Abstract
Epithelial cell dysfunction is postulated as an important component in the pathogenesis of idiopathic pulmonary fibrosis (IPF). Mutations in the surfactant protein C (SP-C) gene (SFTPC), an alveolar type II (AT2) cell–restricted protein, have been found in sporadic and familial IPF. To causally link these events, we developed a knockin mouse model capable of regulated expression of an IPF-associated isoleucine-to-threonine substitution at codon 73 (I73T) in Sftpc (SP-CI73T). Tamoxifen-treated SP-CI73T cohorts developed rapid increases in SftpcI73T mRNA and misprocessed proSP-CI73T protein accompanied by increased early mortality (days 7–14). This acute phase was marked by diffuse parenchymal lung injury, tissue infiltration by monocytes, polycellular alveolitis, and elevations in bronchoalveolar lavage and AT2 mRNA content of select inflammatory cytokines. Resolution of alveolitis (2–4 weeks), commensurate with a rise in TGF-β1, was followed by aberrant remodeling marked by collagen deposition, AT2 cell hyperplasia, α–smooth muscle actin–positive (α-SMA–positive) cells, and restrictive lung physiology. The translational relevance of the model was supported by detection of multiple IPF biomarkers previously reported in human cohorts. These data provide proof of principle that mutant SP-C expression in vivo causes spontaneous lung fibrosis, strengthening the role of AT2 cell dysfunction as a key upstream driver of IPF pathogenesis.
Keywords: Pulmonology
Keywords: Fibrosis, Pulmonary surfactants
Introduction
Idiopathic pulmonary fibrosis (IPF) represents the most common subtype of a larger family of diffuse parenchymal lung diseases (DPLDs) of unknown etiology in adults (1). The incidence of IPF is increasing, is higher in men, and rises significantly with age (2, 3). Family-based studies have led to the identification of rare variants in genes related to surfactant function and telomere biology (4, 5). Molecular signatures of cellular stress, apoptosis, and foreshortened telomeres associated with these genetic variants have also been confirmed in sporadic cases of IPF (4, 6). Many of these genetic variants are also described in pediatric populations as part of the larger spectrum of childhood interstitial lung disease (chILD) (7).
The pathognomonic histology of IPF is the usual interstitial pneumonitis (UIP) lesion composed of temporally and spatially heterogeneous areas of fibroblast/myofibroblast accumulation coupled with extracellular matrix deposition, disruption of alveolar architecture, and subpleural honeycombing (8). The typical clinical experience of patients with IPF is marked by progression of symptoms from cough and dyspnea to end-stage respiratory insufficiency, resulting in lung transplantation or in death within 3–5 years of diagnosis (2, 9). In addition, a subset of IPF patients (incidence ~5%–12% per year) experience an acute clinical deterioration, termed acute IPF exacerbation (AE-IPF), characterized by new radiological infiltrates and worsening respiratory physiology, in which a pattern of diffuse alveolar damage is superimposed on the UIP pathology (2, 10). These events precede precipitous declines in lung function and are associated with extremely poor outcomes (median survival ~3–4 months after event) (2, 11).
The pathogenesis of IPF remains poorly understood and has been a major obstacle in developing effective therapies capable of stabilizing or improving lung function for affected individuals (12). Clinical trials based on antiinflammatory strategies (corticosteroids, immunosuppressive/cytotoxic agents, and cytokine modulation) have revealed low efficacy or even harmful toxicities (13). Two FDA-approved antifibrotic agents (i.e., pirfenidone and nintedanib) have been shown to only slow the rate of pulmonary function deterioration (14, 15). Contributing to this knowledge gap has been the lack of relevant preclinical models that fully recapitulate either the clinical features of IPF or develop UIP histology (12, 16).
Over the past 2 decades, due in part to the aforementioned challenges, a paradigm shift has taken place away from viewing inflammatory cells or the mesenchyme as primary drivers of IPF toward the recognition of a pivotal role for alveolar epithelial cells (AECs) in IPF pathogenesis (17). Specifically, the postulated contribution of alveolar type II (AT2) cell dysfunction to the development of a fibrotic lung phenotype has gained momentum: microfoci of repeated cycles of AEC/AT2 injury in conjunction with release of proinflammatory and profibrotic mediators produce a dysfunctional repair process reminiscent of other fibrotic diseases observed in skin, kidney, and liver (12, 17). Perhaps, the most compelling evidence supporting a role for AT2 cells in these processes is derived from studies linking adult IPF patients or children with chILD with the heterozygous expression of more than 60 mutations in the AT2 cell–restricted gene SFTPC, whose secreted protein product, surfactant protein C (SP-C), modulates the biophysical activity of pulmonary surfactant (reviewed in ref. 5).
Among SFTPC variants described to date, the missense substitution g.1286T>C, resulting in a change of isoleucine to threonine at position 73 in the SP-C proprotein (SP-CI73T), is the most common interstitial lung disease–associated (ILD-associated) SFTPC mutation (18). First reported by our group in 2004 (19), the heterozygous expression of the mutant SFTPCI73T allele (either inherited or de novo) has reappeared in several other cohorts of IPF and chILD patients (20–22). Functionally, we have shown that expression of SFTPCI73T in vitro results in a toxic gain of function initiated by the unique and markedly altered intracellular trafficking of the SP-CI73T proprotein. The deposition of proSP-CI73T in the plasma membrane and subsequent accumulation of misprocessed isoforms within early and late endosomes resulted in a dysfunctional cellular phenotype characterized by a late block in macroautophagy, impaired mitophagy, and defective cellular proteostasis (23, 24). Importantly, the highly disrupted organellar homeostasis observed by transmission electron microscopy (TEM) in SP-CI73T cell lines recapitulated ultrastructural changes in AT2 cells noted on a lung biopsy from an SFTPCI73T ILD patient (24).
The striking cellular phenotype of AT2 cells and cell lines expressing mutant proSP-CI73T and the paucity of relevant preclinical IPF models led us to leverage ILD genetics to develop a knockin murine model capable of inducible expression of mutant SftpcI73T in a dose- and time-dependent fashion. The mechanistic characterization of the resultant SP-CI73T mouse lines that follows herein provides a direct link between previously described cellular and clinical phenotypes that result from SP-CI73T expression while offering proof of principle that expression of mutant SP-CI73T by AT2 cells is sufficient to drive a cascade of events that in humans has been pathognomonic of an IPF/UIP endophenotype. From our results, a tractable preclinical IPF model has emerged that can provide insights into disease pathogenesis, permit direct assessment of the consequences of AE-IPF for parenchymal remodeling, facilitate target identification/validation, serve as a platform for development of novel biomarkers, and be exploited for the assessment of new treatment strategies.
Results
We employed a recombineering strategy with embryonic stem cell transduction/screening to selectively knock in an HA-tagged mouse SP-CI73T sequence into the endogenous mouse Sftpc locus. The targeted allele in the resultant parental line (SP-CI73T-Neo), schematically shown in Figure 1A, retained a PGK-neomycin (PGK-Neo) cassette in intron 1 flanked by short flippase (Flp) recognition target (FRT) sites. F1 crosses of SP-CI73T-Neo chimeras with C57BL/6 mice confirmed germline transmission. The litters of F2 and further generations were full-term, of normal size and sex distribution, represented all genotypes, and exhibited normal postnatal alveolarization (data not shown).
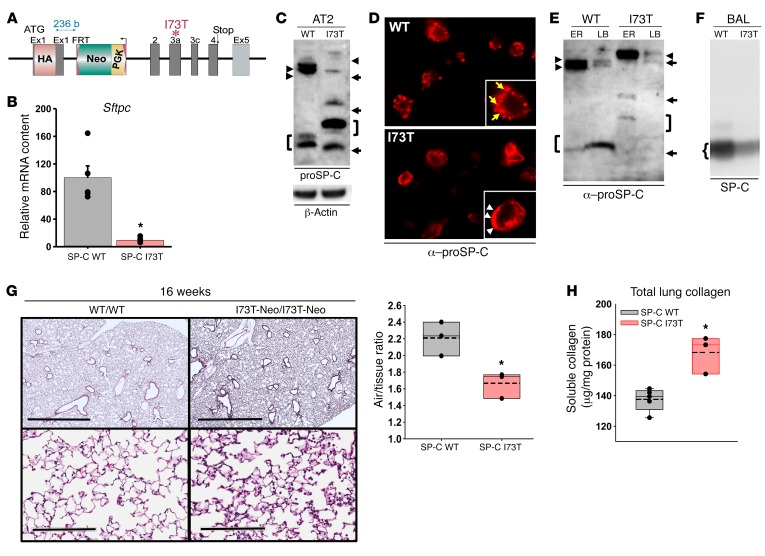
Figure 1. Cellular and histopathological phenotype of the SP-CI73T-Neo founder line.
(A) Schematic of the HA-tagged SftpcI73T knockin allele showing the PGK-Neo cassette in intron 1. (B) qRT-PCR analysis for Sftpc mRNA expression in 12- to 28-week-old WT and homozygous SP-CI73T-Neo/I73T-Neo (SP-C I73T) mice. (C) Western blot of AT2 lysates for proSP-C (20 μg protein/lane). SP-CI73T-Neo/I73T-Neo mice accumulate an HA-tagged primary translation product (arrowheads) and misprocessed isoforms (arrows, right bracket). In WT/WT mice, both the primary translation product (arrowheads) and major processing intermediate (left bracket) were detected. (D) Immunohistochemistry for proSP-C of lung sections from 8-week-old WT and SP-CI73T-Neo/I73T-Neo mice revealed proSP-CI73T expression on AT2 cell plasma membranes (arrowheads); proSP-CWT is expressed in cytosolic vesicles of AT2 cells (arrows). (E) proSP-C Western blot of subcellular fractions enriched in ER or lamellar bodies (LB) from 8-week-old SP-CWT/WT and SP-CI73T-Neo/I73T-Neo mice. ER from each line contained the corresponding proSP-C primary translation product (arrowheads). The major proSP-CWT intermediate (Mr, 6,000) was enriched in LB (left bracket). All aberrantly processed proSP-CI73T isoforms were excluded from SP-CI73T-Neo LB (arrows, right bracket). (F) Western blot of surfactant showing decreased mature SP-C in SP-CI73T-Neo/I73T-Neo mice. (G) H&E-stained sections from 16-week-old mice (scale bars: 2 mm (upper panels); 200 μm (lower panels). Quantitative morphometry using ImageJ expressed as airspace/tissue ratio. (H) Total soluble collagen content in the left lobe from 32-week-old mice. Data represent mean ± SEM with dot plot overlays. *P < 0.05 versus SP-CWT by 2-tailed t test.
This founder line was found to be hypomorphic, with total SftpcI73T mRNA in homozygous SftpcI73T-Neo/I73T-Neo (SP-CI73-Neo/I73-Neo) animals limited to approximately 15% of that of littermates with 2 SftpcWT alleles (Figure 1B).
Expression of SftpcI73T in AT2 cells in vivo results in aberrant posttranslational processing and intracellular accumulation of mutant SP-CI73T proprotein.
Despite low levels of Sftpc mRNA, AT2 cells from SP-CI73T-Neo/I73T-Neo mice accumulated known misprocessed proSP-CI73T isoforms (19, 24–26), suggesting markedly aberrant targeting and/or posttranslational processing (Figure 1C). proSP-CWT immunostaining was consistent with its known localization in AT2 lamellar bodies (27, 28), while HA–proSP-CI73T was prominently mislocalized in the AT2 plasma membrane (Figure 1D). Importantly, the NH2 terminal HA tag does not influence proSP-CI73T mistrafficking. In an experiment utilizing a previously published in vitro model (29), neither the HA nor EGFP tag affected normal anterograde trafficking of proSP-CWT (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI99287DS1) or influenced the mistargeting of proSP-CI73T to the plasma membrane (Supplemental Figure 1, D–F).
Aberrant trafficking of HA–proSP-CI73T in AT2 cells in vivo was corroborated by Western blotting of lamellar body fractions from SP-CI73T-Neo/I73T-Neo mice, wherein levels of mutant proSP-CI73T in this organellar compartment were below the limit of detection (Figure 1E). Interestingly, despite its exclusion from lamellar bodies, an immunoreactive monomeric mature SP-C band (Mr, 3.5–3.7 kDa) was detected in the alveolar compartment (Figure 1F). Quantitatively, surfactant from SP-CI73T-Neo/I73T-Neo lungs contained only 34.5% of mature SP-C found in WT mice despite having mildly elevated levels of SP-B (Supplemental Figure 2). Consistent with prior in vitro data showing intracellular accumulation and increased half-life of proSP-CI73T isoforms (26), the current results support the concept that the aberrant trafficking and posttranslational processing of proSP-CI73T by AT2 cells in vivo result in intracellular accumulation of mutant proSP-C isoforms out of proportion to the lower AT2 SftpcI73T mRNA levels and in inefficient synthesis of mature SP-C.
The hypomorphic SP-CI73T-Neo/I73T-Neo founder line develops diffuse parenchymal lung remodeling.
Constitutive expression of SP-CI73T protein in AT2 cells from this hypomorphic line resulted in development of diffuse parenchymal lung remodeling marked by thickening of alveolar septae beginning at approximately 16 weeks (Figure 1G) that contained trichrome-positive material (Supplemental Figure 3A), with the histological changes persisting up to 40 weeks of age (Supplemental Figure 3B). Evidence of fibrotic remodeling was supported by an elevation in total lung collagen (Figure 1H). A substantial increase in septal cellularity notable for the expansion of proSP-C–positive cells consistent with AT2 cell hyperplasia was found at both 16 and 32 weeks (Supplemental Figure 3, C and D) and accompanied by age-dependent increases in bronchoalveolar lavage fluid (BALF) cellularity (Supplemental Figure 3E).
In parallel with abnormalities in SP-CI73T biosynthesis and aberrant lung remodeling, AT2 cells of SP-CI73T-Neo mice developed ultrastructural abnormalities consistent with an acquired defect in macroautophagy. TEM of lung sections from SP-CI73T-Neo mice revealed hypertrophic AT2 cells containing numerous intracytoplasmic inclusions composed of proteinaceous debris and mitochondria within double membrane vesicles (Supplemental Figure 4A). In autophagic flux assays, AT2 cells isolated from WT mice and treated with the mTOR inhibitor/autophagy activator torin demonstrated increased LC3 turnover (Supplemental Figure 4B). In contrast, SP-CI73T-Neo AT2 cells showed increases in LC3-I/LC3-II and p62 at baseline and failed to respond to torin treatment, phenocopying the late block in macroautophagy we previously observed in vitro in cell lines stably expressing SP-CI73T (24).
Constitutive deletion of intronic PGK-Neo enhances SP-CI73T expression and disrupts lung development.
Due to interference with exonic splicing or impaired transcription efficiency, retained intronic selection cassettes have been used to create murine models with hypomorphic alleles to circumvent toxic gain of function through dampening of gene expression (30, 31). To evaluate the impact of higher levels of SP-CI73T expression on lung homeostasis, we constitutively removed the FRT-flanked PGK-Neo cassette from SftpcI73T-Neo alleles by backcrossing the founder line to a FLP-e recombinase “deleter” line (Figure 2). Consistent with an acquired toxic gain of function, the average litter size of the resultant CFlp-SP-CI73T line was significantly decreased, and no live progeny harboring PGK-Neo–excised SftpcI73T alleles were found (Supplemental Table 1). We also established Mendelian inheritance of SftpcI73T alleles in a large cohort of CFlp-SP-CI73T embryos harvested at E18.5, demonstrating successful PGK-Neo cassette excision (heterozygous or homozygous).
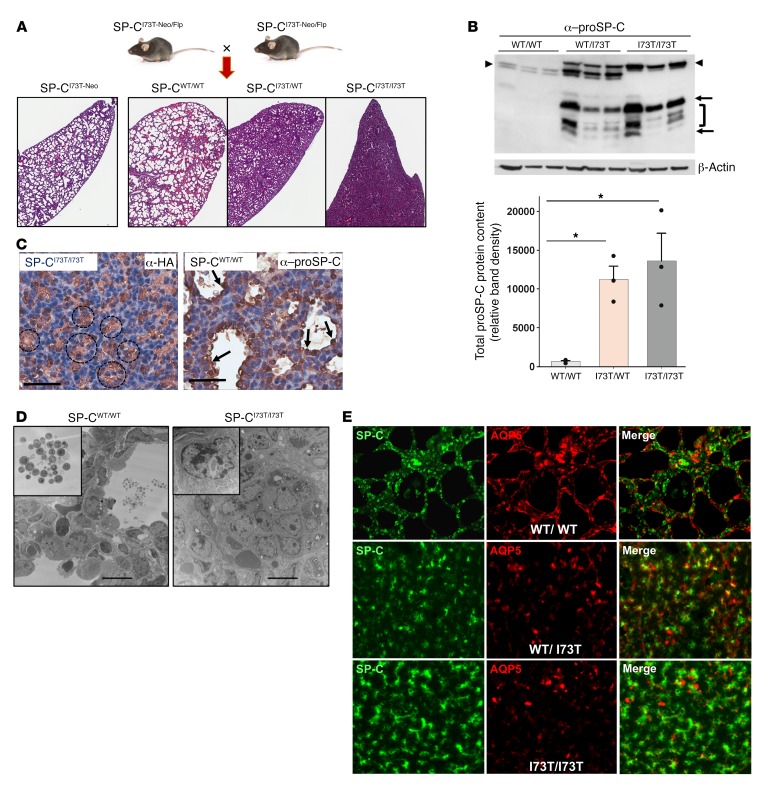
Figure 2. Constitutive deletion of PGK-Neo from SP-CI73T-Neo mice disrupts lung development.
(A) Backcross of SP-CI73T-Neo founder mice to FLP-e recombinase mice generated multiple genotypes of CFlp-SP-CI73T mice, including unexcised (SP-CI73T-Neo), excised (SP-CI73T), or WT alleles. Representative ×1.5 photomicrographs of H&E-stained lungs harvested from E18.5 CFlp-SP-CI73T mice with 1 (SP-CI73T/WT) or 2 (SP-CI73T/I73T) excised SftpcI73T allele(s) reveal a graded arrest of saccular lung development. (B) Immunoblotting of E18.5 WT or Neo-excised CFlp-SP-CI73T lung homogenates for proSP-C with β-actin as a loading control. Densitometric quantitation of aberrant proSP-CI73T (arrows, bracket) and WT proSP-C (arrowheads). *P < 0.05 versus WT/WT by 1-way ANOVA with Tukey’s post hoc test. (C) Staining of E18.5 CFlp-SP-CI73T/I73T embryos for HA revealed tufts of HA+ AT2 cells within obliterated saccules. CFlp-SP-CWT/WT controls showed normal developing saccules lined by proSP-C+ cells. Scale bars: 70 μm. (D) TEM of E18.5 CFlp-SP-CI73T/I73T and CFlp-SP-CWT/WT lungs. SP-CWT/WT saccules contained intraluminal surfactant (inset). CFlp-SP-CI73T/I73T lungs contained disrupted saccules filled with poorly differentiated epithelial cells (inset). Scale bars: 10 μm. (E) Representative ×60 immunofluorescence micrographs of E18.5 lungs from CFlp-SP-CI73T/I73T, CFlp-SP-CI73T/WT, and CFlp-SP-CWT mice stained for proSP-C (green) and AQP5 (red), showing loss of AQP5 and increase in proSP-C staining in CFlp-SP-CI73T animals.
CFlp-SP-CI73T embryos have proven to be informative, as E18.5 lungs carrying SftpcI73T alleles displayed arrested lung morphogenesis in late sacculation in both heterozygous (CFlp-SP-CI73T/WT) and homozygous (CFlp-SP-CI73T/I73T) embryos (Figure 2A). The graded disruption of embryonic lung architecture correlated with an allele-dependent increase in proSP-CI73T expression (Figure 2B). By immunohistochemistry, clumps of HA-tagged proSP-CI73T–expressing epithelia were found obliterating primordial saccules (Figure 2C). TEM of E18.5 CFLP-SP-CI73T lungs confirmed the presence of disrupted saccules containing poorly differentiated epithelium harboring marked ultrastructural changes (Figure 2D). The E18.5 CFlp-SP-CI73T lungs also showed qualitative reductions in aquaporin 5 (AQP5) staining inversely correlated with genotype, suggesting disruption of AT2/AT1 homeostasis (Figure 2E).
Increasing expression of SftpcI73T in adult mice triggers rapid increases in morbidity and mortality from diffuse parenchymal lung injury.
To assess the impact of increasing SP-CI73T expression into a more physiologic range on the postnatal lung, we generated mice with inducible expression of SP-CI73T (IER-SP-CI73TFlp) by crossing SP-CI73T-Neo mice to a Rosa26 ER2-Flp-O line to effect tamoxifen-mediated Flp-O recombinase removal of the inhibitory PGK-Neo cassette from SftpcI73T-Neo alleles (Figure 3A). In the absence of tamoxifen, adult homozygous IER-SP-CI73T/I73TFlp animals retained a phenotype identical to the hypomorphic SP-CI73T-Neo founder line, with low levels of SftpcI73T mRNA, normal litter size, and preserved lung histology prior to 16 weeks of age (data not shown).
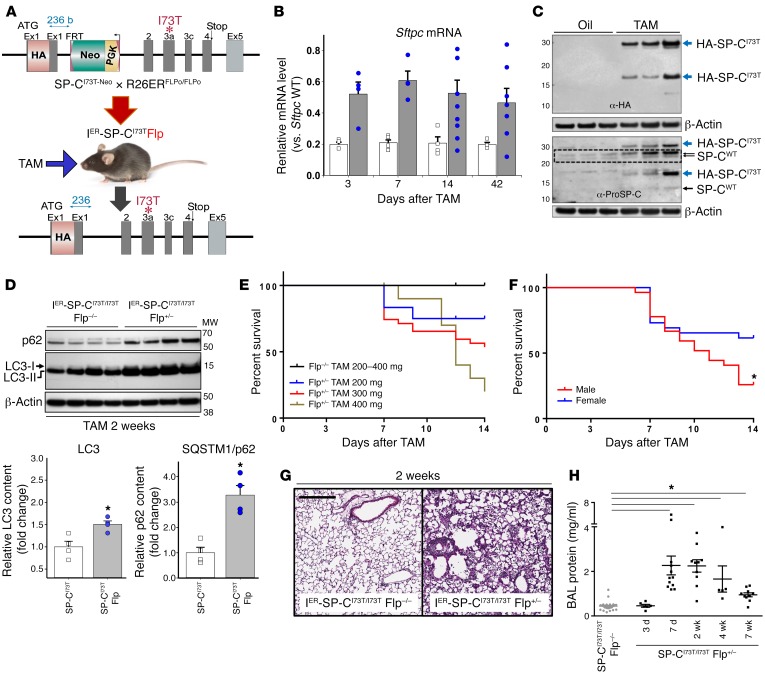
Figure 3. Tamoxifen induction of SftpcI73T expression in IER-SP-CI73TFlp mice causes mortality and lung injury.
(A) IER-SP-CI73TFlp mice were generated by crossing a Rosa26-ER2-Flp-O (deleter) line (R26ERFLPo/FLPo) with SP-CI73T-Neo mice. (B) qRT-PCR analysis of Sftpc mRNA in IER-SP-CI73T/I73TFlp+/– (gray bars) or IER-SP-CI73T/I73TFlp–/– (white bars) mice at 3, 7, 14, and 42 days after tamoxifen (TAM). Normalized data are expressed as percent Sftpc WT (C57BL/6) mRNA standard. *P < 0.05 versus time point control by 2-tailed t test. (C) Top: Anti-HA Western blot of lung homogenates from double-heterozygous IER-SP-CI73T/WTFlp+/– mice 2 weeks after tamoxifen (200 mg/kg) or vehicle (oil) treatment showing upregulation of HA-proSP-CI73T isoforms (blue arrows). Bottom: Reprobing for α–proSP-C showing 22/21-kDa proSP-CWT (dotted rectangle) and 6-kDa proSP-C intermediate (black arrows), plus aberrant proSP-CI73T isoforms (blue arrows). (D) Western blot of AT2 lysates from IER-SP-CI73T/I73TFlp–/– or IER-SP-CI73T/I73TFlp+/– mice 2 weeks after iTAM for LC3 and SQSTM1/p62. Dot plots of densitometry for LC3 and p62 normalized to β-actin. *P < 0.05 versus control by 2-tailed t test. (E) Kaplan-Meier analysis for control IER-SP-CI73T/I73TFlp–/– mice treated with 300 mg/kg iTAM and IER-SP-CI73T/I73TFlp+/– groups treated with 200, 300, or 400 mg/kg iTAM (n = 11–33 / group). Mean survival using death or body weight <75% on 2 consecutive days as end points is shown. P < 0.05 for TAM IER-SP-CI73T/I73TFlp+/– at each iTAM dosage group (200, 300, or 400 mg/kg) versus TAM IER-SP-CI73T/I73TFlp–/– group using log-rank testing. (F) Survival of 8- to 12-week-old male (n =26) and female (n = 29) IER-SP-CI73T/I73TFlp+/– mice treated with 250–300 mg/kg iTAM. *P < 0.05 males versus females. (G) Representative H&E histology from surviving control and IER-SP-CI73T/I73TFlp+/– mice 14 days after 300 mg/kg iTAM. Scale bar: 300 μm. (H) BALF protein content in non-inducible IER-SP-CI73T/I73TFlp–/– or inducible IER-SP-CI73T/I73TFlp+/– mice after iTAM. Data from all time points for the non-inducible control group (gray dots) were combined. *P < 0.05 vs. IER-SP-CI73T/I73TFlp–/– by 1-way ANOVA.
Intraperitoneal tamoxifen (iTAM) administration to double-homozygous IER-SP-CI73T/I73TFlp+/+ mice resulted in rapid increases in SftpcI73T mRNA within 3 days (Figure 3B). Using quantitative real-time PCR (qRT-PCR) to estimate neomycin cassette numbers (32), the efficiency of iTAM-induced Flp-O mediated recombination in purified IER-SP-CI73T/I73TFlp+/+ AT2 cells isolated 1 week after induction was 89.8% ± 2.0% (mean ± SD, n = 4) referenced against tamoxifen-treated IER-SP-CI73T/I73TFlp–/– controls (Supplemental Figure 5). The induction of SftpcI73T mRNA was accompanied by substantial increases in mutant proSP-CI73T protein (Figure 3C) alterations in macroautophagy. Lysates from IER-SP-CI73T/I73TFlp AT2 cells (Flp+/– or Flp+/+) showed significant increases in both LC3-II and p62, consistent with a late block in macroautophagy (Figure 3D) and detectable as early as 1 week after induction (Supplemental Figure 6).
The molecular and cellular changes observed upon induction of SP-CI73T expression were temporally associated with systemic morbidity and mortality. Compared with nonexcised SftpcI73T-Neo allelic controls (i.e., IER-SP-CI73T/I73TFlp–/–), IER-SP-CI73T/I73TFlp+/– mice exhibited increased weight loss and hypoxemia after iTAM (Supplemental Figure 7, A and B). Survival was significantly decreased and was Flp-O recombinase allele– and iTAM dose–dependent, with onset of mortality as early as 5 days in IER-SP-CI73T/I73TFlp+/+ animals (Figure 3E and Supplemental Figure 7C). Notably, mean survival was shorter in IER-SP-CI73T/I73TFlp+/– male animals (10.6 days versus 11.9 for females; P = 0.005) (Figure 3F).
Histologically, IER-SP-CI73TFlp mice developed acute, diffuse lung injury after iTAM (Figure 3G), with time-dependent progression from patchy peribronchial cell infiltrates and interstitial expansion (7 days) to frank parenchymal injury (2 weeks) (Supplemental Figure 7D and Supplemental Figure 8B). Radiographically, bilateral pulmonary infiltrates appeared (Supplemental Figure 8A), accompanied by increases in BALF protein, peaking at days 7–14 (Figure 3H). A transient nadir in BALF SP-B protein occurred (2 weeks) in the setting of preserved AT2 Sftpb mRNA and SP-B protein (Supplemental Figure 9).
SftpcI73T expression in AT2 cells induces polycellular alveolitis.
The induction of AT2 SftpcI73T mRNA and proprotein resulted in the subsequent onset of complex, multiphasic alveolitis (Figure 4A). BALF from iTAM-treated IER-SP-CI73T/I73TFlp+/– mice showed substantial increases in total cell counts beginning by day 7 and peaking 2 weeks after induction, with values 8- to 10-fold less than those of IER-SP-CI73T/I73TFlp–/– controls.
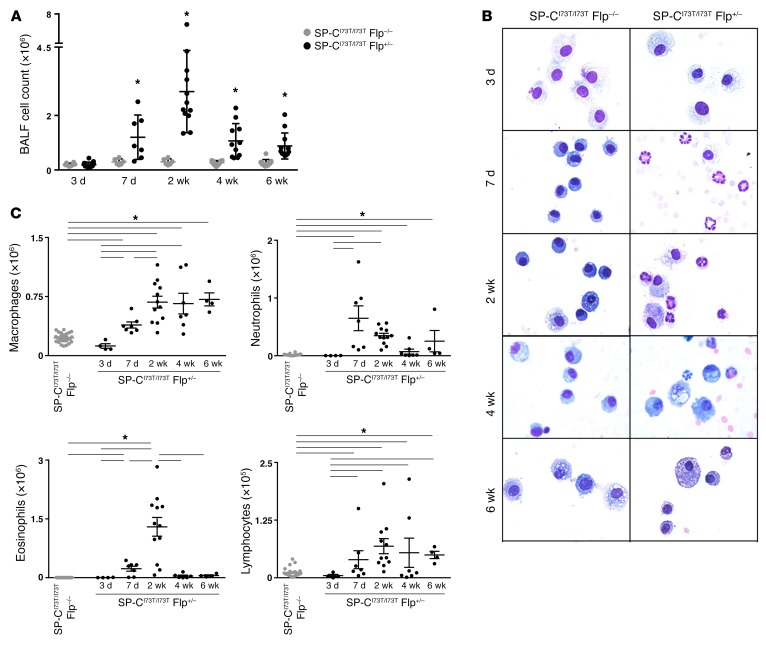
Figure 4. Ontogeny of alveolitis induced by SP-CI73T expression.
(A) Dot plot with group mean ± SEM shown of total BALF cells recovered from IER-SP-CI73T/I73TFlp–/– (control) and IER-SP-CI73T/I73TFlp+/– mice 3, 7, 14, 28, and 42 days after tamoxifen. *P < 0.05 versus corresponding IER-SP-CI73T/I73TFlp–/– group for each time point by 2-tailed t test. (B) Representative Giemsa-stained BALF cytospins from IER-SP-CI73T/I73TFlp+/– and IER-SP-CI73T/I73TFlp–/– groups 3, 7, 14, 28, and 42 days after tamoxifen administration. (C) Dot plots with mean ± SEM of absolute numbers of macrophages, neutrophils, eosinophils, and lymphocytes determined by manual counting (>200 cells per slide). *P < 0.05 versus pooled IER-SP-CI73T/I73TFlp–/– controls by 1-way ANOVA.
After induction, BALF cytospins demonstrated a complex inflammatory cell profile represented by 3 temporally distinct events: an early and sustained macrophage accumulation beginning at 7 days, a spike in polymorphonuclear cells (on day 7), and transient alveolar eosinophilia (peak at 2 weeks) (Figure 4, B and C) confirmed by flow cytometry (Supplemental Figure 10). While total lymphocytes also increased by day 7, the magnitude was 1 log less than the 3 major effector cell populations.
The inflammatory phenotype observed early after PGK-Neo cassette excision was dependent on SftpcI73T allele status but not on expression of Flp-O recombinase. When compared with mice treated with vehicle (oil) , heterozygous IER-SP-CI73T/WTFlp+/– mice receiving maximal doses of tamoxifen developed moderate alveolitis and interstitial histological changes 2 weeks after induction but in the absence of significant weight loss or mortality, suggesting that SftpcI73T allele heterozygosity represents an intermediate phenotype (Supplemental Figure 11). Importantly, iTAM treatment of homozygous Rosa26 ER2-Flp-O mice lacking SftpcI73T-Neo alleles (i.e., IER-SP-CWT/WTFlp+/+) had no effect on weight loss, histology, or BALF cell levels (Supplemental Figure 12). Similarly, in contrast to tamoxifen, vehicle treatment of IER-SP-CI73T/I73TFlp+/– animals failed to elicit substantive increases in weight loss, mortality, or BALF cell numbers (Supplemental Figure 13), indicating that “leakiness” of ER2-Flp-O alleles was not a confounding factor.
When considered in total, the polycellular alveolitis and diffuse parenchymal damage observed after iTAM treatment of IER-SP-CI73TFlp mice was specifically attributable to SP-CI73T expression, was dose dependent, and produced a phenotype with features resembling AE-IPF (10).
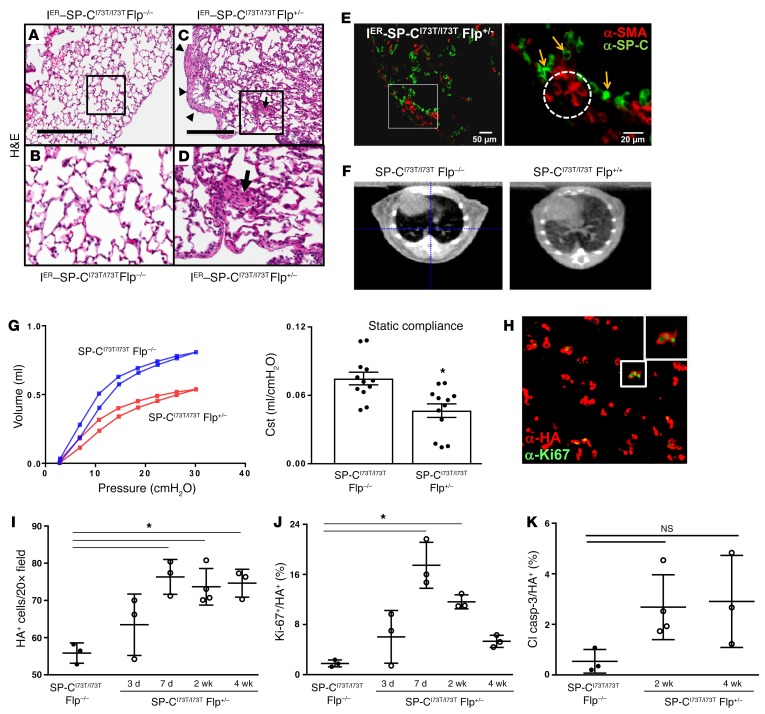
AT2 SftpcI73T expression results in spontaneous fibrotic lung remodeling.
Following the acute injury phase, survivors from IER-SP-CI73T/I73TFlp cohorts showed partial recovery from the nadir in body weight (Supplemental Figure 14A) but went on to develop a fibrotic histological phenotype (Figures 5 and 6). Lung sections from homozygous IER-SP-CI73T/I73T mice obtained 4–6 weeks after induction demonstrated spatially heterogeneous parenchymal remodeling with mesenchymal expansion, and some features of fibroblastic foci, including the accumulation of α–smooth muscle actin–positive (α-SMA–positive) cells adjacent to dilated airspaces lined by hyperplastic AT2 cells (Figure 5, A–E). The histological changes were accompanied by patchy bilateral infiltrates visible on microCT (Figure 5F) and by alterations in lung mechanics. Pressure-volume curves obtained at 4 and 6 weeks following iTAM were shifted down and to the right, consistent with restrictive lung physiology (Figure 5G and Supplemental Figure 14B). Static compliance, maximally decreased at 4 weeks (40%) in the IER-SP-CI73T/I73TFlp+/– cohort, mildly improved at week 6 in parallel with resolution of inflammation (Figure 4).
Figure 5. SftpcI73T expression induces diffuse parenchymal lung remodeling, restrictive lung physiology, and AT2 cell expansion.
H&E sections of control (IER-SP-CI73T/I73TFlp–/–) (A and B) and inducible IER-SP-CI73T/I73TFlp+/– (C and D) mice 6 weeks after 300 mg/kg iTAM showing extensive parenchymal (D, box) and subpleural remodeling (C, arrowheads). On higher power, focal consolidations with features of fibroblastic foci are identified (D, arrow). (E) Double-label immunohistochemistry of 6-week iTAM-treated IER-SP-CI73T/I73TFlp+/– lungs showing focal airspace enlargement lined by proSP-C AT2 cells (arrows) with adjacent α-SMA–positive myofibroblasts (dashed circle). Scale bars: 50 μm (left), 20 μm (right). (F) MicroCT scans of IER-SP-CI73T/I73TFlp–/– and IER-SP-CI73T/I73TFlp+/+ mice at 6 weeks after 200 mg/kg tamoxifen. (G) Control (IER-SP-CI73T/I73TFlp–/–) and IER-SP-CI73T/I73TFlp+/– mice 4 weeks after 250–300 mg/kg tamoxifen administration were subjected to pulmonary function testing. Pooled flow volume loops (left) from each cohort (n = 12) and calculated static compliance (Cst, right) demonstrating restrictive physiology. *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– control. (H) Representative ×20 fluorescence micrograph from IER-SP-CI73T/I73TFlp+/– lung 7 days after iTAM induction stained with anti-HA (red) and Ki-67 (green). (I and J) Sections were analyzed by manual counting. Dot plots with mean ± SEM are shown as the absolute number of HA+ cells per ×20 field (I) or percent of double-positive Ki-67+HA+ cells (J). *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– control by 1-way ANOVA. (K) Sections double stained for α-HA and α–cleaved caspase-3 (Cl Casp-3) were manually counted as for I and J. Dot plots of double-positive (Cl Casp-3/HA+) cells expressed as a percentage of total HA+ AT2 cells are shown with mean and SEM. NS: P > 0.05 versus IER-SP-CI73T/I73TFlp–/– control by 1-way ANOVA.
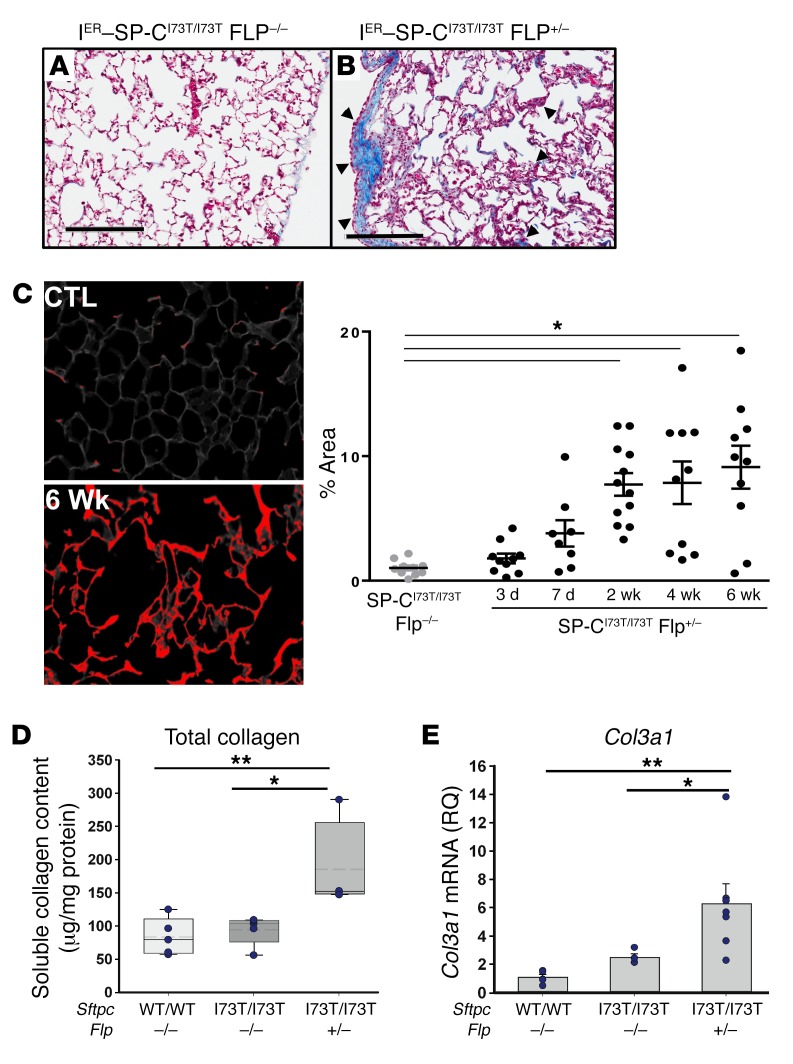
Figure 6. SftpcI73T expression in AT2 cells increases lung collagen content.
Histological sections, homogenates, and RNA were prepared from lungs of IER-SP-CI73T/I73TFlp+/– mice and IER-SP-CI73T/I73TFlp–/– controls 6 weeks after 300 mg /kg tamoxifen. (A and B) Representative trichrome staining showing focal collagen deposition (blue) in both subpleural and parenchymal areas (arrowheads) of IER-SP-CI73T/I73TFlp+/– mice. Scale bars: 300 μm. (C) Representative Picrosirius red stained field from IER-SP-CI73T/I73TFlp+/– and IER-SP-CI73T/I73TFlp–/– (CTL) mice with quantitation performed using ImageJ. Data represent percentage of total section area, with dot plots and mean ± SEM shown. *P < 0.05 versus ER-SP-CI73T/I73TFlp+/– by One-way ANOVA with Tukey’s post hoc test. Scale bars: 50 μm (left), 20 μm (right). (D) Soluble collagen in left lung homogenates measured by Sircol assay. Shown are dot plots with mean ± SEM. The x axis depicts groups by Sftpc allele and Flp allele status. *P = 0.022 for IER-SP-CI73T/I73TFlp+/– versus IER-SP-CI73T/I73TFlp–/– and **P = 0.011 versus untreated WT (WT/WT) group by 1-way ANOVA with Tukey’s post hoc test. (E) The relative quantity (RQ) of Col3a1 mRNA was determined by qRT-PCR and expressed as fold change from untreated WT animals (WT/WT) (mean RQ = 1). x axis depicts groups by Sftpc and Flp allele status. Bar graphs with superimposed dot plots with mean ± SEM are shown. *P = 0.032 for IER-SP-CI73T/I73TFlp+/– versus IER-SP-CI73T/I73TFlp–/–; **P = 0.010 versus WT/WT by 1-way ANOVA with Tukey’s post hoc test.
The focal AT2 cell hyperplasia shown in Figure 5E was part of a larger expansion of AT2 cells in response to SftpcI73T expression. Double-label fluorescence immunohistochemistry for HA–SP-C (AT2 cells) and Ki-67 (proliferation) was performed on iTAM-treated IER-SP-CI73T/I73TFlp+/– mice and controls (IER-SP-CI73T/I73TFlp–/–) (Figure 5H). Manual counting of single- (HA+) and double-positive (Ki-67+HA+) cells revealed a 40% persistent increase in AT2 cell numbers in IER-SP-CI73T/I73TFlp+/– lungs beginning at 7 days that coincided with an early burst in AT2 cell proliferation (Figure 5, I and J). In contrast, consistent with prior in vitro data (33, 34), significant apoptosis was not detected in HA-SP-CI73T–expressing AT2 cells (Figure 5K).
The histological remodeling and restrictive lung physiology were complemented by abnormal lung collagen deposition (Figure 6). Lung sections from tamoxifen-induced IER-SP-CI73T/I73TFlp mice demonstrated increased trichrome staining of peripheral lung parenchyma and subpleural regions (Figure 6, A and B), corroborated by Picrosirius red staining, whereby morphometric quantitation revealed increases in collagen beginning at 2 weeks (Figure 6C). Biochemically, soluble lung collagen content (Figure 6D) and lung Col3a1 mRNA expression (Figure 6E) were also increased.
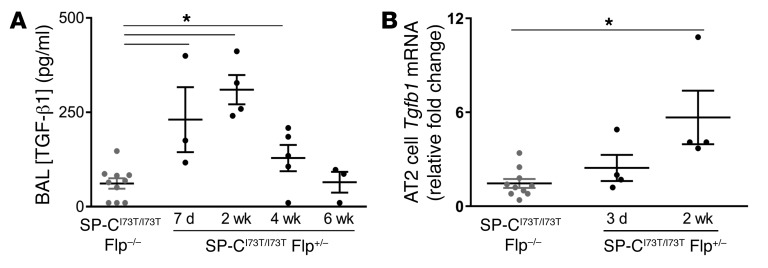
Temporally, the parenchymal remodeling and collagen deposition were preceded by the appearance of the well-known profibrotic mediator TGF-β1, with concentrations peaking at 2 weeks in BALF after iTAM (Figure 7A). By qRT-PCR, isolated AT2 cells but not unsorted BALF cells demonstrated temporal increases in Tgfb1 mRNA expression, suggesting that AT2 cells could contribute early to a profibrotic milieu following SftpcI73T induction mediated in part by TGF-β1 (Figure 7B and Supplemental Figure 15).
Figure 7. BALF TGF-β1 and AT2 Tgfb1 mRNA increase prior to fibrotic remodeling.
(A) TGF-β1 levels in BALF collected at the indicated times after tamoxifen treatment were measured using Luminex. Gray dots represent values from iTAM-treated IER-SP-CI73T/I73TFlp–/– control mice pooled from all 4 time points and black dots values for IER-SP-CI73T/I73TFlp+/– mice at each time point. (B) qRT-PCR for Tgfb1 mRNA expression in AT2 cells isolated from IER-SP-CI73T/I73TFlp+/– mice at 3 and 14 days after tamoxifen (black dots) and iTAM-treated IER-SP-CI73T/I73TFlp–/– control mice pooled from both time points (gray dots). Data expressed as fold change from IER-SP-CI73T/I73TFlp–/– control group are presented as dot plots with mean ± SEM shown. *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– control by 1-way ANOVA with Tukey’s post hoc test.
Importantly, neither lung injury nor fibrotic remodeling was attributable to an adaptive immune response to the epitope tag on HA–proSP-CI73T. Western blotting for HA–proSP-CI73T using sera prepared from bleeds of iTAM-induced IER-SP-CI73T mice failed to detect anti-HA titers (Supplemental Figure 16).
The AT2 epithelium is an upstream driver of early lung injury.
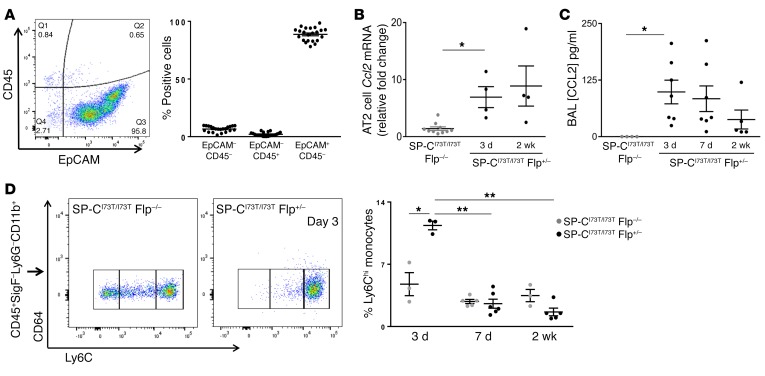
The inducible IER-SP-CI73TFlp model permits detailed assessment of the potential role of dysfunctional SftpcI73T AT2 cells in the ontogeny of events underlying the development of fibrotic lung disease. In highly purified AT2 cells isolated from iTAM-treated cohorts (>95% EpCAM+, proSP-C+) (Figure 8A and Supplemental Figure 17), the rise in BALF alveolar macrophages seen on day 7 (Figure 4A) was preceded by increased Ccl2 mRNA in AT2 cells as early as day 3 (Figure 8B). Commensurate increases both in BALF CCL2 (Figure 8C) and in lung tissue–associated Ly6ChiCD64– monocytes (Figure 8D), a population recognized for its role in modulating fibrotic lung remodeling (35, 36), also occurred.
Figure 8. Ccl2 mRNA expression by SftpcI73T AT2 cells precedes BALF CCL2 and recruitment of Ly6ChiCD64– monocytes in lung tissue.
(A) Flow cytometry analysis for EpCAM (CD326) and CD45 expression in AT2 cells isolated from IER-SP-CI73T/I73TFlp–/– and IER-SP-CI73T/I73TFlp+/– mice. Dot plot showing EpCAM+CD45– (epithelial), EpCAM–CD45+ (immune), and double-negative populations as a percentage of total cells from each preparation (n = 22 animals). (B) qRT-PCR of AT2 RNA for Ccl2 expression (18S-normalized) for IER-SP-CI73T/I73TFlp+/– group expressed as fold change (black dots) over IER-SP-CI73T/I73TFlp–/– controls pooled from both time points (gray dots). *P < 0.05 versus using 1-way ANOVA followed by Tukey’s post hoc test. (C) CCL2 (MCP-1) ELISA of BALF from IER-SP-CI73T/I73TFlp–/– and IER-SP-CI73T/I73TFlp+/– cohorts. Dot plots with mean ± SEM are shown. Controls were pooled from IER-SP-CI73T/I73TFlp–/– at all 3 time points (all less than detectable limit). *P < 0.05 versus control group using 1-way ANOVA followed by Tukey’s post hoc test. (D) Representative flow cytometry plot of lung cell suspensions for CD45+Ly6ChiCD64– populations in IER-SP-CI73T/I73TFlp–/– and IER-SP-CI73T/I73TFlp+/– mice on day 3. Dot plot (3 mice pooled per data point) with mean ± SEM showing relative percentage of Ly6ChiCD11c–CD64–CD11b+CD24–Ly6G– monocytes. Multiple comparisons were made by 1-way ANOVA followed by Tukey’s post hoc test. For Ly6Chi populations, *P < 0.05 for IER-SP-CI73T/I73TFlp+/– versus day 3 IER-SP-CI73T/I73TFlp–/–; **P < 0.05 for 3 day IER-SP-CI73T/I73TFlp+/– group versus IER-SP-CI73T/I73TFlp+/– at 7 or 14 days.
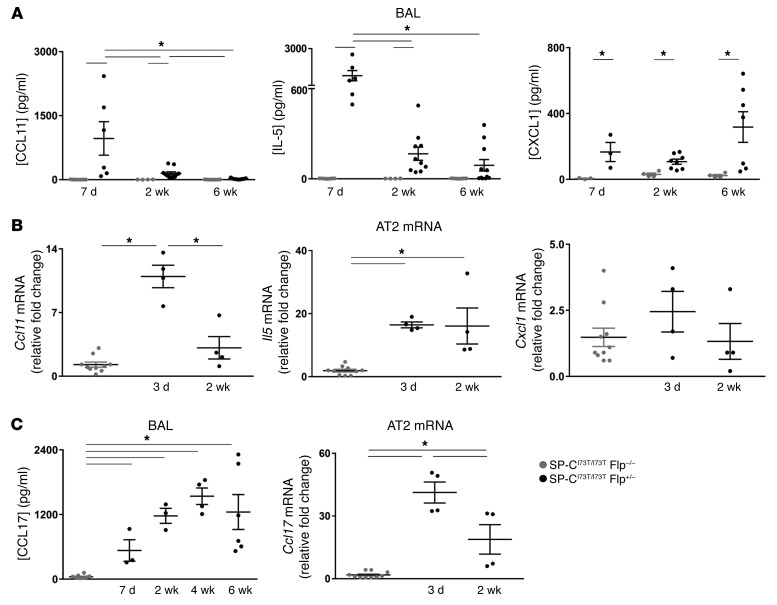
Other chemokines shown to be important for the recruitment of neutrophils and eosinophils demonstrated a similar temporal response. AT2 expression of Ccl11 and IL5 mRNA at 3 days after iTAM induction and increased BALF levels of these eosinophil chemokines by 1 week preceded the spike in BALF eosinophils occurring 2 weeks after induction (Figure 9, A and B). In contrast, while BALF levels of the neutrophil chemoattractant KC (CXCL1/GroA), the functional murine homolog of human IL-8, were also increased by 1 week and remained elevated out to at least 6 weeks after induction, AT2 cells did not exhibit increased Cxcl1 mRNA expression, suggesting that either the CXCL1 response occurs earlier than 3 days after induction or there are additional cellular sources for this chemokine. The BALF cytokine profiles from affected IER-SP-CI73TFlp mice failed to demonstrate significant amounts of IL-4, IL-13, Rantes, TNF-α, IFN-γ, IL-17, IL-1β, MIP1α, or MIP2α (Supplemental Table 2). Furthermore, the elaboration of proinflammatory cytokines in this model was not attributable to the induction of Flp-O recombinase expression (Supplemental Table 3). The lack of a strong consensus Th2 response (e.g., IL-4/IL-13) classically associated with fibrotic lung remodeling led us to consider that other Th2-related CC chemokines could be participating in SP-CI73T–induced injury/fibrosis. CCL17 (TARC) was previously shown to participate in the fibrotic response to exogenous bleomycin in rodents (37) As shown in Figure 9C, early (day 3) increases in AT2 Ccl17 mRNA were followed by sustained elevations in BALF TARC levels from 7 days through 6 weeks.
Figure 9. Cytokine expression profiles of IER-SP-CI73TFlp mice after tamoxifen treatment.
(A) IL-5, CCL11 (eotaxin), and CXCL1 (KC) were measured in BALF of IER-SP-CI73T/I73TFlp–/– (gray dots) and IER-SP-CI73T/I73TFlp+/– (black dots) cohorts at the indicated times after induction. Dot plots with mean ± SEM are shown. *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– or IER-SP-CI73T/I73TFlp+/– as indicated using 1-way ANOVA followed by Tukey’s post hoc test. (B) qRT-PCR for Il5, Ccl11, and Cxcl1 mRNA in isolated AT2 cells. dCT values were normalized to 18S RNA and data expressed as fold change in IER-SP-CI73T/I73TFlp+/– samples (black dots) versus IER-SP-CI73T/I73TFlp–/– controls (gray dots). *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– or 2-week IER-SP-CI73T/I73TFlp+/– groups as indicated by 1-way ANOVA followed by Tukey’s post hoc test. (C) Left: BALF CCL17 (TARC) levels at 1, 2, 4, and 6 weeks after iTAM determined by Luminex assay. Right: qRT-PCR for Ccl17 mRNA expression in AT2 cells from IER-SP-CI73T/I73TFlp+/– (black dots) and IER-SP-CI73T/I73TFlp–/– controls (gray dots) 3 and 14 days after iTAM. Data are presented as dot plots with mean ± SEM shown. *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– or IER-SP-CI73T/I73TFlp+/– using 1-way ANOVA followed by Tukey’s post hoc test.
Together, the effector cell and cytokine patterning observed in IER-SP-CI73TFlp mice recapitulates many of the responses reported in BALF and transcriptomic profiles of lung tissue and cell populations from IPF patients. Furthermore, the ontogeny and selectivity of the observed AT2 cytokine mRNA profiles suggest that epithelial-derived chemokines play a major role in the early recruitment of many of these effector cells to the lungs during the genesis of SP-CI73T–induced fibrosis.
IER-SP-CI73TFlp mice elaborate multiple biomarkers observed in human IPF.
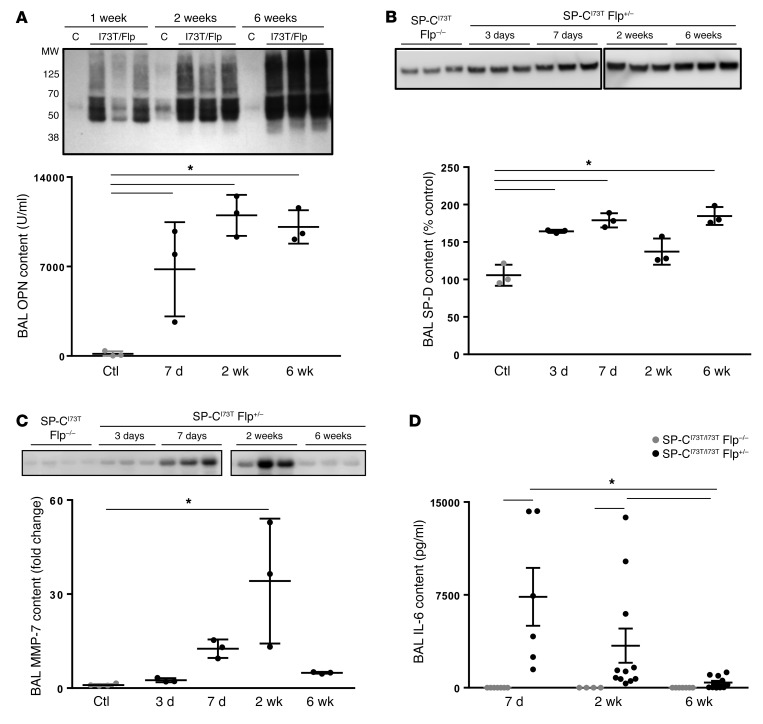
We next evaluated the temporal expression patterns of biomarkers observed in human IPF. As proof of concept, we first focused on 3 analytes (SFTPD, SPP1, MMP7) that recently emerged as part of a multivariate model predictive of an IPF diagnosis in humans (38) (Figure 10, A–C). Osteopontin (OPN), a matricellular protein, was markedly increased in BALF for up to 6 weeks after iTAM treatment (Figure 10A). Surfactant protein D (SP-D) — a multimeric collectin synthesized by the distal airway epithelium that we had shown to be markedly upregulated in multiple preclinical models of lung injury/remodeling (39, 40) — increased in BALF early and remained elevated to 6 weeks (Figure 10B). In contrast, MMP-7, whose cleaved (activated) form plays a role in tissue remodeling, was found to be elevated early, peaking at 2 weeks (Figure 10C).
Figure 10. IER-SP-CI73TFlp mice express IPF biomarkers.
Western blotting of BALF (20 μl/lane) for OPN (A), SP-D (B), and MMP-7 (C) from IER-SP-CI73T/I73TFlp+/– mice obtained at 3 days to 6 weeks after tamoxifen as indicated. Controls (Ctl) consisted of IER-SP-CI73T/I73TFlp–/– mice 2 weeks after tamoxifen treatment. Densitomteric band intensities, expressed as relative units (A), percent control (B), or fold increase (C), are presented as dot plots with mean ± SEM. *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– (CTL) by 1-way ANOVA followed by Tukey’s post hoc test. (D) ELISA for IL-6 content of BALF obtained 1, 2, and 6 weeks after tamoxifen treatment. Dot plots with mean ± SEM are shown for IER-SP-CI73T/I73TFlp+/– mice (black dots) and IER-SP-CI73T/I73TFlp–/– controls (gray dots). At all 3 time points, controls were below the limit of detection. *P < 0.05 versus IER-SP-CI73T/I73TFlp–/– by 1-way ANOVA followed by Tukey’s post hoc test.
BALF levels of IL-6, a pluripotent cytokine found to be elevated in both acute lung injury and chronic parenchymal lung diseases, were massively increased during the early inflammatory phase, peaking at 1 week after iTAM (Figure 10D).
Discussion
Multiple reports of sporadic or inherited mutations in the SFTPC gene appearing in ILD patient cohorts provide a strong rationale for the hypothesis that AT2 cell dysfunction is an important component in the pathogenesis of DPLD in children and adults. However, a direct causal role for the encoded mutant proSP-C protein isoforms in disrupting AT2 homeostasis and any resultant intrinsic cascade of downstream proinflammatory and/or profibrotic events has not to our knowledge been previously demonstrated. In this report, preclinical knockin mouse models expressing ILD-associated SftpcI73T alleles were developed and phenotyped. We have been able to define an ontogeny of events emanating from the mistrafficking of expressed mutant proSP-CI73T protein, specifically alterations in cell quality control (macroautophagy), and a selective profile of chemokines and fibrotic mediators elaborated by the AT2 epithelial cell early after induction of SftpcI73T transcription. The resultant phenotype manifested a spontaneous sequence of acute alveolitis, parenchymal injury, and fibrotic remodeling reflecting multiple features of human IPF not attributable to either an immune response to the HA epitope tag (Supplemental Figure 15) or toxicity from Flp recombinase (Supplemental Figure 12). In total our data offer proof of principle that SP-CI73T expression in AT2 epithelia drives aberrant lung injury and repair.
Prior preclinical murine models used to study the pathogenesis of fibrotic lung diseases such as IPF have been hampered by a variety of issues that complicate feasibility and relevance (reviewed in ref. 16). These include a dependence on injury by exogenous agents (e.g., bleomycin, fluorescein isothiocyanate) to achieve fibrotic endpoints, overexpression of profibrotic genes that may skew mechanistic interpretation (e.g., TGF-β1, TGF-α, IL-13), a failure to develop IPF-like (UIP) histopathology, and/or impracticality imparted by extended lag periods in acquiring fibrotic phenotypes (e.g., ionizing radiation). The expression of rare disease-associated SFTPC mutants using the endogenous mouse Sftpc locus offers several theoretical advantages in modeling human IPF. Among them are that SFTPC expression is limited to AT2 cells (41) and that a case for a dose-dependent toxic gain of function has already been established in humans, as almost 100% of affected patients are heterozygous for mutant SFTPC alleles (42). Indeed, prior attempts at transgenically expressing mutant SP-C isoforms confirmed this dose-dependent toxic gain of function but were limited by an inability to temporally control gene dose to a sufficient extent to yield tractable fibrosis models. Overexpression of an aggregation-prone human SFTPC folding mutant (SP-CΔexon4) using a 13-kb human SFTPC promoter fragment resulted in developmental lethality from disrupted lung morphogenesis (43). Conversely, inducible expression of an ILD mutant SFTPCL188Q transgene only achieved mRNA levels approaching approximately 10% of endogenous alleles and failed to produce spontaneous fibrosis despite increased sensitivity to exogenous bleomycin (44).
Our approach utilizing a strategic knockin of a mutant Sftpc allele extended these prior observations in humans and mouse models by achieving the ability to temporally control expression of mutant Sftpc in a spatially restricted fashion over a full range of gene doses. The founder line, SP-CI73T-Neo, functionally hypomorphic due to the retained intronic PGK-Neo cassette, expressed low basal levels of Sftpc mRNA while acquiring a limited and protracted age-dependent fibrotic phenotype (Figure 1). The inducible homozygous IER-SP-CI73T/I73TFlp line provided a “sweet spot” by permitting postnatal, AT2 cell–specific expression of mutant SftpcI73T mRNA in relative amounts (~60%–70% of WT) approximating those in affected humans carrying a single mutant SFTPC allele (Figure 3B) while developing translationally relevant features of IPF, including a histology with some UIP features and expression of IPF-related chemokines, cytokines, and putative biomarkers. Induction of heterozygous IER-SP-CI73T/WTFlp+/– mice was also informative, as this resulted in mild alveolitis and inflammatory histological changes 2 weeks after induction but failed to yield to frank fibrosis (Supplemental Figure 11). While our qRT-PCR method cannot assess allele-specific expression, based on data from the induced homozygous IER-SP-CI73T/I73TFlp line (Figure 3B), heterozygous IERSP-CI73T/WTFlp animals likely express Sftpc mRNA at levels in the range of approximately 30%–40% of those in WT mice and represent an intermediate phenotype, with some early inflammation that is below the threshold needed to promote late fibrosis.
In contrast to the adult lung, our attempt to constitutively remove PGK-Neo from SftpcI73T alleles was successful but resulted in neonatal lethality from an allele-dependent arrest in lung development (Figure 2). The failure to obtain meaningful numbers of live postnatal heterozygous CFlp-SP-CI73T/WT mice using this strategy contrasts with human SP-CI73T–associated disease, in which the prevailing genotype is heterozygosity and which is associated with live birth and delayed symptom presentation (4, 7, 21). This likely reflects either an enhanced sensitivity of the developing mouse lung epithelia to mutant proSP-CI73T protein and/or the inability of lungs of CFlp-SP-CI73T/WT pups with saccular disruption (Figure 2A) to support the transition from liquid to air breathing, possibly due to the high degree of dependence of the murine lung on postnatal alveolarization (45). Alternatively, differences in genetic background that modify expression of SP-CI73T or injury repair pathways in mice could contribute to perinatal phenotypes.
A significant difference in survival between male and female animals expressing SftpcI73T was found. Notably, the enhanced sensitivity of males occurred in the early (inflammatory) phase after tamoxifen administration (7–14 days) (Figure 3F). Male mice were similarly more susceptible than females to both spontaneous lung fibrosis and low-dose bleomycin injury in a model of tamoxifen-induced telomere dysfunction (46). Despite reanalysis of several of our data sets, we found no significant sex differences in BALF cell numbers, BALF total protein, or lung mechanics (Supplemental Table 4). While tamoxifen has been shown to alter the course of renal fibrogenesis in a model of unilateral ureteral obstruction (47) and to affect myofibroblast differentiation in vitro (48), given the early timing of the mortality coupled with the fact that male rodents were shown to be more sensitive to bleomycin (49) and hyperoxic injuries (50) these events are likely to be tamoxifen and fibrosis phase independent.
The AT2 cell has been affirmed as a key progenitor (stem) cell in the distal lung contributing to a niche that mediates normal alveolar maintenance and repair (51). In preclinical model systems, selective depletion of AT2 cell mass through transgenic, spatially restricted expression of the diphtheria toxin (dT) receptor in AT2 cells followed by exogenous dT administration results in either spontaneous lung fibrosis (52, 53) or enhanced susceptibility to exogenous bleomycin injury (51). Our results contribute to this growing collection of evidence for the critical role of disrupted AT2 cell homeostasis (in either function or numbers) as a proximal component in the pathogenesis of fibrotic lung remodeling. AT2 cell dysfunction manifested as acquired (either age-dependent or tamoxifen-induced) abnormalities in cellular quality control and ultrastructure phenocopied features we found previously in both an SP-CI73T cell line (i.e., proSP-C mistrafficking, a block in macroautophagy, impaired mitophagy) and a patient with ILD expressing the identical mutation (24). The similarities between the SP-CI73T AT2 phenotype and the AT2 phenotype in humans with Hermansky-Pudlak syndrome (HPS) or in mouse models of HPS (24, 54–56), as well as their common association with altered macroautophagy (57), suggest that disrupted AT2 cell quality control represents an important contributing factor for fibrotic lung disease (54, 55, 57). However, because this autophagy block is downstream from initiation (coupled with a lack of effect of torin on LC3 turnover in our model; Supplemental Figure 4B), therapeutic approaches aimed at directly stimulating autophagy flux are potentially problematic. Despite this, a recent preclinical study using manipulation of epithelial endosomal/lysosomal dynamics through modulation of the master regulator of lysosomal biogenesis pathways TFEB in the lung raises the potential for future manipulation of epithelial cell quality control as a targeted therapeutic strategy (58).
Mutations in genes expressed by AT2 cells other than SFTPC and HPS, such as surfactant protein A (SP-A; SFTPA) and the lamellar body lipid transporter ABCA3, have been described in patients with confirmed DPLD (4, 12, 59). Alterations in telomere length in a subgroup of IPF patients with mutations in key telomere-regulatory genes (hTERT, hTR, RTEL1) have been mechanistically confirmed in 2 mouse models wherein disruption of telomere homeostasis in AT2 cells through disruption of telomerase genes (trf1; trf2) results in a fibrotic lung remodeling (46, 60). However, while telomere dysfunction was shown to result in failure of alveolar stemness in vitro (61), the time to development of a fibrotic phenotype in these models is protracted (~6–8 months) or requires a “second hit” (e.g., bleomycin), suggesting that telomere length exerts a smaller effect than SFTPC mutation in driving disease.
Although the exact role of inflammation in IPF remains controversial, some component of an inflammatory response appears to be necessary for fibrogenesis (12). The importance of inflammation in these processes is particularly evident during AE-IPF, a well-known clinical syndrome characterized by the acute onset of significant respiratory deterioration in previously known or concurrently diagnosed IPF characterized by evidence of widespread alveolar abnormalities (10). These events have been attributed to disease acceleration and thus play a major role in IPF mortality. A striking feature of SftpcI73T–driven lung disease in this model has been the rapid development of a multiphasic, polycellular alveolitis in affected animals composed not only of alveolar macrophages but also secondary waves of both neutrophils and eosinophils (Figure 4). The “clinical syndrome” associated with this mouse model is marked temporally by weight loss, respiratory distress, hypoxemia, and radiological pulmonary infiltrates and a high mortality mimicking that of AE-IPF patients (10). Furthermore, the BALF inflammatory cell and cytokine profiles also phenocopy those of IPF patients, in whom increased numbers of eosinophils and neutrophils have been detected (62). While their role in IPF progression has not been completely established, this model represents a platform for further delineation of the individual and collective contributions of inflammatory cell populations in these processes.
Due to the reality of limited or single sampling in IPF patients, defining the temporal events, mechanistic pathways, and molecular mediators underlying either acute exacerbations or fibrogenic responses in the disease process has been challenging. Among several key events defined in the current study using the IER-SP-CI73TFlp model, the elaboration of CCL2/MCP-1 represents one of the most proximal upstream events detectable following induction of SftpcI73T expression. Signatures of this classic monocyte/macrophage chemokine can be detected in both AT2 cells and BALF as early as 3 days after tamoxifen administration, coinciding with the appearance of increased numbers of lung tissue monocytes. Previously we had shown that the onset of pulmonary fibrosis in mouse models of HPS also involved the early expression of MCP-1 by AT2 cells (55). Together with recent studies showing the importance of crosstalk between AT2 cells and macrophages in propagating fibrotic susceptibility in HPS mice, our SftpcI73T mouse models contribute to a growing body of evidence that AT2 cell dysfunction is a proximal component in the pathogenesis of fibrotic remodeling (63).
Perhaps the most striking paradigm shift suggested by data from SP-CI73T mice is the lack of a classic Th2 response previously associated with fibrogenesis. Although overexpression of IL-4 or IL-13 in the lung of transgenic mice confirmed that both cytokines could function as profibrotic mediators (64, 65), we were unable to detect significant amounts of either cytokine in BALF (Supplemental Table 2). While the use of neutralizing antibodies against IL-4/IL-13 signaling afforded protection in preclinical models of lung fibrosis, our data would predict a lack of efficacy in targeting this pathway in human patients with SFTPCI73T mutations, and perhaps more widely in IPF. To date, clinical trials in IPF adopting such an approach have not yet reported efficacy. As an alternative to IL-4/IL-13, we also show here that AT2 cells from SP-CI73T mice elaborate CCL11, IL-5, and TARC (CCL17), which could contribute to the observed alveolar eosinophilia/neutrophilia (Figure 8). Furthermore, both the CCL17/CCR4 axis and IL-5 signaling pathways have been shown to be profibrogenic by recruiting many of these effector cell populations (37, 66, 67). Thus, expression of ILD-related SFTPC mutants in mice could offer a new preclinical platform to sort out well-known chemokine–receptor signaling redundancies and better define which stages of pulmonary fibrosis offer optimal targeting.
Following induction of SftpcI73T expression, we noted changes in BALF levels of a triad of biomarker analytes (SP-D, MMP-7, OPN) previously validated in human IPF (38) (Figure 10, A–C). Another important putative biomarker, IL-6, shown by Collard and colleagues to be elevated during acute exacerbations of IPF (10), was also increased in BALF of IER-SP-CI73Flp mice, with levels peaking at 2 weeks, consistent with the timing of maximal inflammation (Figure 10D). Taken together, the expression of these 4 human IPF biomarkers coupled with the appearance of several other putative biomarker candidates including MCP-1 and CCL17 support the translational relevance of using IER-SP-CI73Flp mice for preclinical biomarker discovery, target identification, and target validation for IPF.
In summary, the present study has shown that expression of an ILD-associated SFTPC mutant in the pulmonary epithelium of mice in an allelic, inducible fashion recapitulates many features of the human IPF phenotype. In addition to providing proof of concept for a critical role for the AT2 cell in distal lung homeostasis, the SP-CI73T mouse models represent a preclinical, translationally relevant platform for exploring IPF disease pathogenesis, identifying IPF biomarkers, and testing novel genetic and pharmaceutical therapeutic approaches.
Methods
Mouse models.
The SP-CI73T founder line (SP-CI73T-Neo) was produced using BAC recombineering strategies to selectively knock in coding sequences for expression of an NH2-terminal HA-tagged murine SP-CI73T into the endogenous mouse Sftpc locus based on a targeting strategy we previously used for generation of an ABCA3E292V line (68). Depicted in Figure 1A, the targeted allele contained an HA tag in exon 1 (the NH2 terminus), an Ile-for-Thr substitution in exon 3a, and a FRT-PGK-gb2-Neo/km-FRT cassette (herein termed PGK-Neo) in intron 1. Additional details of its production are provided in Supplemental Methods.
Constitutive removal of PGK-Neo from SftpcI73T-Neo alleles.
A second line was generated by backcrossing SP-CI73T-Neo mice to a FLP-e recombinase (“deleter”) line [strain B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ; stock 009086; The Jackson Laboratory] and progeny designated CFlp-SP-CI73T. Genotyping was performed as described in Supplemental Methods.
Tamoxifen-inducible model of SftpcI73T expression.
For inducible removal of PGK-Neo from SftpcI73T-Neo alleles, a Flp-O recombinase deleter line, which contained an optimized FLP-e recombinase variant (Flp-O) under control of the mutated ER2 receptor knocked into the Rosa26 locus [strain B6N.129S6(Cg)-Gt(ROSA)26Sortm3 (CAG-FLPo/ERT2)Alj/J], was purchased from the Jackson Laboratory (stock 019016) and crossed with the HA–SP-CI73T-Neo line. Genotyping of the resultant line, termed IER-SP-CI73TFlp, was performed as described in Supplemental Methods.
All mouse strains and genotypes generated for these studies (SP-CI73T-Neo, CFlp-SP-CI73T, and IER-SP-CI73TFlp) were congenic with C57BL/6J. Both male and female animals (aged 8–14 weeks) were utilized in tamoxifen induction protocols. All mice were housed under pathogen-free conditions in an approved barrier facility at the Perelman School of Medicine.
Materials and reagents.
Culture medium was produced by the Cell Center Facility, University of Pennsylvania. Cytological stains used were KwikDiff (Thermo Fisher Scientific) and Giemsa (GS500; Sigma-Aldrich). Tamoxifen (non-pharmaceutical grade) was purchased from Sigma-Aldrich. Except where noted, all other reagents were electrophoretic or immunological grade and purchased from either Sigma-Aldrich or Thermo Fisher Scientific.
Antibodies.
A polyclonal proSP-C antiserum (NPRO-SP-C) raised against the Met10–Glu23 domain of rat proSP-C; polyclonal anti–SP-B (PT3) raised against bovine SP-B; and polyclonal anti–SP-D (antisera 1754) raised against 2 synthetic SP-D peptides were each produced in-house in rabbits and validated as previously published (28, 69, 70). The other antibodies used for these studies were obtained from commercial sources (Supplemental Table 5).
Lung histology and morphometry.
Whole lungs were fixed by tracheal instillation of a 1.5% glutaraldehyde/1.5% paraformaldehyde mixture in 0.15 M HEPES buffer at a constant pressure of 25 cm H2O. 6-μm sections were stained with H&E or Masson’s trichrome stains by the Pathology Core Laboratory of Children’s Hospital of Philadelphia. Morphometric analyses to determine air/tissue ratios as a metric of parenchymal remodeling were performed on H&E sections encompassing multiple lobes and multiple levels. Five to 10 random peripheral lung images per slide were acquired at ×20 magnification and transferred to a computer screen. Total surface area, corrected for the presence of any large airways/bronchovascular bundles, was segmented into tissue and air densities using thresholds assigned in NIH ImageJ software, and air/tissue ratios were determined.
BALF collection and processing.
BALF collected from mice using sequential lavages of lungs with five 1-ml aliquots of sterile saline was processed for analysis as described previously (71). A 200-μl aliquot of each cell-free, “first-return” BALF was removed and stored at –80°C for cytokine analysis.
Cell pellets recovered from centrifugation were resuspended in 1 ml PBS and total cell counts determined using a Z1 Coulter Counter (Beckman Coulter). Differential cell counts were determined manually from cytospins of BALF cell pellets stained with KwikDiff (Thermo Fisher Scientific) or modified Giemsa (Sigma-Aldrich, GS500).
Cytokine analyses: multiplex and ELISA.
Aliquots of first-return, cell-free BALF were analyzed for levels of a panel of cytokines (MCP-1, MIP1α, Mip2, Rantes, TNF-α, eotaxin [CCL-11], IL-4, IL-13, IL-17, IL-5, IL-6, IL-10, GM-CSF, IFN-γ, IL-1β, KC [GroA]) using the Luminex platform (panel MCYTOMAG-70K-17) by the Human Immunology Core at the Perelman School of Medicine. TGF-β1 and CCL17 (TARC) were measured separately, also using Luminex kits (panel TGFBMAG-64K and MECY2MAG-73K). For select cohorts, MCP-1 was measured using a CCL2 ELISA kit (DY479-05, R&D Systems).
Immunohistochemistry and immunofluorescence.
Immunohistochemical staining of paraffin-embedded lung sections was performed with primary antisera at the indicated dilutions as described previously (19). Immunoreactivity was visualized using a commercial horseradish peroxidase–based method (Vectastain Elite Rabbit IgG ABC Kit, PK6101; Vectastain DAB Substrate Kit, SK410; Vector Laboratories). Developed sections were counterstained with Mayer’s hematoxylin.
Immunofluorescence staining of lung sections and coverslips was performed with combinations of primary antisera and secondary Alexa Fluor–conjugated anti-IgG (Supplemental Table 5) as described previously (27). Fluorescence images were viewed on an Olympus I-70 inverted fluorescence microscope, captured with a Hamamatsu 12-bit coupled-charged camera, and processed using Metamorph version 7.8.4.0 (Universal Imaging).
Quantitative determination of total, proliferating, and apoptotic AT2 cells was performed using double-label fluorescence immunostaining of lung sections with anti-HA and either anti–cleaved caspase-3 or anti–Ki-67. Five consecutive ×20 images were obtained from 2 different lobes (10 peripheral lung fields per section). AT2 cell number was determined by manual counting of HA-positive cells and reported as average number of AT2 cells per ×20 field. Proliferative (percentage of Ki-67/HA double-positive AT2 cells per total HA-positive cells) and apoptotic (percentage cleaved caspase-3/HA double-positive cells per total HA-positive cells) indices were determined by manual counting.
Mouse AT2 cell isolation.
Mouse AT2 cells were isolated as previously reported (55). Briefly, perfused mouse lungs were digested with Dispase (BD Biosciences), mechanically dissociated using a McIlwain tissue chopper (Metrohm USA) and treated with 20 μg/ml DNAase I (Sigma-Aldrich). Negative selection of mesenchymal cells (fibroblasts) was done by differential adherence on plastic culture dishes. CD45-positive cells were removed by negative selection using simultaneous incubation with a Dynabeads Untouched Mouse T Cells Kit (11413D) and Dynabeads Mouse DC Enrichment Kit (11429D; Thermo Fisher Scientific). Recovered cells were collected and flash frozen at –80°C or placed into primary culture. Subaliquots were analyzed for purity by flow cytometry for EpCAM, CD45, and viability (Supplemental Table 5). Purity was also determined by immunostaining preparations adhered overnight to 10% Matrigel-coated coverslips using DAPI in combination with primary antisera for HA, proSP-C, NKX2.1, and/or vimentin (Supplemental Table 5). Manual counts in five ×20 fields per sample were done, and purity was defined as number of HA+ cells (AT2 cells) divided by total nuclei.
Preparation of subcellular fractions and surfactant.
Subcellular fractions enriched in lamellar bodies or endoplasmic reticulum (microsomes) were prepared from whole lungs using upward flotation on sucrose density gradients as described previously (27).
Large-aggregate (LA) surfactant fractions were prepared from cell-free BALF by centrifugation at 20,000 g for 60 minutes at 4°C as described previously (71).
Total protein content of BALF, surfactant, cells, and tissue fractions was assayed by the Bradford method as previously published (71).
SFTPC expression plasmids.
For in vitro proSP-C trafficking studies, A549 cells (ATCC CCL-185) were transiently transfected with a family of previously validated SFTPC expression vectors encoding for untagged, HA epitope–tagged, or EGFP-fused WT (SFTPCWT) or mutant (SFTPCI73T) isoforms as previously published (23, 24, 29, 33, 72).
SDS-PAGE and immunoblotting.
SDS-PAGE using Novex Bis-Tris gels (NP0301, Thermo Fisher Scientific) and immunoblotting of PVDF membranes with primary antisera (Supplemental Table 5) followed by species-specific horseradish peroxidase–conjugated secondary antisera were performed as previously published (24, 33). Bands detected by enhanced chemiluminescence (ECL2 80196 Thermo Fisher Scientific or WesternSure 926-95000, LI-COR) were acquired by exposure to film or direct scanning using an LI-COR Odyssey Fc Imaging Station and quantitated using the manufacturers’ software.
RNA isolation and qRT-PCR.
cDNA was prepared from RNA template purified from whole lung or AT2 RNA using the RETROscript Kit. Quantitative single-plex PCRs (qRT-PCR) were performed in triplicate using Taq polymerase and TaqMan RT-PCR kits (Applied Biosystems, Thermo Fisher Scientific) on an ABI Prism 7900 system and a QuantStudio 7 Flex Real-Time PCR System. Commercial primer sets for mouse genes are listed in Supplemental Table 6.
Determination of lung collagen content.
Total acid soluble collagen content in harvested left lungs was determined using the Sircol assay (Biocolor) according to the manufacturer’s instructions as described previously (55).
Picrosirius red staining.
Staining for collagen was performed using the Picrosirius Red Stain Kit following the manufacturer’s instructions (Polysciences Inc.). Digital morphometric measurements were performed using ImageJ using 10 random fields of peripheral and central lung (without large airways) for each section and analyzed at a final magnification of ×400. The mean area of each lung field in each section staining for Picrosirius red was calculated and expressed as a percentage of total section area (73).
Multichannel flow cytometry.
Blood-free perfused lungs were digested in DMEM + 5% FBS + 2 mg/ml collagenase D (catalog 11088866001, Roche), passed through 70-μm nylon mesh to obtain single-cell suspensions, and then mixed with Gibco RBC Lysis Buffer (A10492-01, Thermo Fisher Scientific). Cell pellets collected by centrifugation were resuspended in PBS + 0.1% sodium azide and aliquots removed for determination of cell number using a NucleoCounter (New Brunswick Scientific). Cells were blocked with anti–mouse CD16/32 antibody (Fc block, eBioscience), followed by incubation with antibody mixtures (or isotype controls) and conjugated viability dye (see Supplemental Table 5).
Fixed cells were analyzed with an LSR Fortessa (BD Biosciences). Cell populations were defined, gated, and analyzed with FlowJo software (FlowJo LLC). Immune populations were identified by forward and side scatter followed by doublet discrimination of CD45+ viable cells and a FACS strategy modified from Misharin et al. (74). Ly6Chi monocytes were identified as SigFloLy6G–CD11b+CD11c–CD64–. The relative percentages of neutrophils (Ly6G+) and eosinophils (SigFhiLy6G–) present among CD45+ viable BALF cells were analyzed to confirm cytospin staining.
Statistics.
All data are presented with dot plots and group mean ± SEM unless otherwise indicated. Statistical analyses were performed with GraphPad Instat or SigmaStat 13 (Systat Inc.). Student’s t test (1 or 2 tailed as appropriate) was used for data describing 2 groups. Multiple group comparisons were performed by ANOVA using post hoc testing as indicated. Survival analyses were performed using log-rank testing. In all cases, statistical significance was considered at P values less than 0.05.
Study approval.
Mice were housed in pathogen-free facilities, and studies were performed according to protocols approved by the IACUC of the Perelman School of Medicine at the University of Pennsylvania.
Author contributions
MFB and SM developed the concept. MFB, SM, SIN, YT, and AV designed the experiments. YT and MK performed in vivo animal experiments; SIN, AV, JK, SJR, SJ, MB, VN, MFB, and SM performed in vitro experiments and endpoint analyses for in vivo studies; MFB, SIN, JK, and AV analyzed data, generated figures, and interpreted results; SIN and MFB drafted the manuscript; MFB, AV, YT, and SM edited the manuscript. All authors reviewed and approved the final version prior to submission.
Supplementary Material
Acknowledgments
The authors thank Costas Koumenis, Richard Davis IV, and the staff of the SAARP Core Facility in the Department of Radiation Oncology, University of Pennsylvania (supported by NIH S10 RR026587) for assistance in acquiring lung CT images. We thank Hasina Outtz-Reed for assistance with pulse oximetry measurements. MFB is an Albert M. Rose Established Investigator of the Pulmonary Fibrosis Foundation. We thank Edward E. Morrisey and Susan Guttentag for helpful discussions. This work was supported by VA Merit Review I01BX001176 (MFB), NIH R01 HL119436 (MFB), and NIH R01 HL129150 (SM); JK was supported by NIH 2T32 HL007586. No conflicts of interest, financial or otherwise, are declared by the authors. The content of this article does not represent the views of the US Department of Veterans Affairs or the United States Government.
Version 1. 06/19/2018
In-Press Preview
Version 2. 08/13/2018
Electronic publication
Version 3. 08/31/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(9):4008–4024.https://doi.org/10.1172/JCI99287.
See the related Commentary at Lung injury and fibrosis induced by a mutant form of surfactant protein C.
Contributor Information
Shin-Ichi Nureki, Email: nureki@oita-u.ac.jp.
Yaniv Tomer, Email: yaniv.tomer@gmail.com.
Alessandro Venosa, Email: avenosa@mail.med.upenn.edu.
Jeremy Katzen, Email: Jeremy.Katzen@uphs.upenn.edu.
Scott J. Russo, Email: sjrusso@pennmedicine.upenn.edu.
Sarita Jamil, Email: sjamil@sas.upenn.edu.
Matthew Barrett, Email: mab5@princeton.edu.
Vivian Nguyen, Email: vivianlxnguyen@gmail.com.
Meghan Kopp, Email: mkopp223@gmail.com.
Surafel Mulugeta, Email: mulugeta@pennmedicine.upenn.edu.
Michael F. Beers, Email: mfbeers@pennmedicine.upenn.edu.
References
- 1.Travis WD, et al. An official American Thoracic Society/European Respiratory Society statement: Update of the international multidisciplinary classification of the idiopathic interstitial pneumonias. Am J Respir Crit Care Med. 2013;188(6):733–748. doi: 10.1164/rccm.201308-1483ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King TE, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378(9807):1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 3.Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2006;174(7):810–816. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 4.Kropski JA, Lawson WE, Young LR, Blackwell TS. Genetic studies provide clues on the pathogenesis of idiopathic pulmonary fibrosis. Dis Model Mech. 2013;6(1):9–17. doi: 10.1242/dmm.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulugeta S, Nureki S, Beers MF. Lost after translation: insights from pulmonary surfactant for understanding the role of alveolar epithelial dysfunction and cellular quality control in fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol. 2015;309(6):L507–L525. doi: 10.1152/ajplung.00139.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korfei M, et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178(8):838–846. doi: 10.1164/rccm.200802-313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurland G, et al. An official American Thoracic Society clinical practice guideline: classification, evaluation, and management of childhood interstitial lung disease in infancy. Am J Respir Crit Care Med. 2013;188(3):376–394. doi: 10.1164/rccm.201305-0923ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzenstein AL, Myers JL. Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med. 1998;157(4 Pt 1):1301–1315. doi: 10.1164/ajrccm.157.4.9707039. [DOI] [PubMed] [Google Scholar]
- 9.Flaherty KR, et al. Clinical significance of histological classification of idiopathic interstitial pneumonia. Eur Respir J. 2002;19(2):275–283. doi: 10.1183/09031936.02.00182002. [DOI] [PubMed] [Google Scholar]
- 10.Collard HR, et al. Acute exacerbation of idiopathic pulmonary fibrosis. An International Working Group report. Am J Respir Crit Care Med. 2016;194(3):265–275. doi: 10.1164/rccm.201604-0801CI. [DOI] [PubMed] [Google Scholar]
- 11.Song JW, Hong SB, Lim CM, Koh Y, Kim DS. Acute exacerbation of idiopathic pulmonary fibrosis: incidence, risk factors and outcome. Eur Respir J. 2011;37(2):356–363. doi: 10.1183/09031936.00159709. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell TS, et al. Future directions in idiopathic pulmonary fibrosis research. An NHLBI workshop report. Am J Respir Crit Care Med. 2014;189(2):214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idiopathic Pulmonary Fibrosis Clinical Research Network, Raghu G, Anstrom KJ, King TE, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366(21):1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuyts WA, Kolb M, Stowasser S, Stansen W, Huggins JT, Raghu G. First data on efficacy and safety of nintedanib in patients with idiopathic pulmonary fibrosis and forced vital capacity of ≤50 % of predicted value. Lung. 2016;194(5):739–743. doi: 10.1007/s00408-016-9912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King TE, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 16.B Moore B, Lawson WE, Oury TD, Sisson TH, Raghavendran K, Hogaboam CM. Animal models of fibrotic lung disease. Am J Respir Cell Mol Biol. 2013;49(2):167–179. doi: 10.1165/rcmb.2013-0094TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selman M, Pardo A. Revealing the pathogenic and aging-related mechanisms of the enigmatic idiopathic pulmonary fibrosis. an integral model. Am J Respir Crit Care Med. 2014;189(10):1161–1172. doi: 10.1164/rccm.201312-2221PP. [DOI] [PubMed] [Google Scholar]
- 18.Cameron HS, et al. A common mutation in the surfactant protein C gene associated with lung disease. J Pediatr. 2005;146(3):370–375. doi: 10.1016/j.jpeds.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Brasch F, et al. Interstitial lung disease in a baby with a de novo mutation in the SFTPC gene. Eur Respir J. 2004;24(1):30–39. doi: 10.1183/09031936.04.00000104. [DOI] [PubMed] [Google Scholar]
- 20.Abou Taam R, et al. Familial interstitial disease with I73T mutation: A mid- and long-term study. Pediatr Pulmonol. 2009;44(2):167–175. doi: 10.1002/ppul.20970. [DOI] [PubMed] [Google Scholar]
- 21.Crossno PF, et al. Identification of early interstitial lung disease in an individual with genetic variations in ABCA3 and SFTPC. Chest. 2010;137(4):969–973. doi: 10.1378/chest.09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Moorsel CH, et al. Surfactant protein C mutations are the basis of a significant portion of adult familial pulmonary fibrosis in a dutch cohort. Am J Respir Crit Care Med. 2010;182(11):1419–1425. doi: 10.1164/rccm.200906-0953OC. [DOI] [PubMed] [Google Scholar]
- 23.Beers MF, et al. A nonaggregating surfactant protein C mutant is misdirected to early endosomes and disrupts phospholipid recycling. Traffic. 2011;12(9):1196–1210. doi: 10.1111/j.1600-0854.2011.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins A, et al. A non-BRICHOS SFTPC mutant (SP-CI73T) linked to interstitial lung disease promotes a late block in macroautophagy disrupting cellular proteostasis and mitophagy. Am J Physiol Lung Cell Mol Physiol. 2015;308(1):L33–L47. doi: 10.1152/ajplung.00217.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galetskiy D, Woischnik M, Ripper J, Griese M, Przybylski M. Aberrant processing forms of lung surfactant proteins SP-B and SP-C revealed by high-resolution mass spectrometry. Eur J Mass Spectrom (Chichester) 2008;14(6):379–390. doi: 10.1255/ejms.957. [DOI] [PubMed] [Google Scholar]
- 26.Stewart GA, et al. 4-Phenylbutyric acid treatment rescues trafficking and processing of a mutant surfactant protein-C. Am J Respir Cell Mol Biol. 2012;47(3):324–331. doi: 10.1165/rcmb.2012-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beers MF, Kim CY, Dodia C, Fisher AB. Localization, synthesis, and processing of surfactant protein SP-C in rat lung analyzed by epitope-specific antipeptide antibodies. J Biol Chem. 1994;269(32):20318–20328. [PubMed] [Google Scholar]
- 28.Beers MF, Lomax C. Synthesis and processing of hydrophobic surfactant protein C by isolated rat type II cells. Am J Physiol. 1995;269(6 Pt 1):L744–L753. doi: 10.1152/ajplung.1995.269.6.L744. [DOI] [PubMed] [Google Scholar]
- 29.Wang WJ, Mulugeta S, Russo SJ, Beers MF. Deletion of exon 4 from human surfactant protein C results in aggresome formation and generation of a dominant negative. J Cell Sci. 2003;116(Pt 4):683–692. doi: 10.1242/jcs.00267. [DOI] [PubMed] [Google Scholar]
- 30.Fritsch A, et al. A hypomorphic mouse model of dystrophic epidermolysis bullosa reveals mechanisms of disease and response to fibroblast therapy. J Clin Invest. 2008;118(5):1669–1679. doi: 10.1172/JCI34292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holzenberger M, et al. A targeted partial invalidation of the insulin-like growth factor I receptor gene in mice causes a postnatal growth deficit. Endocrinology. 2000;141(7):2557–2566. doi: 10.1210/endo.141.7.7550. [DOI] [PubMed] [Google Scholar]
- 32.Mancini C, Messana E, Turco E, Brussino A, Brusco A. Gene-targeted embryonic stem cells: real-time PCR assay for estimation of the number of neomycin selection cassettes. Biol Proced Online. 2011;13:10. doi: 10.1186/1480-9222-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguire JA, Mulugeta S, Beers MF. Multiple ways to die: delineation of the unfolded protein response and apoptosis induced by surfactant protein C BRICHOS mutants. Int J Biochem Cell Biol. 2012;44(1):101–112. doi: 10.1016/j.biocel.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulugeta S, Nguyen V, Russo SJ, Muniswamy M, Beers MF. A surfactant protein C precursor protein BRICHOS domain mutation causes endoplasmic reticulum stress, proteasome dysfunction, and caspase 3 activation. Am J Respir Cell Mol Biol. 2005;32(6):521–530. doi: 10.1165/rcmb.2005-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons MA, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med. 2011;184(5):569–581. doi: 10.1164/rccm.201010-1719OC. [DOI] [PubMed] [Google Scholar]
- 36.Misharin AV, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214(8):2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belperio JA, et al. The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J Immunol. 2004;173(7):4692–4698. doi: 10.4049/jimmunol.173.7.4692. [DOI] [PubMed] [Google Scholar]
- 38.White ES, et al. Plasma surfactant protein-D, matrix metalloproteinase-7, and osteopontin index distinguishes idiopathic pulmonary fibrosis from other idiopathic interstitial pneumonias. Am J Respir Crit Care Med. 2016;194(10):1242–1251. doi: 10.1164/rccm.201505-0862OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casey J, et al. Alveolar surfactant protein D content modulates bleomycin-induced lung injury. Am J Respir Crit Care Med. 2005;172(7):869–877. doi: 10.1164/rccm.200505-767OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atochina EN, Beck JM, Scanlon ST, Preston AM, Beers MF. Pneumocystis carinii pneumonia alters expression and distribution of lung collectins SP-A and SP-D. J Lab Clin Med. 2001;137(6):429–439. doi: 10.1067/mlc.2001.115220. [DOI] [PubMed] [Google Scholar]
- 41.Korfhagen TR, et al. Cis-acting sequences from a human surfactant protein gene confer pulmonary-specific gene expression in transgenic mice. Proc Natl Acad Sci U S A. 1990;87(16):6122–6126. doi: 10.1073/pnas.87.16.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogee LM, Dunbar AE, Wert S, Askin F, Hamvas A, Whitsett JA. Mutations in the surfactant protein C gene associated with interstitial lung disease. Chest. 2002;121(3 Suppl):20S–21S. doi: 10.1378/chest.121.3_suppl.20s. [DOI] [PubMed] [Google Scholar]
- 43.Bridges JP, Wert SE, Nogee LM, Weaver TE. Expression of a human surfactant protein C mutation associated with interstitial lung disease disrupts lung development in transgenic mice. J Biol Chem. 2003;278(52):52739–52746. doi: 10.1074/jbc.M309599200. [DOI] [PubMed] [Google Scholar]
- 44.Lawson WE, et al. Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc Natl Acad Sci U S A. 2011;108(26):10562–10567. doi: 10.1073/pnas.1107559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pozarska A, et al. Stereological monitoring of mouse lung alveolarization from the early postnatal period to adulthood. Am J Physiol Lung Cell Mol Physiol. 2017;312(6):L882–L895. doi: 10.1152/ajplung.00492.2016. [DOI] [PubMed] [Google Scholar]
- 46.Povedano JM, Martinez P, Flores JM, Mulero F, Blasco MA. Mice with pulmonary fibrosis driven by telomere dysfunction. Cell Rep. 2015;12(2):286–299. doi: 10.1016/j.celrep.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 47.Falke LL, Broekhuizen R, Huitema A, Maarseveen E, Nguyen TQ, Goldschmeding R. Tamoxifen for induction of Cre-recombination may confound fibrosis studies in female mice. J Cell Commun Signal. 2017;11(2):205–211. doi: 10.1007/s12079-017-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carthy JM, et al. Tamoxifen inhibits TGF-β-mediated activation of myofibroblasts by blocking non-Smad signaling through ERK1/2. J Cell Physiol. 2015;230(12):3084–3092. doi: 10.1002/jcp.25049. [DOI] [PubMed] [Google Scholar]
- 49.Redente EF, et al. Age and sex dimorphisms contribute to the severity of bleomycin-induced lung injury and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2011;301(4):L510–L518. doi: 10.1152/ajplung.00122.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lingappan K, Jiang W, Wang L, Couroucli XI, Moorthy B. Sex-specific differences in hyperoxic lung injury in mice: role of cytochrome P450 (CYP)1A. Toxicology. 2015;331:14–23. doi: 10.1016/j.tox.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sisson TH, et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am J Respir Crit Care Med. 2010;181(3):254–263. doi: 10.1164/rccm.200810-1615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia O, et al. Targeted type 2 alveolar cell depletion. a dynamic functional model for lung injury repair. Am J Respir Cell Mol Biol. 2016;54(3):319–330. doi: 10.1165/rcmb.2014-0246OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young LR, et al. The alveolar epithelium determines susceptibility to lung fibrosis in Hermansky-Pudlak syndrome. Am J Respir Crit Care Med. 2012;186(10):1014–1024. doi: 10.1164/rccm.201207-1206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atochina-Vasserman EN, et al. Early alveolar epithelial dysfunction promotes lung inflammation in a mouse model of Hermansky-Pudlak syndrome. Am J Respir Crit Care Med. 2011;184(4):449–458. doi: 10.1164/rccm.201011-1882OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakatani Y, et al. Interstitial pneumonia in Hermansky-Pudlak syndrome: significance of florid foamy swelling/degeneration (giant lamellar body degeneration) of type-2 pneumocytes. Virchows Arch. 2000;437(3):304–313. doi: 10.1007/s004280000241. [DOI] [PubMed] [Google Scholar]
- 57.Ahuja S, et al. MAP1LC3B overexpression protects against Hermansky-Pudlak syndrome type-1-induced defective autophagy in vitro. Am J Physiol Lung Cell Mol Physiol. 2016;310(6):L519–L531. doi: 10.1152/ajplung.00213.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hidvegi T, et al. Enhancing autophagy with drugs or lung-directed gene therapy reverses the pathological effects of respiratory epithelial cell proteinopathy. J Biol Chem. 2015;290(50):29742–29757. doi: 10.1074/jbc.M115.691253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bullard JE, Wert SE, Whitsett JA, Dean M, Nogee LM. ABCA3 mutations associated with pediatric interstitial lung disease. Am J Respir Crit Care Med. 2005;172(8):1026–1031. doi: 10.1164/rccm.200503-504OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naikawadi RP, et al. Telomere dysfunction in alveolar epithelial cells causes lung remodeling and fibrosis. JCI Insight. 2016;1(14):e86704. doi: 10.1172/jci.insight.86704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alder JK, et al. Telomere dysfunction causes alveolar stem cell failure. Proc Natl Acad Sci U S A. 2015;112(16):5099–5104. doi: 10.1073/pnas.1504780112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pesci A, Ricchiuti E, Ruggiero R, De Micheli A. Bronchoalveolar lavage in idiopathic pulmonary fibrosis: what does it tell us? Respir Med. 2010;104(Suppl 1):S70–S73. doi: 10.1016/j.rmed.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 63.Young LR, et al. Epithelial-macrophage interactions determine pulmonary fibrosis susceptibility in Hermansky-Pudlak syndrome. JCI Insight. 2016;1(17):e88947. doi: 10.1172/jci.insight.88947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rankin JA, et al. Phenotypic and physiologic characterization of transgenic mice expressing interleukin 4 in the lung: lymphocytic and eosinophilic inflammation without airway hyperreactivity. Proc Natl Acad Sci U S A. 1996;93(15):7821–7825. doi: 10.1073/pnas.93.15.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee CG, et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1) J Exp Med. 2001;194(6):809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trujillo G, O’Connor EC, Kunkel SL, Hogaboam CM. A novel mechanism for CCR4 in the regulation of macrophage activation in bleomycin-induced pulmonary fibrosis. Am J Pathol. 2008;172(5):1209–1221. doi: 10.2353/ajpath.2008.070832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huaux F, Liu T, McGarry B, Ullenbruch M, Xing Z, Phan SH. Eosinophils and T lymphocytes possess distinct roles in bleomycin-induced lung injury and fibrosis. J Immunol. 2003;171(10):5470–5481. doi: 10.4049/jimmunol.171.10.5470. [DOI] [PubMed] [Google Scholar]
- 68.Beers MF, et al. Aberrant lung remodeling in a mouse model of surfactant dysregulation induced by modulation of the Abca3 gene. Ann Anat. 2017;210:135–146. doi: 10.1016/j.aanat.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao Y, Tao JQ, Bates SR, Beers MF, Haczku A. IL-4 induces production of the lung collectin surfactant protein-D. J Allergy Clin Immunol. 2004;113(3):439–444. doi: 10.1016/j.jaci.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 70.Beers MF, Bates SR, Fisher AB. Differential extraction for the rapid purification of bovine surfactant protein B. Am J Physiol. 1992;262(6 Pt 1):L773–L778. doi: 10.1152/ajplung.1992.262.6.L773. [DOI] [PubMed] [Google Scholar]
- 71.Jain D, et al. SP-D-deficient mice are resistant to hyperoxia. Am J Physiol Lung Cell Mol Physiol. 2007;292(4):L861–L871. doi: 10.1152/ajplung.00145.2006. [DOI] [PubMed] [Google Scholar]
- 72.Maguire JA, Mulugeta S, Beers MF. Endoplasmic reticulum stress induced by surfactant protein C BRICHOS mutants promotes proinflammatory signaling by epithelial cells. Am J Respir Cell Mol Biol. 2011;44(3):404–414. doi: 10.1165/rcmb.2009-0382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henderson NC, et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19(12):1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49(4):503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.