expression array data revealed decreased markers of T cell activation in all recurrent tumors (Supplemental Figure 8, A and B). This was supported by analysis of the data through the use of IPA (Qiagen; https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis), where 11 of the top 20 canonical pathways showed a blunting of immune response or inflammatory processes in the recurrent tumors (Supplemental Figure 8C). Interestingly, the recurrent ATCs also showed GSEA evidence of increased polycomb repressor complex 1 (PRC1) activity as compared with the primary tumors (Supplemental Figure 8, A and B). All ATC relapses exhibited a signature of BMI-1, a core PRC1 component that regulates cell stemness and has been implicated in the pathogenesis of numerous cancer types.
Met amplification drives ATC recurrences after BRAF silencing.
The sensitivity of the dox-independent ATC recurrences to CKI prompted us to explore whether these tumors had acquired a de novo somatic mutation in a gene encoding a MAPK pathway effector. We first performed targeted sequencing of hotspots in mouse Nras, Hras, Kras, and Braf and of the entire coding sequences of Eif1ax, Mek1, and Mek2. One of 8 relapses negative for expression of myc-tagged BRAFV600E harbored an activating mutation of Hras (HrasQ61K), but no mutations were found in any of the others. We then performed whole-exome sequencing of 11 recurrent tumors that were found to have a high MAPK transcriptional output and/or demonstrated sensitivity to MEK inhibition. Mutation calling algorithms identified 67,575 somatic alterations among the 11 recurrent tumors. Of these, 271 were found to be clonal (mutant allele frequency at least one-quarter the tumor purity), nonsynonymous, and within coding regions or within a splice site (Supplemental Table 3, data set 1). Each alteration was found in a single sample, except for Cdv3, Fnbp1, Map3k5, and Olfr521, which were each identified in 2 samples. None of these recurrent alterations had a known direct connection with MEK/ERK activation. Analysis of the 271 altered genes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) showed the genes to be significantly enriched in DAVID terms related to antigen processing, guanylate-binding protein, calcium ion transport, and autoimmune thyroid disease (Supplemental Table 4).
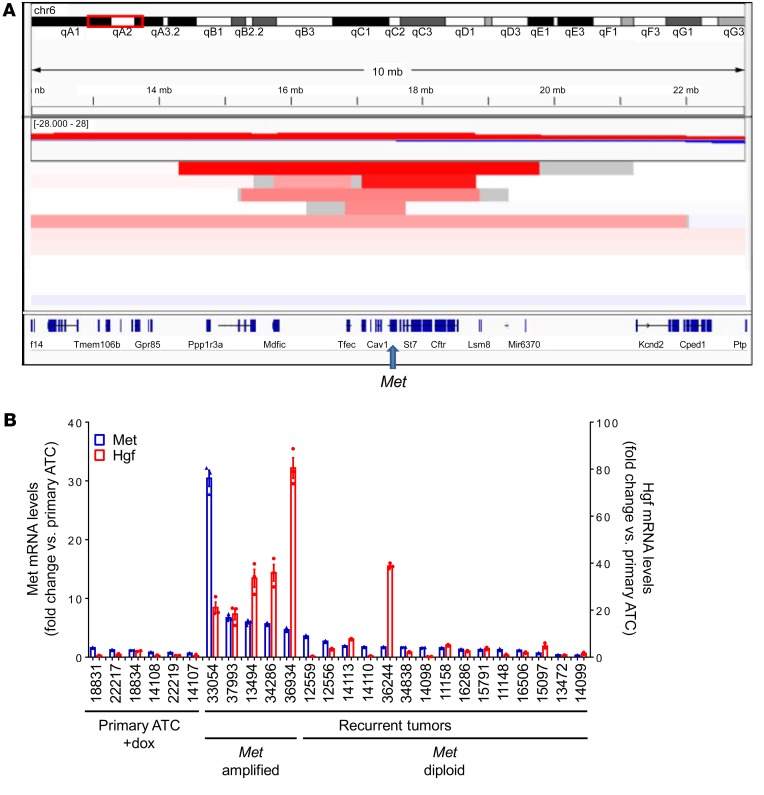
We also mined the data for copy number changes and found only 2 loci altered in more than 2 recurrent tumors. These included a focal loss of chromosome 11, which corresponds to the floxed Trp53 allele, and a focal amplification on chromosome 6, present in 5 of 11 samples. Examination of regions altered in recurrent tumors without the chromosome 6 amplification did not show any other amplification events of recognized cancer drivers (Supplemental Table 5). The minimal region of chromosome 6 amplification contained only 7 genes: Tfec, Tes, Cav2, Cav1, Met, Capza2, and St7 (Figure 4A). Of these we focused on Met, as it has been reported to play a role in acquired (17) and adaptive (18, 38) resistance to BRAFV600E inhibition in BRAFV600E-mutant melanoma models (38, 39). Supplemental Table 6 summarizes the main drivers identified in the recurrent tumors.
Figure 4. Met amplification and Hgf overexpression in recurrent tumors.
(A) IGV of copy number changes determined by FACET from exome sequencing reads of recurrent ATCs. Focal amplification of chromosome 6 was seen in 5 of 11 tumor relapses. The minimal common amplicon includes Tfec, Tes, Cav2, Cav1, Met, Capza2, and St7. (B) Quantitative RT-PCR demonstrates overexpression of Met and Hgf in recurrent tumors with Met amplification. Bars represent the fold change in expression of Met and Hgf compared with primary ATCs. Quantitative RT-PCR for primary ATCs and recurrent tumors were run in triplicate.
We confirmed that Met was overexpressed in the relapsed tumors with Met amplification (Figure 4B). Interestingly, the Met ligand Hgf was also selectively overexpressed in the Met-amplified recurrences, but not in the recurrent ATCs that were diploid for Met (Figure 4B). Cell lines derived from relapses with Met amplification retained Met and Hgf overexpression compared with cell lines derived from primary ATCs or from recurrences that were Met diploid (Figure 5A). Met-amplified lines were exquisitely sensitive to the growth inhibitory effects of the highly specific Met inhibitor PF-04217903, whereas primary ATC and non–Met amplified cells were resistant (Figure 5B). Similar findings were observed with the FDA-approved Met inhibitor crizotinib (Supplemental Figure 9, A and B). PF-04217903 (Figure 5C) or crizotinib (Supplemental Figure 9C) inhibited Met phosphorylation only in the Met-amplified tumor cells, and blocked MAPK and PI3K signaling. Knockdown of Hgf in a Met-amplified cell line with a shRNA that reduced Hgf mRNA levels by 70% decreased pMET, pERK, and pAKT, supporting a role for cell-autonomous Hgf overexpression in the acquired resistance to loss of BRAFV600E (Figure 5D).
Figure 5. Met amplification and Hgf overexpression mediate ATC recurrence after suppression of BRAF.
(A) Met and Hgf expression in cell lines derived from primary ATC and recurrent tumors with and without Met amplification. Quantitative RT-PCR for all cell lines was run in triplicate. (B) Left: Dose response of the selective Met inhibitor PF-04217903 on growth of cell lines derived from primary and recurrent tumors with or without Met amplification. Cells were counted after 6 days of exposure to drug or vehicle. Right: Corresponding IC50 for PF-04217903 in the indicated cell lines. Each dose was run in triplicate. Bars represent the average of 2–3 experiments. (C) Western blots of primary and recurrent tumor cell lines 1 hour after addition of the indicated dose of PF-04217903. Similar results were obtained in a second experiment. (D) Western blots of recurrent tumor cell line with Met amplification stably infected with a scrambled or Hgf shRNA at the indicated time after removing Hgf.
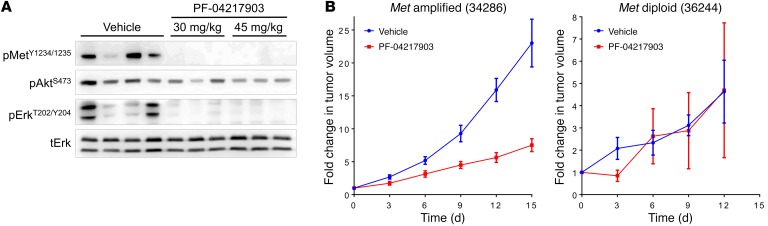
We next tested the activity of PF-04217903 in mouse allografts of Met-diploid and Met-amplified recurrent ATC cells. A single dose of 30 mg/kg or 45 mg/kg PF-04217903 by oral gavage was equally effective at inhibiting Met phosphorylation and downstream signaling in vivo in the allograft of the Met-amplified line (Figure 6A). Accordingly, PF-04217903 caused significant regression (P = 0.005 at day 15) of the Met-amplified ATC tumors, but was ineffective in the Met-diploid allografts (Figure 6B).
Figure 6. Allografts from Met-amplified cell lines are sensitive to Met inhibition.
(A) Western blots for the indicated protein from allografts of a Met-amplified cell line treated with vehicle or the indicated dose of PF-04217903 for 2 hours. (B) Allograft tumor volumes at the indicated times after starting daily treatment with 45 mg/kg PF-04217903. Tumor volume is the average of 5 animals. Similar results were obtained in a second experiment.
Discussion
Murine BRAF/p53 ATCs recapitulate the histological characteristics and aggressive biological behavior of the human disease, despite lacking many of the other genetic lesions associated with human ATCs (e.g., mutations in the telomerase [TERT] gene promoter, genes encoding effectors in the PI3K signaling pathway, SWI/SNF complex subunits, and histone methyltransferases, among others) (5, 6). This does not negate the potential importance of those defects in determining the full complexity of human ATC, but points to the sufficiency of BRAF activation in the context of Tp53 homozygous loss to phenocopy key features of the disease, including spindle cell histology with extensive macrophage infiltration, rapid tumor growth, invasiveness, and metastatic potential.
Levels of activated RAS are low in thyroid cancers with BRAFV600E mutation, preventing RAF dimerization (40). Consequently, BRAFV600E signals as a monomer, and thus bypasses key negative feedback effects that dampen the output of the pathway by disrupting RAF dimerization (41). Because of this, the output of the MAPK pathway, measured through an integrated score derived from quantifying mRNA levels of genes regulated by RAF-MEK-ERK activation, is higher in thyroid PTCs driven by BRAFV600E as compared with those with RAS mutations or receptor tyrosine kinase fusions (30, 42). Interestingly, mouse models of BRAF-driven ATCs have even greater MAPK transcriptional output than BrafV600E PTCs (31, 32). In the present study, the ATCs had higher expression of the BRAF transgene, which may explain this effect. However, that was not the case in a previously reported mouse model of ATC driven by a Cre-activated latent BrafV600E allele in the setting of Trp53 loss (TPOCreER; BrafCA/+/Trp53Δex2-10/Δex2-10 or TBP) (32), which like human ATCs also had increased MAPK transcriptional output. Loss of p53 has been shown to activate RAF-MEK-ERK signaling in a RAS-independent manner (43), which may also contribute to a higher MAPK output. In the absence of Trp53, BRAF-mutant cells can likely bypass the senescence or toxicity caused by the markedly increased MAPK flux (44, 45), which paradoxically could arise in part as a consequence of Trp53 loss. The high MAPK signaling flux may also cause resistance to inhibitors of the pathway. Consistent with this, clones of TRP53-mutant non–small cell lung cancer cells with mutant BRAF gene amplification arise in response to selective pressure induced by treatment with ERK inhibitors (46).
The objective of this study was to identify mechanisms of acquired resistance to inhibition of oncogenic BRAF in ATCs, as there is currently no data on genetic events associated with resistance to RAF or MEK inhibitors in this disease. The BRAF/p53 ATCs were unresponsive to the RAF kinase inhibitor PLX4720, consistent with results in the TBP mouse (32). Primary resistance could be due to several mechanisms, including relief of negative feedback on signaling effectors upstream of RAF, which in human ATC cell lines is mediated in part by activation of receptor tyrosine kinases, primarily of HER3/HER2 heterodimers (12). As adaptive resistance to small-molecule RAF inhibitors is particularly prominent in thyroid cancer cells, we opted to use a mouse model that enabled genetic inactivation of the oncogene after cancer development. The intent was to fully extinguish the driver of the disease, and in response we saw near-universal tumor regression, which in many mice was quite durable. The tumor remnants consisted of cells with a benign morphology in most cases, or with a papillary-like phenotype. Macrophage infiltration is a hallmark of ATC. Interestingly, this was markedly decreased shortly after dox withdrawal, suggesting that signals driven by the oncoprotein help trigger their recruitment and/or proliferation (47). This study was not designed to explore the impact of the interventions on the immune landscape of the tumors, but it is also noteworthy that recurrent tumors manifested a decrease in T cell–mediated immunity based on their gene expression signatures.
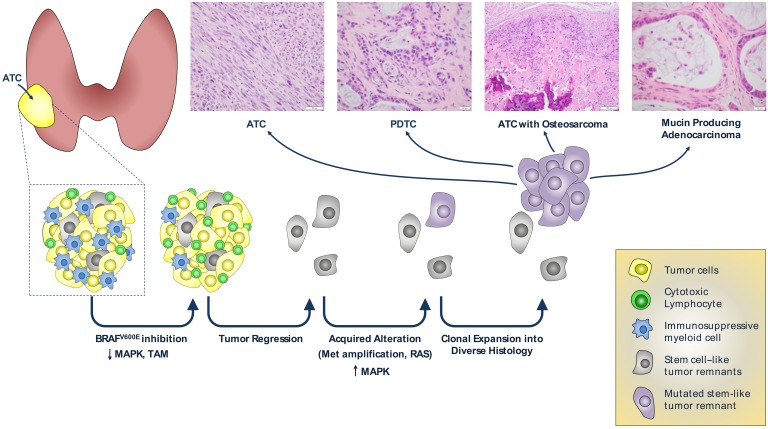
Two of the highest expression signatures associated with disease recurrence pointed to genes regulated by BMI-1 and MEL18, which are components of PRC1. The polycomb group of proteins assembles into 2 major multisubunit complexes, which have distinct histone tail–modifying activities. PRC1 complexes are histone H2A E3 ubiquitin ligases, whereas PRC2 trimethylates the lysine 27 residue of histone H3. These chromatin modifications primarily cause gene silencing of targets important for the maintenance of cell identity and differentiation. These data suggest that recurrences arising after BRAF suppression originate from cells endowed with stem cell–like or progenitor properties, which may explain their propensity to transdifferentiate to cells with diverse lineage characteristics (Figure 7).
Figure 7. Model for acquired resistance of BRAF-ATCs to inhibition of the oncogenic driver.
ATCs are heavily infiltrated by immune suppressive tumor-associated macrophages. ATCs regress upon inhibition of BRAFV600E expression or signaling, which is associated with a decrease in immunosuppressive myeloid cells. Recurrent disease is driven by mutations that reactivate MAPK signaling, primarily by Met amplification and concurrent cell-autonomous overexpression of the Met ligand Hgf. The recurrent cancer clones likely arise from cells with stem-like properties, and give rise to ATCs that exhibit focal transdifferentiation to cells with other lineage properties. The images depicting the different histologies of the recurrent tumors are those shown in Figure 1 or Supplemental Figure 5.
Most of the recurrences could not be accounted for by reactivated expression of the BRAF transgene, and yet retained a striking dependence on MAPK signaling for growth and viability. Met amplification was the most prevalent defect associated with disease recurrence, and generated a de novo vulnerability to MET kinase inhibitors. Activation of MET has been shown to cause adaptive resistance to RAF kinase inhibition in melanoma cells and in 2 thyroid cancer cell lines (38, 39, 48–50). There is also clinical evidence that MET overexpression can arise in melanomas (17) and colorectal cancers (51) resistant to RAF or RAF plus EGFR inhibition, respectively, although the dependency on MET for viability was only demonstrated in the latter.
MET activation as a cause of RAF kinase resistance has been shown to be secondary to stromal expression of the MET ligand HGF in cancer cell lines cocultured with fibroblasts, and in melanoma models in vivo (38, 52). The Met-amplified ATC recurrences reported here differ in one important respect, in that these tumor clones consistently evolved to express high levels of Hgf in a cell-autonomous fashion, pointing to a requirement for autocrine activation of the pathway to drive disease recurrence.
These data should be interpreted with the understanding that in this model oncogenic BRAF is expressed under the control of a heterologous promoter system, and that suppression of BRAFV600E expression is not feasible in patients. As opposed to tumors driven by mutant EGFR, KIT, RET, BCR-ABL, and other oncoproteins, resistance to pharmacological targeting of BRAFV600E is not associated with selection for gatekeeper or other secondary mutations of the oncogene (53). RAF inhibitors can select for expression of a truncated splice variant of BRAFV600E that lacks the RAS-binding domain and that dimerizes constitutively, rendering it resistant to the drug (20). Other than this particular event, which could not occur with the design of our model, we posited that other resistance mechanisms would likely be informative and relevant to the biology of the disease. We believe this was validated by our discovery of recurrences driven by HrasG12V and by recurrent Met amplifications, and in the latter, by their dependence on Hgf-Met signaling for viability. Hence, when profound blockade of MAPK signaling is attained pharmacologically, as has been shown with combined RAF and MEK inhibition (13), the acquired resistance mutations that arise may well be consistent with those observed in this experimental model.
Methods
Experimental animals.
We crossed TPO-Cre (24), LSL-rtTA_GFP (25), Trp53fl (26), and tetO-mycBRAFV600E (54) mice to create the quadruple transgenic line (BRAF/p53), which results in thyroid-specific loss of p53 and dox-inducible expression of a myc-tagged BRAFV600E (Figure 1A). Mouse lines were obtained from the following sources: TPO-Cre (from Shioko Kimura, NIH); Trp53fl, NCI Mouse Repository; LSL-rtTA-GFP, The Jackson Laboratory (stock number 005670). To induce thyroid-specific expression of BRAFV600E, mice were fed dox-impregnated chow (2,500 ppm, Envigo). Mice were in mixed genetic backgrounds. Genotypes were determined by PCR using primers described in Supplemental Table 7.
Histology and immunohistochemistry.
Tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Sections 5-μm thick were stained with H&E. Histological diagnosis was performed by a thyroid pathologist (RG) blinded to the genotype and the treatment status of the animal. IHC was performed by the MSK Molecular Cytology Core Facility with the following antibodies on paraffin-embedded sections: pERK (Cell Signaling, 4370), Myc-Tag (Abcam, ab9132), and Iba1 (Wako, 019-1974).
Thyroid imaging.
High-resolution MRI of the transgenic mouse thyroid tumors was done on a 200 MHz Bruker 4.7T Biospec scanner equipped with a 400 mT/m gradient coil (Bruker Biospin MRI GmbH). Mice were anesthetized with 1% isoflurane gas during scanning, and monitored using a small animal physiological monitoring system for respiration (SA Instruments Inc.). All the scanning was done by a custom-built 26 mm internal diameter (ID) birdcage resonator capable of uniform quadrature radiofrequency (RF) excitation and acquisition (Stark Contrast MRI Coils Research Inc.). T2-weighted scout images along 3 orthogonal orientations were first acquired for animal positioning. Mouse thyroid tumor images were then acquired using T2-weighted fast spin-echo RARE sequence (Rapid Acquisition with Relaxation Enhancement) with a coronal slice 0.4-mm thick, field of view (FOV) 21 mm × 20 mm, with a spatial resolution of 82 mm × 104 mm. We used the following acquisition parameters: time of repetition (TR), 2 seconds; time of echo (TE), 45 milliseconds; RARE factor 8, with an average of 40 scans and 32 minutes of acquisition time. H&E staining confirmed that the tumors occupied the entire thyroid gland, therefore the volume of the entire thyroid gland was determined before and after treatment using the formula: V = d × (sum (Ai) – 0.5 × (A1+An)), where d is the slice thickness and Ai (i = 1 through n) is the tumor area in each slice.
Real-time reverse transcription PCR.
Thyroid lobes were surgically removed and immediately placed in liquid N2. RNA was isolated using TRIzol (Invitrogen) or RNAeasy (Affymetrix) and 100–500 ng was reverse transcribed with SuperScript III (Invitrogen) in the presence of random hexamers to generate cDNA. Quantitative reverse transcription PCR (RT-PCR) was done using Power SYBR Green PCR Master Mix (Applied Biosystems) and primer pairs (Supplemental Table 7) for β-actin, myc-BRAFV600E, rtTA, total BRAF, Met, and Hgf. The cycle threshold values for β-actin and the target genes were determined with a 7500 real-time PCR instrument (Applied Biosystems) and used to calculate the normalized relative expression using the QGENE program (32).
Cell line generation and growth curves.
Primary ATCs and relapsed tumors were collected, washed with 1× PBS, minced, and placed in digestion medium (MEM with 112 U/ml collagenase type I, 1.2 U/ml dispase and pen/strep). Minced tissues were incubated at 37°C for 45–60 minutes and then washed twice with Coon’s F2 containing 0.5% bovine brain extract (BBE, Hammond Cell Tech) and pen/strep/glutamine (PSG, Gemini Bio Products). Cells were resuspended in Coon’s F12 containing 0.5% BBE and PSG and then plated in CellBIND plates (Corning). After culturing for 7–10 days, the culture medium was changed to growth medium (Coon’s F12 containing 5% FBS from Omega Scientific, and 0.5% BBE and PSG). Cells were passaged for at least 1 month prior to being used in experiments. Thyroid cancer cells from this model should express GFP as a result of thyroid-specific expression of rtTAIRESGFP (Figure 1A), which was confirmed by FACS in all cell lines prior to being used in experiments.
Viral particles with the mirE Hgf shRNAs were generated as previously described (55). In brief, a 97-mer oligonucleotide containing the Hgf shRNA was amplified and cloned into the SREP (pRRL) miR-E recipient vector. Viral particles were produced by transient transfection of 293FT cells using Lipofectamine 2000 (Invitrogen), psPAX2, pMD2.G, and pcSUPER-DGBR8 plasmids. Mouse cell lines were incubated with viral particles in the presence of 10 μg/ml polybrene (Santa Cruz) overnight. After recovering for 24 hours in complete media, cells were selected in growth medium with 1 μg/ml puromycin and 10 ng/ml Hgf. To investigate effects of Hgf knockdown on Met signaling, cells were plated in CellBIND plates and allowed to attach overnight. Cells were then fed with FBS-free growth medium with Hgf and incubated for 24 hours, then with FBS-free growth medium with or without Hgf, and collected at the indicated times for Western blotting.
For cell growth assays, 40,000 to 100,000 cells were plated in Coon’s F12 with 5% FBS, and 0.5% BBE and PSG in CellBIND 6-well plates in triplicate. The next day, medium was switched to Coon’s F12 with 0.1% BSA and PSG (2 μg/ml dox was included in primary ATC lines unless indicated) and incubated for 1–2 days before adding drugs (day 0). Cells were then incubated for 6 days with one medium change and counted using the Vi-Cell series cell viability analyzer (Beckman Coulter). IC50 values were calculated by nonlinear regression using Prism v6.05 (GraphPad Software).
Western blotting.
Mouse thyroid cancer cell lines were plated in CellBIND plates in Coon’s F12 with 5% FBS, 0.5% BBE and PSG, and incubated for 24–48 hours. Medium was then switched to Coon’s F12 with 0.1% BSA and PSG and incubated for 24–48 hours. Met and MAPK pathway inhibitors were added directly to culture 1 hour prior to collection. Cells were then washed with ice-cold PBS and harvested by scraping and centrifugation (1000 g for 4 minutes at 4°C). The cell pellets were resuspended in a lysis buffer consisting of 10 mM Tris-HCl (pH 7.5), 5 mM ETDA, 4 mM EGTA, 1% Triton X100, protease inhibitor cocktail (Sigma-Aldrich), and phosphatase inhibitor cocktail I and II (Sigma-Aldrich). Cells were placed on ice for 10 minutes and then lysed by passing through a p200 tip. Cell debris were removed by centrifugation (18,000 g for 15 minutes at 4°C) and supernatant was collected. Frozen tissues were placed in lysis buffer and then ground with a Polytron homogenizer. Lysates were centrifuged to remove debris (18,000 g for 15 minutes at 4°C ) and supernatant was collected. Protein concentration for all lysates was determined using the MicroBCA kit (Thermo Fisher Scientific). Protein lysate (10–50 μg) was subjected to SDS-PAGE and transferred to PVDF membranes (Millipore).The membranes were probed with the indicated antibody and the target protein was detected by incubating with species-specific HRP-conjugated IgGs (Santa Cruz) and then with enhanced chemiluminescence reagent (Amersham Biosciences). Images were captured using the KwikQuant Imager (Kindle Biosciences). The following antibodies were used: pMET (D26), pAKT(9271), pERK (9101), ERK (9102), pMEK (9121), and anti-mouse HRP (7074S) (all from Cell Signaling); β-actin (A2228) (Sigma-Aldrich); and anti-rabbit HRP (sc-2031) (Santa Cruz).
Mouse microarray hybridizations and analysis.
Expression profiling was performed on flash-frozen tissue from normal mouse thyroid or thyroid cancers from the indicated mouse models using the Affymetrix Mouse430A array, as recommend by the manufacturer (GEO accession number GSE118022). Arrays were normalized by the robust multiarray average method, and genes were considered to be differentially expressed if the FDR adjusted P value was less than 0.05 and fold change was greater than 2 or less than –2. Previously published expression array data from mouse (GSE55933) and human (GSE33630 and GSE76039) PTCs and ATCs driven by BrafV600E (5, 32, 33) were downloaded from the NCBI’s Gene Expression Omnibus Database (GEO). Normalization and differential expression were done using Partek Genomic Suites.
Transcriptional scores.
The MAPK transcriptional score is based on a core set of 52 transcripts identified through their regulation by a selective MEK inhibitor in a panel of BRAFV600E-mutant melanoma cell lines (29). These transcriptional targets were used by the TGCA to measure differences in the MAPK output of papillary thyroid cancers driven by distinct oncogenic driver mutations (30). We identified the mouse homologues of these transcripts, 45 of which were present on the Affymetrix Mouse430A array chip (Supplemental Table 1). The MAPK output score for each sample was determined by taking the inverse log of the average log-fold change for the 45 transcripts. The T cell score was based on the mouse homologue of core enrichment genes common to the GSEA gene sets IL15_UP.V1_UP and IL2_UP.V1_UP (56). The PRC1 score used the mouse homologue of the core enrichment genes common to the MEL18_DN.V1_UP and BMI1_DN.V1_UP gene sets (ref. 57 and Supplemental Table 1). Scores were determined by the inverse log of the average log-fold change for these transcripts. For all these analyses, if multiple probes for a single transcript were present, the probe with the largest standard deviation between samples was used to calculate the score.
Whole-exome sequencing.
Samples were prepared for whole-exome sequencing according to the manufacturer’s instructions. PCR amplification of the libraries was carried out for 6 cycles in the precapture step and for 8 cycles after capture. Samples were barcoded and run on a Hiseq 2500/4000 in a 100 bp/100 bp paired-end run, using the TruSeq SBS Kit v3 (Illumina). The average coverage was 300× for the tumor samples and 116× for the normal samples. The data processing pipeline for detecting variants in Illumina HiSeq data was as follows. First, the FASTQ files were processed to remove any adapter sequences at the end of the reads using cutadapt (v1.6) (https://github.com/marcelm/cutadapt/commit/25fa1e828b0f737dc43b7aec9f29582fbcb0245e). The files were then mapped using the BWA mapper (bwa mem v0.7.12; bio-bwa.sourceforge.net). After mapping, the SAM files were sorted and read group tags were added using the Picard tools (Broad Institute). After sorting in coordinate order, the compressed SAMs (BAMs) were processed with Picard MarkDuplicates. The marked BAM files were then processed using the Genome Analysis Toolkit (v3.2, Broad Institute) according to best practices for tumor-normal pairs. They were first realigned using the InDel realigner (Broad Institute) and then the base quality values were recalibrated with the BaseQRecalibrator (Broad Institute). Somatic variants were then called in the processed BAMs using muTect (v1.1.7, Broad Institute) for single nucleotide variants (SNVs) and haplotype caller plus custom postcall filtering to get somatic events. FACETS was used to determine tumor purity and allele-specific copy number analysis (58). The full pipeline is available online at https://github.com/soccin/BIC-variants_pipeline
Allografts.
Cells were trypsinized, resuspended in Coon’s F12 with 5% FBS, 0.5% BBE and PSG, and injected into the right flank of nude mice (5 × 106 cells per mouse). Treatment with PF-04217903 by oral gavage was commenced when allografts reached 200 to 400 mm3. Tumor volume was measured 3 times per week using calipers. Allografts were collected 2 hours after final dosing and aliquots were immediately placed into liquid N2 or 4% PFA.
Drug administration.
CH5126766 (CKI) was from Chugai Pharmaceutical Company, PLX4720 from Plexxikon, and crizotinib and PF-04217903 purchased from Selleck Chemicals. For in vivo experiments, CKI was dissolved in 2 hydroxypropyl-β-cyclo-dextrin and administered once daily by oral gavage at a dose of 1.5 mg/kg. A stock solution of PF-04217903 was dissolved in DMSO and then diluted in PBS to final concentration and administered to mice once daily by oral gavage at a dose of 45 mg/kg. PLX4720 was administered via PLX4720-impregnated chow (12). In these mice, dox was given through drinking water (2 mg/ml). For all in vitro experiments, inhibitors were dissolved in DMSO and then diluted to final concentration in culture medium.
Statistics.
P values were calculated using an unpaired, 2-tailed, Student’s t test (GraphPad Software Prism v6.05). A P value of less than 0.05 was considered significant. All data are mean ± SEM unless otherwise indicated.
Study approval.
All animal studies were reviewed and approved by the IACUC of Memorial Sloan Kettering Cancer Center, New York, New York, USA.
Author contributions
JAK and JAF designed the research studies. JAK, KAL, KYC, and FV conducted the experiments. JAK, JAF, KAL, RG, and NDS analyzed the data. JAK, JAF, and KAL wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01-CA50706, R01-CA072597, P50-CA172012, P30-CA008748; the Lefkovsky Family Foundation; the Linn Family Fund; the Society of Memorial Sloan Kettering; and the Byrne Fund. We thank the Integrated Genomics, Bioinformatics, Molecular Cytology, Comparative Pathology, Small Animal Imaging, and Mouse Genetics Core Facilities at MSKCC for support of this project.
Version 1. 07/10/2018
In-Press Preview
Version 2. 08/20/2018
Electronic publication
Version 3. 08/31/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(9):4086–4097.https://doi.org/10.1172/JCI120966.
Contributor Information
Jeffrey A. Knauf, Email: Knaufj@mskcc.org.
Kathleen A. Luckett, Email: luckettk@mskcc.org.
Kuen-Yuan Chen, Email: dtsurg51@gmail.com.
Francesca Voza, Email: franvoza@gmail.com.
Nicholas D. Socci, Email: soccin@mskcc.org.
Ronald Ghossein, Email: ghosseir@mskcc.org.
James A. Fagin, Email: faginj@mskcc.org.
References
- 1.Foote RL, et al. Enhanced survival in locoregionally confined anaplastic thyroid carcinoma: a single-institution experience using aggressive multimodal therapy. Thyroid. 2011;21(1):25–30. doi: 10.1089/thy.2010.0220. [DOI] [PubMed] [Google Scholar]
- 2.Ricarte-Filho JC, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009;69(11):4885–4893. doi: 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikiforova MN, et al. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J Clin Endocrinol Metab. 2003;88(11):5399–5404. doi: 10.1210/jc.2003-030838. [DOI] [PubMed] [Google Scholar]
- 4.Namba H, et al. Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J Clin Endocrinol Metab. 2003;88(9):4393–4397. doi: 10.1210/jc.2003-030305. [DOI] [PubMed] [Google Scholar]
- 5.Landa I, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126(3):1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunstman JW, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015;24(8):2318–2329. doi: 10.1093/hmg/ddu749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falchook GS, et al. BRAF inhibitor dabrafenib in patients with metastatic BRAF-mutant thyroid cancer. Thyroid. 2015;25(1):71–77. doi: 10.1089/thy.2014.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim AM, et al. BRAF inhibition in BRAFV600E-positive anaplastic thyroid carcinoma. J Natl Compr Canc Netw. 2016;14(3):249–254. doi: 10.6004/jnccn.2016.0030. [DOI] [PubMed] [Google Scholar]
- 9.Marten KA, Gudena VK. Use of vemurafenib in anaplastic thyroid carcinoma: a case report. Cancer Biol Ther. 2015;16(10):1430–1433. doi: 10.1080/15384047.2015.1071734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosove MH, Peddi PF, Glaspy JA. BRAF V600E inhibition in anaplastic thyroid cancer. N Engl J Med. 2013;368(7):684–685. doi: 10.1056/NEJMc1215697. [DOI] [PubMed] [Google Scholar]
- 11.Hyman DM, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montero-Conde C, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3(5):520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subbiah V, et al. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J Clin Oncol. 2018;36(1):7–13. doi: 10.1200/JCO.2017.73.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4(1):80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugo W, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162(6):1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DB, et al. Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51(18):2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizos H, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20(7):1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 20.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29(22):3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18(6):683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusakabe T, Kawaguchi A, Kawaguchi R, Feigenbaum L, Kimura S. Thyrocyte-specific expression of Cre recombinase in transgenic mice. Genesis. 2004;39(3):212–216. doi: 10.1002/gene.20043. [DOI] [PubMed] [Google Scholar]
- 25.Belteki G, et al. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33(5):e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 27.Ryder M, Ghossein RA, Ricarte-Filho JC, Knauf JA, Fagin JA. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr Relat Cancer. 2008;15(4):1069–1074. doi: 10.1677/ERC-08-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caillou B, et al. Tumor-associated macrophages (TAMs) form an interconnected cellular supportive network in anaplastic thyroid carcinoma. PLoS One. 2011;6(7):e22567. doi: 10.1371/journal.pone.0022567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106(11):4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franco AT, et al. Thyrotrophin receptor signaling dependence of Braf-induced thyroid tumor initiation in mice. Proc Natl Acad Sci U S A. 2011;108(4):1615–1620. doi: 10.1073/pnas.1015557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFadden DG, et al. p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proc Natl Acad Sci U S A. 2014;111(16):E1600–E1609. doi: 10.1073/pnas.1404357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomás G, et al. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene. 2012;31(41):4490–4498. doi: 10.1038/onc.2011.626. [DOI] [PubMed] [Google Scholar]
- 34.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105(13):5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi PS, et al. Lymphomas that recur after MYC suppression continue to exhibit oncogene addiction. Proc Natl Acad Sci U S A. 2011;108(42):17432–17437. doi: 10.1073/pnas.1107303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapoor A, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishii N, et al. Enhanced inhibition of ERK signaling by a novel allosteric MEK inhibitor, CH5126766, that suppresses feedback reactivation of RAF activity. Cancer Res. 2013;73(13):4050–4060. doi: 10.1158/0008-5472.CAN-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caenepeel S, et al. MAPK pathway inhibition induces MET and GAB1 levels, priming BRAF mutant melanoma for rescue by hepatocyte growth factor. Oncotarget. 2017;8(11):17795–17809. doi: 10.18632/oncotarget.14855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dougherty MK, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17(2):215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 42.Fagin JA, Wells SA., Jr Biologic and clinical perspectives on thyroid cancer. N Engl J Med. 2016;375(23):2307. doi: 10.1056/NEJMc1609116. [DOI] [PubMed] [Google Scholar]
- 43.Drosten M, et al. Loss of p53 induces cell proliferation via Ras-independent activation of the Raf/Mek/Erk signaling pathway. Proc Natl Acad Sci U S A. 2014;111(42):15155–15160. doi: 10.1073/pnas.1417549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di MR, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 45.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 46.Xue Y, et al. An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat Med. 2017;23(8):929–937. doi: 10.1038/nm.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryder M, et al. Genetic and pharmacological targeting of CSF-1/CSF-1R inhibits tumor-associated macrophages and impairs BRAF-induced thyroid cancer progression. PLoS One. 2013;8(1):e54302. doi: 10.1371/journal.pone.0054302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohrbeck L, et al. Hepatocyte growth factor renders BRAF mutant human melanoma cell lines resistant to PLX4032 by downregulating the pro-apoptotic BH3-only proteins PUMA and BIM. Cell Death Differ. 2016;23(12):2054–2062. doi: 10.1038/cdd.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vergani E, et al. Identification of MET and SRC activation in melanoma cell lines showing primary resistance to PLX4032. Neoplasia. 2011;13(12):1132–1142. doi: 10.1593/neo.111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byeon HK, et al. c-Met-mediated reactivation of PI3K/AKT signaling contributes to insensitivity of BRAF(V600E) mutant thyroid cancer to BRAF inhibition. Mol Carcinog. 2016;55(11):1678–1687. doi: 10.1002/mc.22418. [DOI] [PubMed] [Google Scholar]
- 51.Pietrantonio F, et al. MET-driven resistance to dual EGFR and BRAF blockade may be overcome by switching from EGFR to MET inhibition in BRAF-mutated colorectal cancer. Cancer Discov. 2016;6(9):963–971. doi: 10.1158/2159-8290.CD-16-0297. [DOI] [PubMed] [Google Scholar]
- 52.Fedorenko, Wargo JA, Flaherty KT, Messina JL, Smalley KSM. BRAF inhibition generates a host-tumor niche that mediates therapeutic escape. J Invest Dermatol. 2015;135(12):3115–3124. doi: 10.1038/jid.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lovly CM, Shaw AT. Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin Cancer Res. 2014;20(9):2249–2256. doi: 10.1158/1078-0432.CCR-13-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakravarty D, et al. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. J Clin Invest. 2011;121(12):4700–4711. doi: 10.1172/JCI46382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5(6):1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 56.Marzec M, et al. Differential effects of interleukin-2 and interleukin-15 versus interleukin-21 on CD4+ cutaneous T-cell lymphoma cells. Cancer Res. 2008;68(4):1083–1091. doi: 10.1158/0008-5472.CAN-07-2403. [DOI] [PubMed] [Google Scholar]
- 57.Wiederschain D, et al. Contribution of polycomb homologues Bmi-1 and Mel-18 to medulloblastoma pathogenesis. Mol Cell Biol. 2007;27(13):4968–4979. doi: 10.1128/MCB.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.