Abstract
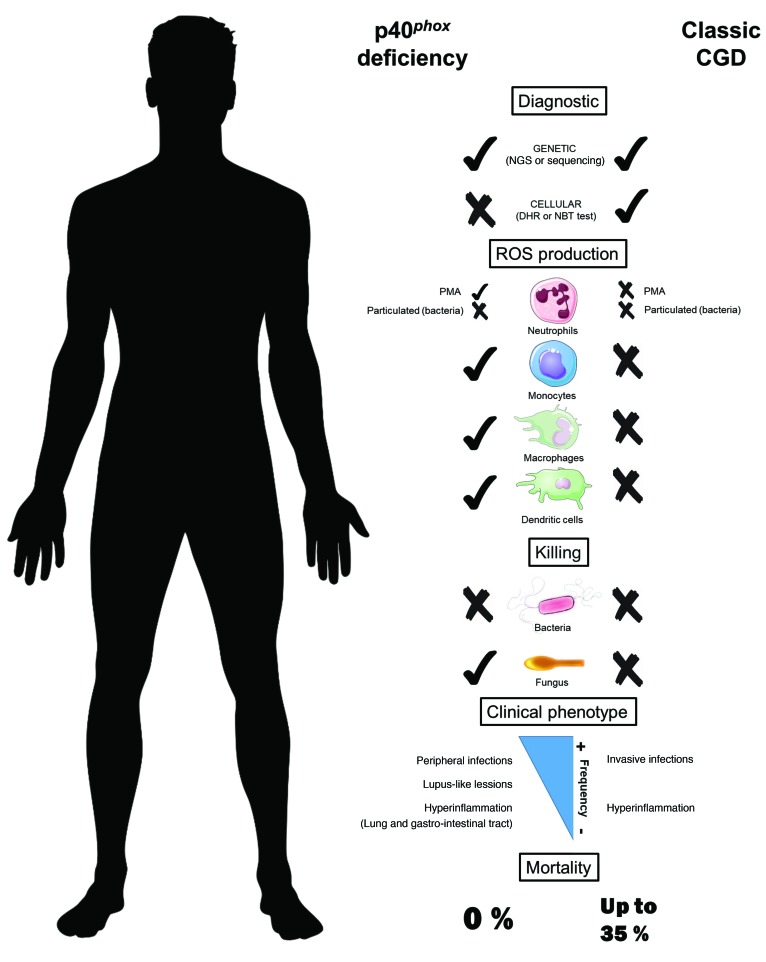
Biallelic loss-of-function (LOF) mutations of the NCF4 gene, encoding the p40phox subunit of the phagocyte NADPH oxidase, have been described in only 1 patient. We report on 24 p40phox-deficient patients from 12 additional families in 8 countries. These patients display 8 different in-frame or out-of-frame mutations of NCF4 that are homozygous in 11 of the families and compound heterozygous in another. When overexpressed in NB4 neutrophil-like cells and EBV-transformed B cells in vitro, the mutant alleles were found to be LOF, with the exception of the p.R58C and c.120_134del alleles, which were hypomorphic. Particle-induced NADPH oxidase activity was severely impaired in the patients’ neutrophils, whereas PMA-induced dihydrorhodamine-1,2,3 (DHR) oxidation, which is widely used as a diagnostic test for chronic granulomatous disease (CGD), was normal or mildly impaired in the patients. Moreover, the NADPH oxidase activity of EBV-transformed B cells was also severely impaired, whereas that of mononuclear phagocytes was normal. Finally, the killing of Candida albicans and Aspergillus fumigatus hyphae by neutrophils was conserved in these patients, unlike in patients with CGD. The patients suffer from hyperinflammation and peripheral infections, but they do not have any of the invasive bacterial or fungal infections seen in CGD. Inherited p40phox deficiency underlies a distinctive condition, resembling a mild, atypical form of CGD.
Keywords: Genetics, Immunology
Keywords: Inflammatory bowel disease, Macrophages, Neutrophils
Introduction
Chronic granulomatous disease (CGD) is a recessive primary immunodeficiency (PID) caused by loss-of-function (LOF) mutations of autosomal or X-linked genes encoding 5 components of the phagocyte-reduced NADPH oxidase complex (1). This enzyme catalyzes the production of ROS in phagocytes. CGD was first described in 1954 (2), the first functional disorder of phagocytes was identified in 1966 (3), and the first genetic etiology of CGD, which is X-linked recessive (XR), was discovered in 1986 in the first positional identification of a PID gene (4, 5). The other genetic etiologies proved to be autosomal recessive (AR) (6). Patients with CGD suffer from recurrent, life-threatening, invasive infections with specific bacteria and fungi (7–10). Severe parasitic infections have rarely been reported in CGD patients, who also seem to have no particular predisposition to severe viral illnesses (11). Systemic hyperinflammation, including granulomatous lesions of the respiratory and gastrointestinal (GI) tracts in particular, is also common (12–14). Patients with CGD may initially present with unexplained granulomatosis, which is associated with a poor prognosis, (13, 15). Autoimmune conditions, such as systemic lupus erythematosus, are rare in patients with CGD (16). The XR (gp91phox [phagocyte oxidase]) form of CGD is clinically more severe than the AR p47phox-deficient form (7, 9, 17). AR p22phox and p67phox deficiencies appear to be as severe as XR CGD (18, 19). Patients are routinely placed on prophylactic antimicrobial therapy, and the only curative treatment widely available is allogeneic hematopoietic stem cell transplantation (HSCT) (20).
The NADPH oxidase in human phagocytes is a complex of at least 5 subunits. The membrane-bound component is a heterodimer consisting of gp91phox and p22phox, encoded by the X-linked CYBB gene and the autosomal CYBA gene, respectively. This heterodimer is known as flavocytochrome b558 and is the catalytic core of the NADPH oxidase complex (21). The second component is a cytosolic heterotrimer composed of p40phox, p47phox, and p67phox, which form the regulatory part of the NADPH oxidase and are encoded by NCF4, NCF1, and NCF2, respectively (22). The membrane-bound and cytosolic components do not interact in steady-state conditions. Upon phagocyte activation, after the phagocytosis of a bacterium or fungus, for example, the 2 components and a small GTPase (Rac1 and/or Rac2) are activated and assemble on the phagosomal membrane to produce superoxide (O2–) in the phagosome (23). This O2– is subsequently converted into hydrogen peroxide (H2O2) and other ROS, which together contribute to the destruction of the phagocytized microorganisms. LOF mutations of CYBB underlie the XR form of CGD, whereas LOF mutations of CYBA, NCF1, and NCF2 underlie the AR forms. Hypomorphic mutations of some of these genes have been shown to underlie variant CGD, in which residual activity does not differ between cell types (24). Hypomorphic CYBB mutations, which are more deleterious in monocyte-derived macrophages (MDMs) than in peripheral phagocytes, have been shown to underlie Mendelian susceptibility to mycobacterial disease (MSMD) (25).
Much less is known about the fifth component, p40phox, as biallelic mutations of NCF4 have only been described once before in a young boy with severe colitis (26). p40phox has 3 domains: PX, PB1, and SH3 (27, 28). During phagocytosis, upregulation of the membrane phospholipid phosphatidylinositol 3-phosphate [PI(3)P] and subsequent high-affinity binding of the p40phox PX domain enhance NADPH oxidase activity (21, 29). The p40phox-deficient patient is compound heterozygous for a premature stop codon and a missense mutation in the PX domain that compromises binding to PI(3)P, which results in the impairment of neutrophil phagocytosis–induced oxidase activity (26). As in classic CGD neutrophils, intracellular oxidant production after stimulation with serum-opsonized zymosan (SOZ), IgG beads, or serum-opsonized Staphylococcus aureus is impaired in the patient’s neutrophils, and S. aureus killing is also defective (20, 26, 30). However, in this patient, unlike in classic CGD patients, the production of O2– by neutrophils in response to stimulation with PMA or formyl-methionyl-leucyl-phenylalanine (fMLF) is normal (26). The killing of S. aureus by neutrophils is also impaired in p40phox–deficient and CGD mice (31). However, p40phox-deficient murine neutrophils display highly impaired ROS production upon stimulation with PMA or heat-killed S. aureus, but not SOZ (32). Moreover, gp91phox- and p47phox-KO mice (33–35) seem to have a less severe form of inflammatory colitis than do p40phox-KO mice (36, 37). The cellular and clinical overlap and differences between human p40phox deficiency and classic CGD are largely unknown. Here, we describe the characteristics of 24 patients from 12 families in 8 countries with biallelic mutations of NCF4.
Results
Biallelic NCF4 mutations in 24 individuals from 12 families in 8 countries.
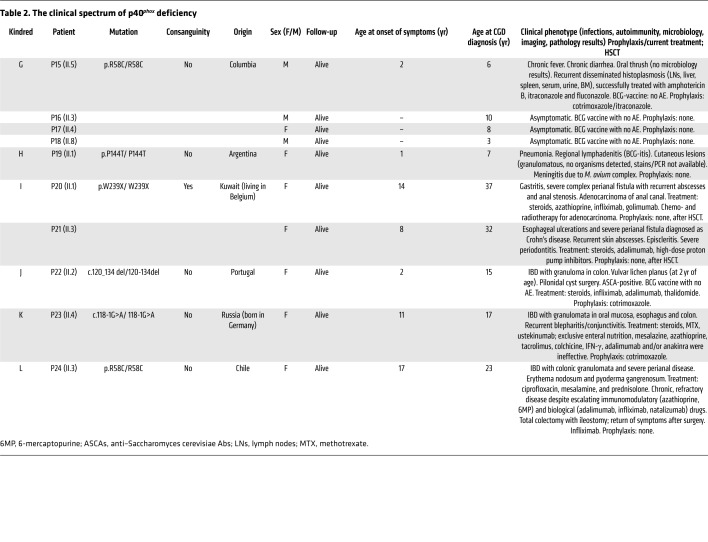
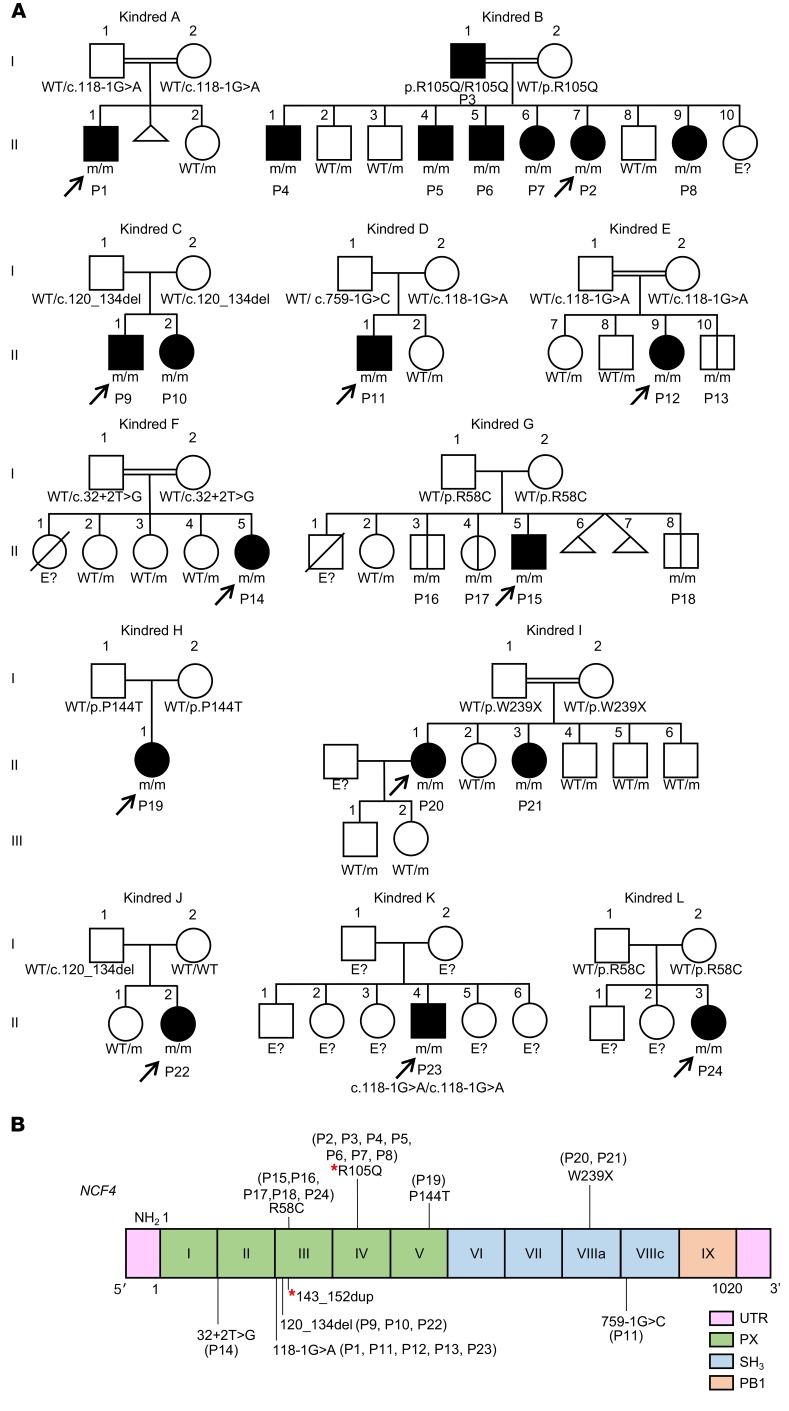
We studied 20 patients from 12 families with clinical features suggestive of CGD (kindreds A–L) (Tables 1 and 2 and Figure 1A). The families originated from Pakistan (kindreds A, B, E, and F), Portugal (kindreds C and J), Australia (kindred D), Colombia (kindred G), Argentina (kindred H), Kuwait (kindred I), Russia (kindred K), and Chile (kindred L). Whole-exome sequencing (WES) or targeted next-generation sequencing (NGS) with a panel of PID genes identified homozygous variants of the NCF4 gene in patients from 11 families: 2 variants affecting essential splice sites (c.118-1G>A in kindreds A, E, and K and c.32+2T>G in kindred F); 3 missense variants (c.314G>A in kindred B, resulting in p.R105Q; c.172C>T in kindreds G and L, resulting in p.R58C; and c.430C>A in kindred H, resulting in p.P144T); 1 nonsense variant (c.716G>A in kindred I, resulting in p.W239X); and 1 in-frame deletion (c.120_134del in kindreds C and J). Finally, we detected compound heterozygous splice-site variants in 1 family (c.118-1G>A and c.759-1G>C in kindred D). All variants were verified by Sanger sequencing. In 11 kindreds, the familial segregation of the alleles was consistent with an AR trait. Kindred J displayed uniparental isodisomy (UPD), as both copies of chromosome 22 were inherited from the father (Figure 1A and ref. 38). Four asymptomatic individuals were shown to carry biallelic mutations (kindreds E and G) (Figure 1A). None of the variants, other than p.R58C (minor allele frequency [MAF] = 0.001), was found in public databases (Exome Aggregation Consortium [ExAC], Human Gene Mutation Database [HGMD], gnomAD, and Biomarker Recognition and Validation Online [BRAVO]) or in our in-house WES database (~4,500 WES). Furthermore, combined annotation-dependent depletion (CADD) scores predicted all mutations to be deleterious, as these scores were above or close to the mutation significance cutoff (MSC) (ref. 24 and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI97116DS1). The 3 missense mutations affect residues located in the PX domain of the p40phox protein (Figure 1B). These residues are highly conserved in other species, suggesting that they have an important function in the p40phox protein (Supplemental Figure 1B). We searched for a founder effect accounting for the presence of the c.120_134del, c.118-1G>A, and p.R58C mutations in different families. The patients homozygous for these mutations were genotyped with the genome-wide array 6.0. The c.120_134del mutation in 2 Portuguese families (kindreds C and J) is within a common haplotype of 884 kb and 1,926 SNPs, demonstrating a founder effect. We dated the most recent common ancestor (MRCA) to 175 years ago (CI: 50–650 years). The c.118-1G>A variant in patients from Pakistan (kindreds A and E) is within a common haplotype of 337 kb and 136 SNPs. The MRCA of these kindreds was dated to 1,950 years ago (CI: 625–8,075 years). The patient from Russia (kindred K) also shared a common haplotype of 152 kb and 41 SNPs in common with the 2 Pakistani patients, suggesting a common ancestor for these 3 patients, and the MRCA was estimated to have lived 6,275 years ago (CI: 2,475–19,000 years) (39). Finally, the c.172C>T (p.R58C) mutation, present in 2 Latin American kindreds, was within a common haplotype of 122 kb and 47 SNPs. The MRCA of these kindreds was dated to 4,425 years ago (CI: 1,375–16,025 years). Collectively, these data suggest that 24 individuals from 12 kindreds in 8 countries suffer from inherited p40phox deficiency, including 20 symptomatic and 4 asymptomatic individuals (Figure 1A).
Table 1. The clinical spectrum of p40phox deficiency.
Table 2. The clinical spectrum of p40phox deficiency.
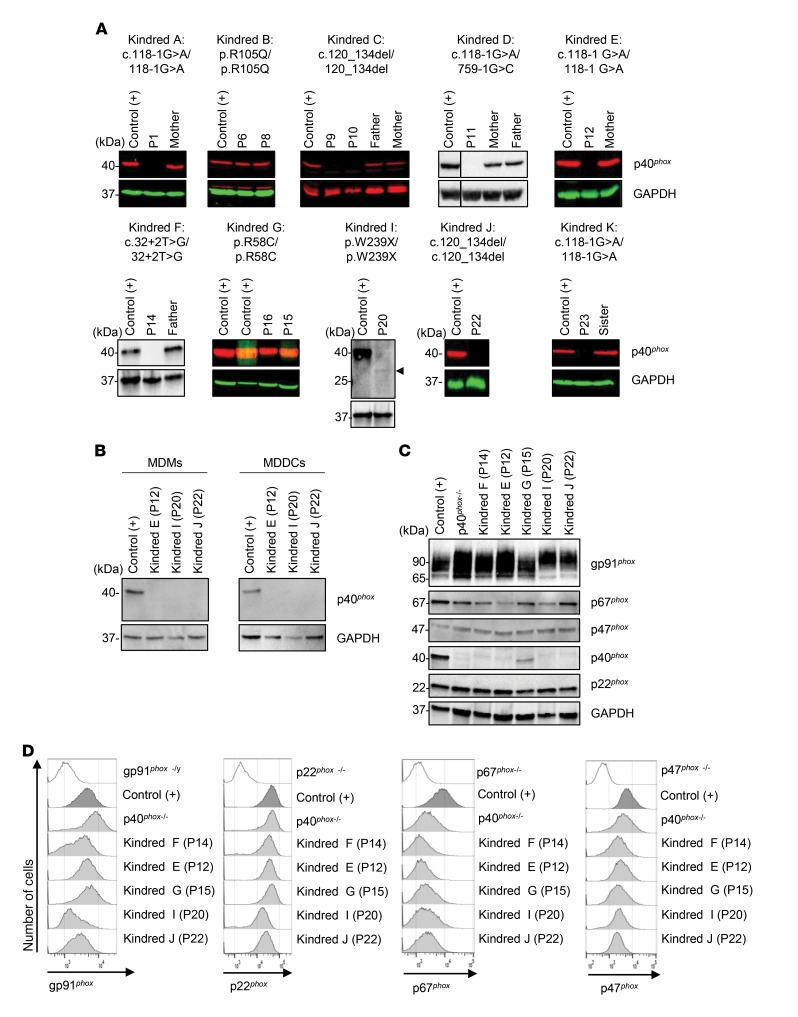
Figure 1. Identification of NCF4 mutations.
(A) Pedigrees of 12 families showing allele segregation. The index cases are indicated by an arrow, and “E?” indicates an unknown genotype. Black or white symbols represent individuals with or without clinical manifestations, respectively. Triangles represent pregnancies not carried to term. m, mutated. (B) Schematic diagram of the structure of the NCF4 gene and p40phox protein, consisting of 9 exons and 3 protein domains, respectively. Newly discovered and previously reported mutations of the NCF4 gene are noted according to their localization. Red asterisks indicate mutations previously reported by Matute et al. (26).
Impaired NCF4 mRNA expression in the patients’ cells.
We measured NCF4 transcript levels in cells from the patients and healthy controls by quantitative real-time RT-PCR (qPCR). EBV-transformed B cell lines (EBV–B cells) and neutrophils homozygous for c.32+2T>G (patient 14 [P14]) produced no NCF4 transcripts detectable by qPCR (Supplemental Figure 2A) or cDNA PCR (data not shown). These findings suggest that transcripts carrying this mutation are subjected to nonsense-mediated mRNA decay. For P12 (c.118-1G>A), P15 (p.R58C), P20 (p.W239X), and P22 (c.120_134del), we found that NCF4 transcript levels in EBV–B cells and neutrophils were lower than those in 10 unrelated controls (Supplemental Figure 2A). Cells homozygous for c.118-1G>A or c.120_134del produced transcripts of lower molecular weight (MW), suggesting the occurrence of alternative splicing. Using TOPO-TA cloning, we detected 3 aberrantly spliced variants for c.118-1G>A, all of which were predicted to encode truncated proteins, and we identified a fourth minor transcript (6%) with a large in-frame deletion, c.118_198del (Supplemental Figure 2B). For c.120_134del, most of the transcripts carried a 5-codon deletion (p.F41_V45del) at the beginning of exon 3, and another 3 aberrantly spliced variants were predicted to encode truncated proteins (Supplemental Figure 2B). Finally, for the c.759-1G>C allele, we performed exon trapping. We obtained 3 spliced variants: 1 with the retention of 37 bp of intron 8a (63%), 1 in which all of intron 8a was retained (26%), and 1 with the retention of 16 bp of intron 8a (11%) (Supplemental Figure 2C).
The mutant NCF4 alleles are deleterious in EBV–B cells.
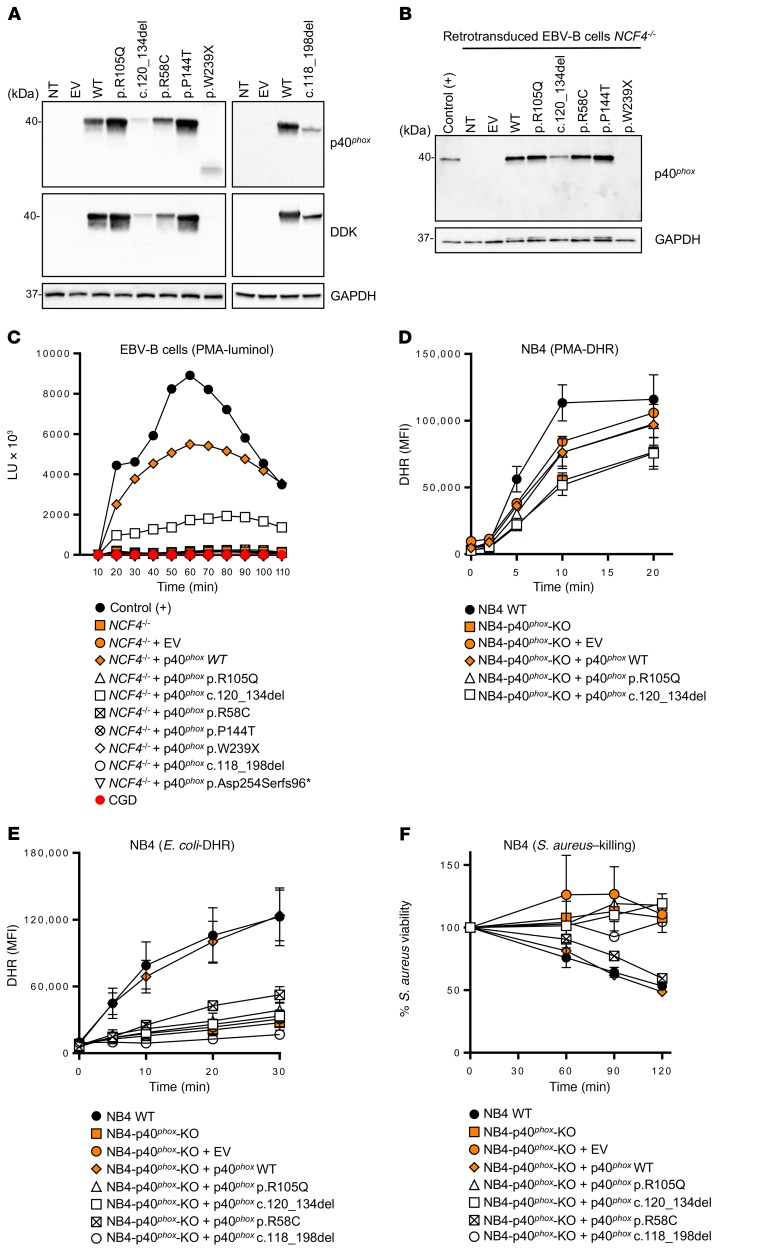
We assessed the potential impact of all individual NCF4 alleles. We used HEK293T cells for analyses of p40phox protein expression, whereas, for analyses of p40phox function, we used EBV–B cells from P14 (homozygous for c.32+2T>G), designated NCF4–/–, as they produced no detectable NCF4 mRNA. We first evaluated protein expression by transfecting HEK293T cells with expression vectors carrying C terminally Flag-tagged WT or mutant NCF4 cDNAs (R58C, R105Q, P144T, W239X, c.118_198del, and c.120_134del). Immunoblot analyses revealed that the WT, R58C, R105Q, and P144T cDNAs encoded a single, full-length 40-kDa protein that was produced in large amounts. By contrast, cells transfected with the in-frame deletions c.118_198del and c.120_134del produced proteins with a lower MW, at much lower abundance. Finally, the protein encoded by W239X cDNA was not detected with an anti-Flag Ab, whereas a truncated protein of approximately 25 kDa gave a low-intensity signal with an anti-p40phox Ab (Figure 2A). We investigated the mode of functioning of the mutant alleles (LOF or hypomorphic) in EBV–B cells, which express all components of the NADPH oxidase complex and produce ROS (25, 26, 40). We transduced NCF4–/– EBV–B cells from P14 with retroviruses for the overexpression of missense (R58C, R105Q, and P144T), in-frame deletion (c.120_134del), and nonsense (W239X) cDNAs. Western blotting confirmed that the missense proteins were expressed normally, whereas c.120_134del was produced in small amounts, and the W239X protein was undetectable (Figure 2B). We then evaluated ROS production in NCF4–/– EBV–B cells by measuring O2– levels after PMA stimulation. EBV–B cells transduced with the R58C, R105Q, P144T, c.118_198del, p.Asp254Serfs96*, and W239X alleles produced no detectable O2–. By contrast, retrotransduction with the c.120_134del variant led to a small, reproducible increase in O2– production (Figure 2C). No H2O2 was generated following the retrovirus-mediated transduction of NCF4–/– EBV–B cells with any of the mutant alleles in these conditions (Supplemental Figure 3A). These data suggest that 6 of the 7 alleles tested were LOF, with the remaining allele (c.120_134del) being severely hypomorphic, at least under these experimental conditions.
Figure 2. p40phox levels and NADPH oxidase activity of the NCF4-mutated alleles.
(A) Western blot of total protein extracts from HEK293T cells expressing NCF4 cDNAs. The upper-panel data were obtained with a polyclonal anti-p40phox Ab, and the lower-panel data were obtained with an anti–DDK (Flag) Ab. An Ab against GAPDH was used as a loading control. (B) Western blot of total protein extracts from healthy control and NCF4–/– EBV–B cells retrovirally transduced with NCF4 cDNAs. A polyclonal anti-p40phox Ab was used, with an Ab against GAPDH as the loading control. NT, not transduced. (C) Production of O2– by NCF4–/– EBV–B cells retrovirally transduced with NCF4 cDNAs upon PMA stimulation. The results are representative of 2 independent assays. LU, luminescence units. (D) DHR reaction of NB4 p40phox-KO cells stimulated with PMA or (E) with E. coli. (F) Killing activity of NB4 p40phox-KO cells infected with S. aureus (n = 2–3). Data represent the mean ± SEM (n ≥3) or the mean only (n <3) and were analyzed using a 2-tailed Mann-Whitney U test. EV, empty vector.
The mutant NCF4 alleles are deleterious in NB4 cells.
We then generated CRISPR-Cas9–KO p40phox NB4 cells, which we allowed to differentiate into neutrophil-like cells. NADPH oxidase activity and S. aureus killing capacity were assessed in p40phox WT cells, NB4-p40phox–KO cells, and in NB4-p40phox–KO cells reconstituted with alleles R105Q, R58C, c.120_134del, or c.118_198del or WT p40phox. We observed no decrease in PMA-induced extracellular H2O2 release or intracellular H2O2 generation in NB4-p40phox–KO cells, as was the case with cells reconstituted with any of the mutant alleles, consistent with the results obtained with neutrophils from the previously reported p40phox-deficient patient (Figure 2D, Supplemental Figure 3B, and ref. 26). Assessments of extracellular H2O2 release in response to serum-opsonized E. coli and A. fumigatus hyphae showed that NADPH oxidase activity was impaired in NB4-p40phox–KO cells (Supplemental Figure 3B). This defect was restored by reconstitution with WT p40phox or the R58C allele, but not with the other 3 mutant alleles (Supplemental Figure 3B). Nevertheless, for E. coli–induced intracellular DHR oxidation, only WT p40phox restored the NADPH oxidase activity (Figure 2E). Moreover, S. aureus killing was impaired in NB4-p40phox–KO cells as well as in cells reconstituted with the mutant alleles, except for the R58C allele (Figure 2F). These experiments suggest that the R105Q, c.120_134del, and c.118_198del alleles are LOF in terms of ROS production and S. aureus killing in NB4 cells. In contrast, the R58C allele is hypomorphic in the same experimental conditions. Collectively, these data indicate that all the NCF4 alleles tested are LOF, with the exception of c.120_134del in EBV–B cells and R58C in NB4 cells, at least upon overexpression.
Impaired p40phox expression in the patients’ phagocytes.
We studied the expression of endogenous p40phox by Western blotting in neutrophils, monocytes, MDMs, and monocyte-derived dendritic cells (MDDCs). We detected no p40phox protein in neutrophils from P1 (c.118-1G>A), P9 or P10 (c.120_134del), P11 (c.118-1G>A/c.759-1G>C), P12 (c.118-1G>A), P14 (c.32+2T>G), P22 (c.120_134del), or P23 (c.118-1G>A). By contrast, neutrophils from P6 and P8 (R105Q) and from P15 and P16 (R58C) had normal levels of p40phox protein. Neutrophils from P20 (W239X) contained a lower-MW protein present at low abundance (Figure 3A), which was consistent with the truncated protein observed upon W239X overexpression in HEK293T cells (Figure 2A). Western blotting also revealed that the levels of the other components of the NADPH oxidase complex in the patients’ neutrophils were normal (Supplemental Figure 4). Finally, we detected no p40phox protein in MDMs or MDDCs from P12, P20, or P22, (Figure 3B). Thus, these mutations result in a loss of p40phox expression in various types of myeloid cells. Collectively, these data are consistent with those obtained for the overexpression of mutant cDNAs in recipient cells.
Figure 3. p40phox and NADPH oxidase subunit expression in p40phox-deficient cells.
(A) Western blot of total protein extracts from the neutrophils of healthy controls, patients, and heterozygous relatives. Abs against p40phox protein were used. Actin or GAPDH protein was used as a loading control. (B) Western blot of total protein extracts from MDMs and MDDCs from a healthy control and p40phox-deficient patients. Detection was done with a polyclonal anti-p40phox Ab, and an Ab against GAPDH was used as a loading control. (C) Western blot of total extracts from EBV–B cells from a healthy control, p40phox-deficient patients, and XR and AR CGD patients. Abs against p40phox protein or gp91phox, p22phox, p67phox, and p47phox were used. Actin or GAPDH protein was used as a loading control. (D) Intracellular detection of gp91phox, p22phox, p67phox, and p47phox in EBV–B cells from a healthy control, p40phox-deficient patients, and XR and AR CGD patients. The results shown are representative of 2 independent assays. p40phox–/– indicates a patient reported in a previous study (26).
Impaired p40phox expression in the patients’ EBV–B cells.
We performed Western blotting to study the expression of the NADPH oxidase subunits in EBV–B cells (26). We observed a complete loss of expression of the p40phox protein in EBV–B cells from P12, P14, P20, and P22. By contrast, p40phox was detected, but at low abundance, in cells from P15 (Figure 3C). The other subunits of the NADPH oxidase complex, gp91phox, p22phox, and p47phox, were normally expressed in the cells of all patients, as shown by Western blotting and flow cytometry with intracellular staining (Figure 3, C and D) and by comparing with expression levels in gp91phox-, p22phox-, and p47phox-deficient patients. However, the expression of p67phox, which interacts with p40phox, was affected in the EBV–B cells (Figure 3D), but not the neutrophils (Supplemental Figure 4). These findings indicate that p40phox is involved in stabilizing p67phox levels in EBV–B cells, but not in neutrophils.
Residual to normal levels of NADPH oxidase activity in p40phox-deficient neutrophils exposed to soluble inducers.
We studied the impact of NCF4 mutations on NAPDH oxidase activity by evaluating ROS production in patients’ cells. The PMA-induced DHR test, which measures intracellular H2O2 generation, is widely used on neutrophils for the diagnosis of classic CGD (1). This test showed a wide range of intracellular ROS production activity in p40phox-deficient neutrophils, from completely normal oxidase activity to residual levels of activity, but always markedly higher than those in classic CGD cells (Figure 4A). We observed a similar pattern at several time points (Supplemental Figure 5A). The release of H2O2 and production of O2– were evaluated after stimulation with PMA, platelet-activating factor (PAF) plus fMLF, or SOZ. We found that H2O2 generation by the patients’ neutrophils was normal or subnormal, whereas neutrophils from patients with classic CGD produced no H2O2 or O2– (Supplemental Figure 4C). Thus, neutrophils from p40phox-deficient patients had normal or impaired, but not abolished, ROS production as shown by extracellular detection tests, contrasting sharply with the results for neutrophils from patients with classic CGD.
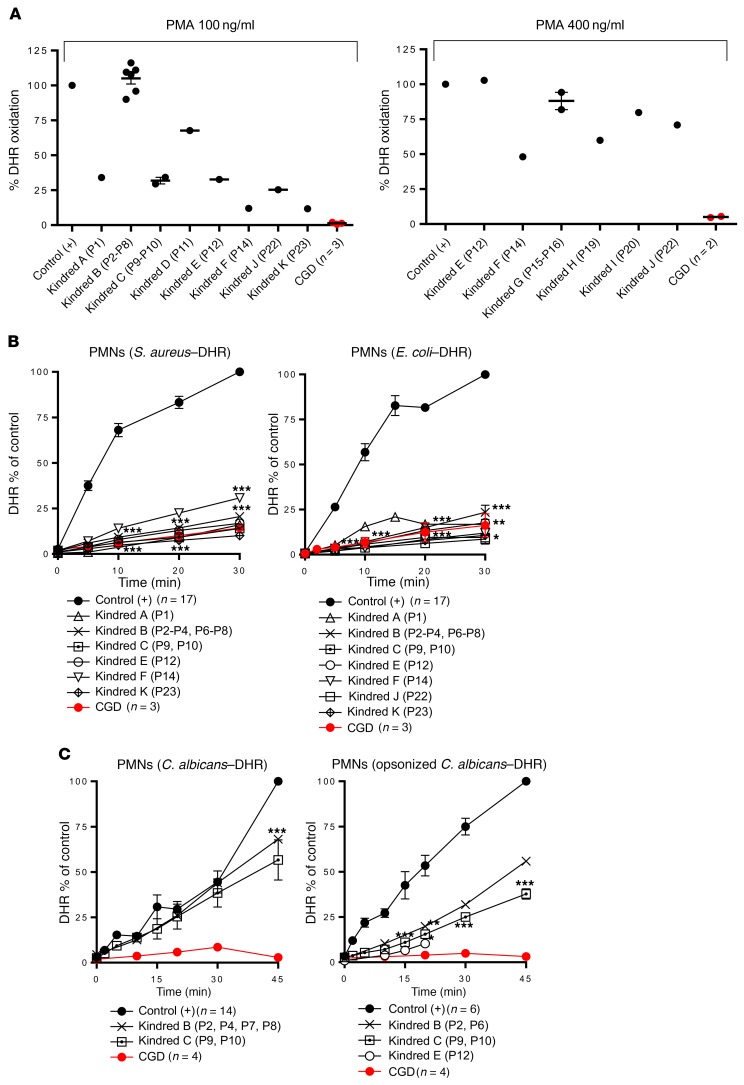
Figure 4. NADPH oxidase activity in p40phox-deficient neutrophils.
(A) Intracellular ROS production by DHR in neutrophils from healthy controls (n = 37), p40phox-deficient patients (n = 17), and CGD patients (n = 5) upon PMA stimulation (100 ng PMA/ml and 400 ng/μl). (B) Particle-induced DHR oxidation in neutrophils from healthy controls (n = 17 and n = 23), p40phox-deficient patients (n = 12 and n = 14), and CGD patients (n = 3 and n = 6) upon stimulation with S. aureus (left) or E. coli (right). (C) DHR oxidation in neutrophils from healthy controls (n = 14 and n = 6), p40phox-deficient patients (n = 6 and n = 5), and CGD patients (n = 4 and n = 3) upon addition of C. albicans, with (left) or without (right) opsonization. PMNs, polymorphonuclear neutrophils. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed Mann-Whitney U test.
Impaired NADPH oxidase activity in p40phox-deficient neutrophils exposed to particulate inducers.
As previously reported in p40phox-KO mice, ROS production in neutrophils in response to particulate stimuli depends on the recruitment of p40phox protein to the endosomes (31). We therefore evaluated intracellular and extracellular ROS production by the patients’ neutrophils following stimulation with serum-opsonized S. aureus, E. coli, A. fumigatus hyphae, or C. albicans. Particle-induced intracellular DHR oxidation after neutrophil stimulation with S. aureus or E. coli was severely impaired in all patients tested, as in neutrophils from patients with classic CGD (Figure 4B). By contrast, DHR oxidation in neutrophils stimulated with unopsonized or serum-opsonized C. albicans conidia was only partially impaired in the p40phox-deficient patients, whereas a more pronounced defect was observed in the neutrophils from patients with classic CGD (Figure 4C). Moreover, extracellular H2O2 release by the neutrophils of p40phox-deficient patients in response to S. aureus, E. coli, or A. fumigatus was only partially impaired (Supplemental Figure 5D). Finally, oxygen consumption was slightly impaired in the cells of all p40phox-deficient patients tested after stimulation with either soluble (PMA) or particulate (E. coli, SOZ) stimuli (Supplemental Figure 5E). We found that neutrophils lacking gp91phox had no activity at all. Heterozygous carriers of p40phox deficiency displayed no defects of ROS production (data not shown). These findings confirm that p40phox plays an essential role in the NADPH oxidase activity of neutrophils upon stimulation with bacteria and, to a lesser extent, fungi, suggesting the translocation of at p40phox to assist in activation of the NADPH oxidase complex in response to these microorganisms.
Normal NADPH oxidase activity in other myeloid cells from the patients.
We demonstrated that p40phox expression in some patients was equally impaired in different types of phagocytes (Figure 3). We analyzed the effects on NADPH oxidase activity in these cells by evaluating ROS production in monocytes triggered by PMA-induced DHR oxidation. Monocytes from 6 patients (P12, P15, P16, P19, P20, and P22) responded similarly to cells from healthy controls (Supplemental Figure 5B). We further investigated the NADPH oxidase activity of MDMs, as previously described (25, 41). MDMs from healthy controls, P12, P20, and P22 had normal or low levels of H2O2 after stimulation, depending on the stimulus, whereas MDMs from a patient with classic CGD released no H2O2 (Figure 5A). Finally, we assessed NADPH oxidase activity in MDDCs, again evaluating H2O2 release. We found that MDDCs from the 3 patients tested (P12, P20, and P22) released normal amounts of H2O2, as shown by comparison with MDDCs from healthy controls, and unlike MDDCs from classic CGD patients (Figure 5B). Overall, these data suggest that p40phox is redundant in monocytes, MDMs, and MDDCs in terms of the NAPDH oxidase activity triggered by PMA. Further experiments assessing particle-induced ROS production by monocytes, MDMs, and MDDCs are required to determine the extent of this redundancy.
Figure 5. NADPH oxidase activity in p40phox-deficient mononuclear cells.
(A) Extracellular H2O2 production by MDMs from healthy controls (n = 5), p40phox-deficient patients (n = 3), and CGD patients (n = 1) upon PMA stimulation (with and without IFN-γ priming). (B) Extracellular H2O2 production by MDDCs from healthy controls (n = 5), p40phox-deficient patients (n = 3), and CGD patients (n = 1) upon PMA stimulation (with and without LPS priming). (C) Production of O2– (left) and release of H2O2 (right) by EBV–B cells from healthy controls (n = 2), p40phox-deficient patients (n = 6), and CGD patients (n = 1) upon PMA stimulation, assessed on the basis of luminol bioluminescence or Amplex Red test. The results are representative of 2 independent assays. Data represent the mean ± SEM. NS, no stimulation.
Impaired NADPH oxidase activity in EBV–B cells from patients.
We tested EBV–B cells from the p40phox-deficient patients studied here, patients with classic CGD, and the first reported patient with p40phox deficiency (42). In luminol bioluminescence assays, EBV–B cells from P12, P14, P15, and P20 displayed severely impaired NADPH oxidase activity (Figure 5C). By contrast, EBV–B cells from P22 (with c.120_134del) displayed residual luminescence, consistent with the results obtained following the retrovirus-mediated transduction of NCF4–/– EBV–B cells with the c.120_134del allele (Figure 2C). This allele is, therefore, also constitutively hypomorphic. We then evaluated the release of H2O2 in EBV–B cells from P12, P14, and P20. H2O2 detection was completely negative, as in cells from classic CGD patients. By contrast, EBV–B cells from P22 and P15 displayed a residual release of H2O2 (Figure 5C). Finally, EBV–B cells from P12, P14, P15, and P20 were transduced with a p40phox WT cDNA. The rescue of p40phox protein expression was confirmed by immunoblotting before functional assays (Supplemental Figure 6A). Production of O2– in response to PMA stimulation was restored in the transduced EBV–B cells (Supplemental Figure 6B). These experiments with EBV–B cells demonstrate that p40phox is not redundant for NADPH oxidase complex activity in these cells. They also confirm that 1 of the 8 NCF4 alleles is severely hypomorphic.
Impaired killing of Staphylococcus, but not of fungi, by the patients’ cells.
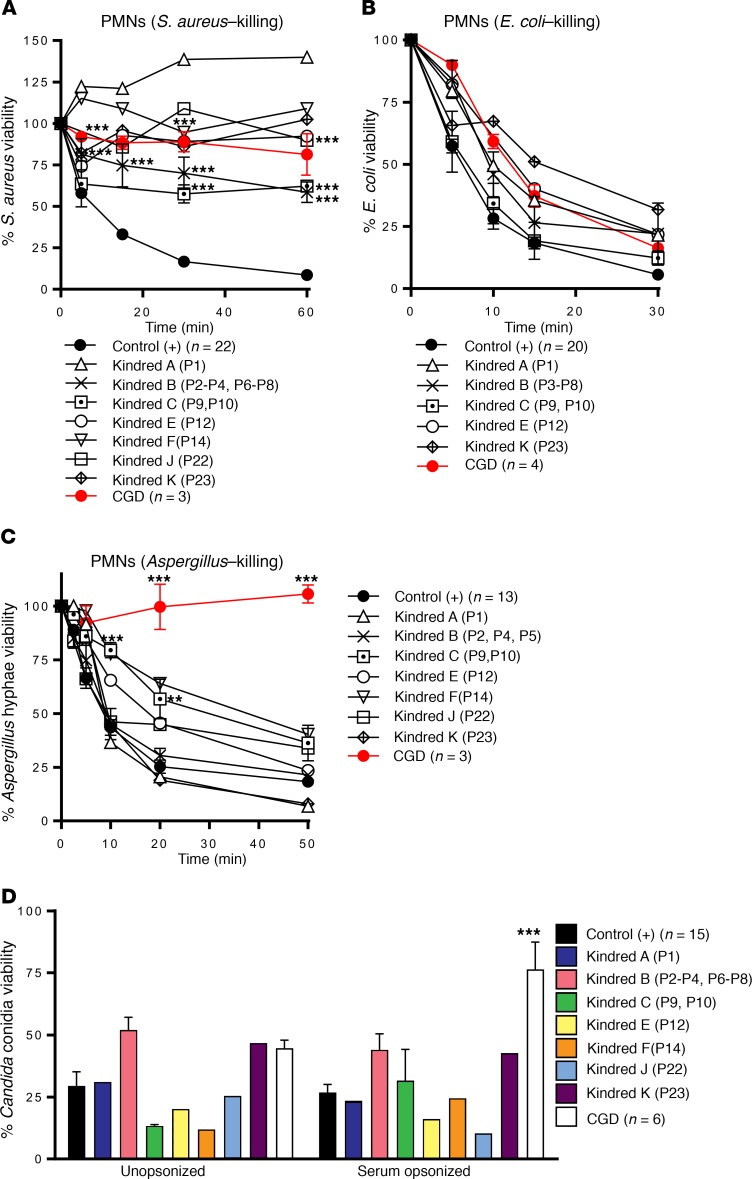
We then assessed the ability of the patients’ neutrophils to kill various microorganisms that commonly cause disease in patients with classic CGD. We found that the phagocytosis of serum-opsonized E. coli, S. aureus, C. albicans, and SOZ by the patients’ neutrophils was similar to that by neutrophils from healthy controls and patients with classic CGD (data not shown). Nonoxidative phagocyte killing involves the release of the proteolytic content of granules into the phagosome or at the neutrophil plasma membrane. In the p40phox-deficient neutrophils tested, protease release from the azurophilic granules (elastase, cathepsin G) in the dye-quenched–BSA (DQ-BSA) assay did not differ from that of neutrophils from healthy controls (data not shown) or patients with classic CGD (43). We therefore analyzed the phagocyte-mediated killing of different species of bacteria and fungi. The killing of S. aureus by the patients’ neutrophils was markedly impaired for all the patients tested, similar to that observed for the classic CGD neutrophils (Figure 6A and ref. 44). E. coli killing was unaffected in the patients’ neutrophils, as in those from patients with classic CGD (Figure 6B and ref. 45). By contrast, the killing of A. fumigatus was normal or subnormal, whereas neutrophils from patients with classic CGD were completely incapable of killing these hyphae (Figure 6C). C. albicans killing by the patients’ neutrophils was similar to that by the neutrophils of healthy control cells when C. albicans was opsonized, whereas neutrophils from patients with classic CGD were unable to kill this fungus (Figure 6D). Moreover, we observed that inhibition of the germination and hyphenation of Candida conidia were similar for neutrophils from the patients and neutrophils from healthy controls (Supplemental Figure 7). Finally, we evaluated the formation of neutrophil extracellular traps (NETs), a process for which ROS have been shown to be essential (46). We assessed NET formation in response to PMA in 3 patients from kindreds A, B, and C. Only neutrophils from kindred B (expressing p40phox protein) produced NETs. We detected no NET production in the other p40phox-deficient families tested (without p40phox protein expression) or for gp91phox-deficient CGD neutrophils (Supplemental Figure 8). These findings suggest that NET formation is dependent on p40phox expression. Collectively, these data indicate that p40phox-deficient neutrophils have normal fungus-killing capacities, unlike classic CGD neutrophils.
Figure 6. Pathogen killing activity by neutrophils from p40phox-deficient patients.
(A) S. aureus viability was measured in infected neutrophils from healthy controls (n = 22), p40phox-deficient patients (n = 13), and CGD patients (n = 3). (B) E. coli viability was measured in infected neutrophils from healthy controls (n = 20), p40phox-deficient patients (n = 10), and CGD patients (n = 4). (C) A. fumigatus hypha viability was measured at different time points with infected neutrophils from healthy controls (n = 13), p40phox-deficient patients (n = 13) and CGD patients (n = 3). (D) C. albicans conidial viability was measured with infected neutrophils from healthy controls (n = 15), p40phox-deficient patients (n = 12), and CGD patients (n = 6). Data represent the mean ± SEM (n ≥3) or the mean only (n <3). (A–C) **P < 0.01 and ***P < 0.001, by 2-tailed Mann-Whitney U test. (D) ***P < 0.001, by 2-tailed Mann-Whitney U test, and P < 0.0063, by 2-tailed Mann-Whitney U test with Bonferroni’s correction for multiple comparisons.
Inflammatory lesions in patients with p40phox deficiency.
Deficiency of p40phox is not characterized by the invasive bacterial and fungal infections seen in patients with classic CGD, but mostly by inflammatory manifestations as well as autoimmunity and peripheral infections (Tables 1 and 2 and Figure 7A). We identified 20 symptomatic patients, of whom 10 (50%) presented with skin inflammation (P1, P2, P3, P5, P6, P7, P8, P11, P19, and P24), with lupus-like cutaneous lesions (P1, P6, and P8) or discoid lupus (P3, P7, and P11). We found inflammatory, granulomatous GI manifestations in 10 patients (50%), and these included oral ulcers (P1 and P10), abscesses/granulomas (P1, P2, P3, P4, P5, P6, P7, P8, P9, P10, P11, P23, and P24), periodontitis (P9 and P21), gingivitis or gum inflammation (P9, P10, P20, P21), esophagitis (P21 and P23), gastritis (P20), Crohn-like inflammatory bowel disease (IBD) (P1, P11, P21, P22, P23, and P24), and perianal abscesses/fistula (P11, P20, P21, and P24). P24 suffered from chronic and severe IBD requiring immunosuppressive treatment and a total colectomy. Some lesions were probably both infectious and inflammatory, as is often seen in CGD. Three patients (15.7%) (P12, P14, and P19) reportedly suffered from pulmonary infections, including P12, who was later diagnosed with interstitial lung disease that responded well to systemic steroids rather than antibiotics, suggesting an autoinflammatory component. Ab evaluations were performed in some patients (n = 7) suffering from exacerbated inflammation. P1, P4, P5, P7, P8, and P12 were negative for antinuclear Abs (ANAs). By contrast, P14 was positive for ANAs and antineutrophil cytoplasmic Abs (ANCAs), suggesting autoimmune activity in this patient.
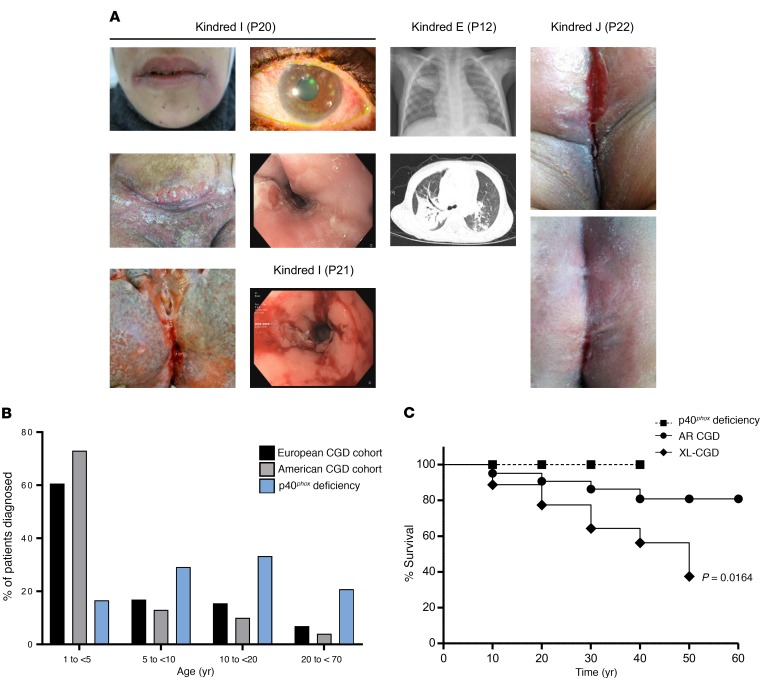
Figure 7. Clinical manifestation and outcome of p40phox-deficient patients.
(A) P20 images show cheilitis, episcleritis, severe chronic cutaneous lesions, and esophagitis. P21 images show esophagitis. P12 images show multifocal consolidations and infiltrates in both the lungs on x-ray and thorax on CT scan. P22 images show vulvar lichen planus before (top) and after (bottom) steroid treatment. (B) Distribution of age at diagnosis of CGD in the American (ref. 7) and European (ref. 9) cohorts and of the p40phox-deficient patients in this study. (C) Kaplan-Meier survival curve compiling survival data on 363 CGD patients (n = 240 XR CGD and n = 123 AR CGD) from refs. 7 and 19 and on the p40phox-deficient patients studied here.
Peripheral but not invasive infections in patients with p40phox deficiency.
We investigated the possible infectious causes of pulmonary manifestations in the cohort. No pathogen was identified in P19. Cutaneous infections were reported in 8 patients (42%) (P1, P4, P6, P7, P9, P11, P21, and P23), 3 of whom had at least 1 proven episode of S. aureus infection (P1, P11, and P23). One patient (P15) had two episodes of disseminated histoplasmosis, which was successfully treated with antifungal drugs and steroids. Ten patients from six different families were vaccinated against tuberculosis with live bacillus Calmette-Guérin (BCG) (Tables 1 and 2). No adverse effects were reported in 9 patients, whereas P19 developed local lymphadenitis (BCG-itis). This same patient subsequently suffered from meningitis caused by Mycobacterium avium. Interestingly, this patient was also heterozygous for a dominant-negative LOF mutation of STAT1, which is responsible for MSMD (47). Four asymptomatic children, aged 1, 3, 8, and 10 years at diagnosis of p40phox deficiency, were found to have homozygous NCF4 mutations (P13, P16, P17, and P18), demonstrating incomplete clinical penetrance of the disease, at least until adolescence. In summary, p40phox deficiency conferred a variable phenotype marked by noninvasive peripheral infections, mostly of the skin and lungs, and inflammation, mostly of the GI tract (Tables 1 and 2).
Favorable clinical outcome of p40phox-deficient individuals.
Inherited p40phox deficiency differs from classic CGD in terms of cellular phenotype and clinical manifestations, but also in terms of clinical outcome (Supplemental Table 1). The mean age at symptom onset was 6 years (range: 1–17 years), later than what was reported for symptom onset in most patients with classic CGD (Supplemental Table 2). The mean age at diagnosis (for patients with symptoms) was 15.3 years (range: 1–46 years), whereas classic CGD is typically diagnosed before the age of 5 years (Figure 7B and Supplemental Table 2). All patients included in this study are currently alive and are between the ages of 1 and 47 years (mean: 15.3 years), whereas a mortality of close to 4% per year has been reported for classic CGD (6, 17), with the highest mortality rates during the first 2 decades of life. The Kaplan-Meier curves for the survival of the largest classic CGD cohort and this p40phox-deficient cohort differed significantly when comparing XR-CGD and p40phox deficiency (P = 0.0164, Mantel-Cox analysis) (Figure 7C and Supplemental Table 2). Fourteen patients were prescribed antibiotic prophylaxis. The treatments administered for hyperinflammation included corticosteroids, azathioprine, methotrexate, infliximab, adalimumab, golimumab, ustekinumab, and hydroxychloroquine. Given the severity of clinical manifestations, 4 patients underwent allogeneic HSCT (P11, P14, P20, and P21), resulting in the resolution of symptoms in all these patients. Overall, p40phox deficiency is a severe condition, but its clinical outcome is nevertheless better than that of classic CGD.
Discussion
We report the molecular, cellular, and clinical characteristics of 24 patients from 12 families with biallelic mutations of the NCF4 gene, which encodes the NADPH oxidase p40phox protein. Deficiency of p40phox is caused by homozygous (23 patients, 11 families) or compound heterozygous (1 patient) mutations in families from 8 countries. We identified 8 previously undescribed alleles of the NCF4 gene, 6 of which are completely LOF and caused complete p40phox deficiency in the corresponding patients. Upon overexpression, the allele c.120_134del is hypomorphic in EBV–B cells, and the allele R58C is hypomorphic in NB4 cells. However, we detected no clinical difference between the patient bearing these variants and the other patients. We further characterized phagocytic cells from the p40phox–deficient patients and compared them with those of patients with other molecular causes of CGD to define more clearly the contribution of human p40phox to NADPH oxidase activity. Interestingly, NCF4 biallelic mutations were found to result in a cellular phenotype related to but clearly different from that of other forms of CGD. In particular, the PMA-induced DHR oxidation test widely used for the diagnosis of CGD gave normal or just below normal values for patients with inherited p40phox deficiency. Diagnosis of p40phox deficiency can be achieved using serum-opsonized E. coli as a stimulus in routine DHR- or Amplex Red–based tests, which must be performed in a diagnostic laboratory with expertise in analysis of NADPH oxidase activity. A genetic approach based on Sanger sequencing or NGS of NCF4 might also facilitate the diagnosis of p40phox deficiency.
The clinical phenotype of patients with p40phox deficiency is also related to but different from that of patients with classic CGD. The patients in this cohort had peripheral infections (mostly staphylococcal) and hyperinflammation (mostly granulomatous lesions of the GI tract and skin). Nevertheless, the frequency of inflammatory lesions is lower than in patients with classic CGD (48). A few of the patients showed autoimmunity, but, strikingly, none had the invasive bacterial and fungal infections commonly seen in patients with classic CGD (7–9, 18). Consistent with these findings, the clinical outcome of p40phox deficiency is better than that of classic CGD. No deaths were recorded in this cohort of patients aged 1 to 46 years, whereas classic CGD–related mortality may reach 35% in its XR and 15% in its AR forms in the first 3 decades of life (10, 17). Moreover, 4 patients remained asymptomatic at 1 to 10 years of age, and the age at clinical onset ranged from 1 to 17 years in patients with symptoms. The incomplete clinical penetrance observed at 1 to 10 years of age also contrasts strongly with observations for classic CGD (7–9, 18). Thus, the healthy siblings of probands should be tested immunologically and genetically. Curative and preventive treatments for CGD are based on prolonged treatment with regimens of antibiotics, antifungal drugs, exogenous IFN-γ, and, increasingly, allogeneic HSCT. Decisions concerning allogeneic HSCT for patients with p40phox deficiency should be made on a case-by-case basis, given the milder overall phenotype and greater variability between patients. Likewise, the immediate initiation of antifungal prophylaxis may not be required, whereas treatment with immunosuppressive agents may be indicated.
One patient with disseminated histoplasmosis (P15) was included in this cohort. However, to our knowledge, no patient with CGD has ever been reported to suffer from this fungal disease. This situation is reminiscent of that of tuberculosis, which is actually much more common than initially thought in CGD patients living in areas of endemic disease (49). Its pathogenesis has been attributed to defects of NADPH oxidase activity in mononuclear phagocytes (25). The normal NADPH oxidase activity of the p40phox-deficient mononuclear phagocytes tested suggests that histoplasmosis may actually be due to the disruption of NADPH oxidase activity in tissue-resident macrophages and DCs, which were not tested in patients with classic CGD or p40phox deficiency (50). Another patient in this cohort (P19) had BCG and M. avium diseases. BCG disease has been reported in CGD patients, but cases of M. avium complex infection have rarely been reported in this population. Moreover, these 2 mycobacterial infections are probably not related to p40phox deficiency in P19, because this patient also carries a dominant-negative STAT1 mutation responsible for MSMD (47). Finally, 1 of the patients in our cohort (P20) had adenocarcinoma, possibly related to chronic Crohn’s colitis, although other factors may also be involved.
Other clinical features of the patients can be understood in light of their cellular phenotypes. In vitro S. aureus killing by neutrophils from patients with p40phox deficiency or classic CGD was severely impaired, whereas E. coli killing by neutrophils from these patients was similar to that by neutrophils from healthy controls. S. aureus killing is a ROS-dependent process, whereas E. coli killing is relatively ROS independent (45, 51). The absence of invasive S. aureus infections in the p40phox-deficient patients was unexpected and remains unexplained, as both in vitro S. aureus–induced oxidase activity and S. aureus killing are as severely impaired in these patients as in those with classic CGD, who do suffer from invasive S. aureus infections. The almost normal killing of fungi by neutrophils from p40phox-deficient patients, even though the killing of Candida and Aspergillus is ROS dependent (52, 53), is of direct clinical relevance and contrasts with the situation in patients with classic CGD (54). The lack of invasive fungal infections is highly consistent with our in vitro biochemical findings for phagocytic cells from p40phox-deficient patients. Residual NADPH oxidase activity was generally higher in p40phox-deficient cells than in classic CGD cells, and in vitro fungal killing was largely unaffected. This situation contrasts with that for classic CGD, in which invasive fungal infections are among the most feared and life-threatening complications, despite antimicrobial prophylaxis (10), and for which fungus killing is impaired in vitro. Given the importance of p40phox-PX interactions with PI(3)P in the phagosome for the NADPH oxidase activity elicited by bacteria, the lack of effect on C. albicans–induced ROS production and killing is surprising. Normal fungal killing in these patients may also result from the residual fungus-induced NADPH oxidase activity observed, suggesting a p40phox-independent mechanism for ROS production that is preferentially triggered by fungi. This implies that ROS production would have to exceed a certain threshold for effective fungal killing. Further studies of late phagosome formation in relation to p40phox localization and ROS formation might clarify the differences in S. aureus and C. albicans killing, which diverge in patients with p40phox deficiency, but not in those with classic CGD.
Interestingly, our studies of monocytes, MDMs, and MDDCs suggest that these cell types are not dependent on p40phox for PMA-induced NADPH oxidase activity. This situation contrasts with that for MSMD-causing gp91phox mutations, which disrupt NADPH oxidase activity in MDMs but not in other phagocytes (neutrophils, monocytes, and MDDCs) (25, 41). The conservation of NADPH oxidase activity may explain the milder infectious phenotype of p40phox-deficient patients than of patients with classic CGD, including the absence of invasive bacterial and fungal infections. This feature may be specific to human cells, as p40phox-mutant mouse macrophages (either KO or R58A mice) have impaired NADPH oxidase activity upon stimulation with a physiological stimulus (36). The PX domain of p40phox is thought to have a high affinity for and specificity of binding to PI(3)P, a lipid on phagosomal membranes, leading to enhanced NADPH oxidase activity. Further studies are required to evaluate NADPH oxidase activity in phagocytic cells other than neutrophils in patients with p40phox deficiency or classic CGD. In this respect, approaches involving the derivation of different types of phagocytes from induced pluripotent stem cells may be particularly effective (55).
Excessive granulomatous inflammation of the skin and GI tract was the most common clinical phenotype observed in this cohort of patients. Systemic lupus–like symptoms are rare in patients with CGD (7, 12, 20) and in the p40phox-deficient patients reported here. Enhanced inflammation has also been reported in mice with p40phox deficiency (36, 37, 56). Mouse p40phox has been reported to be required for the expression of glycan structure–modifying enzymes, which may underlie the recruitment of neutrophils to resolve intestinal inflammation and facilitate wound healing (37). Mouse macrophages with p40phox p.R58A/R58A or KO mutations also produce smaller than normal amounts of ROS (36). The excessive inflammation seen in classic CGD and p40phox deficiency indicates that ROS play a role in immune regulation (12, 57). Patients with p40phox deficiency display at least as much inflammation as those with classic CGD, but have higher levels of ROS production. The ROS produced by neutrophils may contribute to the clearance of apoptotic cells or cell debris, and their deficiency may contribute to inflammation in various ways, including aberrant efferocytosis, for example (58–62). Alternatively, the inflammation observed in classic CGD and p40phox deficiency may be due to abnormal ROS production by circulating B cells. This possibility was already suggested in the first report describing a p40phox-deficient patient (42). Our results also suggest that NADPH activity is defective in all p40phox-deficient EBV–B cells tested, confirming that p40phox is not redundant in human B cells. An understanding of the excessive inflammation in patients with classic CGD or p40phox deficiency will require further studies of the respiratory burst in various other cell types, including tissue-resident DCs and macrophages, and in different types of circulating B cells.
Methods
Cell lines.
NB4 cells were obtained from DSMZ (catalog ACC 207). HEK293T cells were obtained from the American Tissue Culture Collection (ATCC) (catalog CRL-3216).
Ion torrent sequencing, WES, copy number variation, and Sanger sequencing.
We extracted genomic DNA from blood samples with the iPrep PureLink gDNA Blood Kit and iPrep Instruments (Life Technologies, Thermo Fisher Scientific). An AmpliSeq Custom Panel (Thermo Fisher Scientific) was used in families A–E, J, and K. A targeted NGS panel for PID was used in kindred I (63) and another for IBD was used in kindred J (64).
WES was performed with 3 μg DNA from kindreds F, G, H, and L. Exome capture was performed with the SureSelect Human All Exon 72 Mb Kit (Agilent Technologies). SNP arrays for CNV or ancestral founder effect were used with the Genome-Wide Human SNP Array 6.0 (Affymetrix) in kindreds A, C, E, G, J, K, and L.
For confirmation of the mutations detected and the analysis of familial segregation for kindred members, we amplified the flanking regions of the variants obtained, including part of the intronic regions for all NCF4 exons studied. Primers and conditions are described in detail in Supplemental Table 3.
NADPH oxidase functional assays.
NADPH oxidase activity on neutrophils and NB4 cells was assessed by measuring extracellular H2O2 release with an Amplex Red Kit (Life Technologies, Thermo Fisher Scientific). Cells were stimulated with PMA (100 ng/ml), PAF (1 μM) followed by fMLF (1 μM) (all from Sigma-Aldrich), E. coli (OD600 0.1, strain ML-35 in the presence of 2 mM azide), and S. aureus (OD600 0.1, strain Oxford in the presence of 2 mM azide) or A. fumigatus hyphae (clinical isolate). We used 0.25 × 106 cells/ml, and the experiments were performed as previously described (65). The same method was used for the MDMs, MDDCs, and EBV–B cells, except that 400 ng/ml PMA was also added to the cells. The above-mentioned reagents and E. coli, S. aureus, and A. fumigatus strains were used in all experiments. Particles were opsonized with 10% (v/v) pooled human serum. MDMs and MDDCs were obtained by differentiation as previously described (25, 41). MDMs were cultured for 16 to 18 hours with 1 mg/ml purified protein derivatives (PPDs) (tuberculin PPD Batch RT49, Statens Serum Institute) or 5 × 103 IFN-γ IU/ml (Imukin, Boehringer Ingelheim) and then washed and activated by incubation for 30 minutes with PMA, or left unactivated. MDDCs were treated with LPS (1 μg/ml Salmonella minnesota, Sigma-Aldrich), alone or immediately before treatment with PMA.
The production of intracellular ROS by neutrophils, monocytes, and NB4 cells was analyzed with DHR (Thermo Fisher Scientific), as previously described (66). We added 0.5 μM DHR in DMSO to 2.5 × 106 neutrophils. PMA (100 ng/ml), serum-opsonized E. coli or S. aureus (OD600 0.1 in the presence of 2 mM azide), C. albicans (OD625 0.1, strain SC5314), or SOZ (0.1 mg/ml) was then added. At the desired time points, samples were added to STOP buffer (0.5% [w/v] PFA, 1% [w/v] BSA, and 20 mM NaF in PBS), and the amount of rhodamine-1,2,3 (mean MFI [mean fluorescence intensity]) was assessed by flow cytometry using a FACSCanto II/LSR (BD Biosciences) and expressed as the percentage of rhodamine-1,2,3 produced by healthy control cells.
ROS production by monocytes was analyzed by assessing DHR after stimulation with PMA (400 ng/ml) in kindreds E, F, G, H, and J, using a Galios FACS Analyzer (Beckman Coulter) and an LSR Fortessa Cell Analyzer (BD). DHR is expressed as a percentage of the control, and MFI values were normalized against the healthy control.
Generation of NB4 p40phox-KO cells.
The Optimized CRISP Design Tool (http://crispr.mit.edu/) was used (Zhang laboratory) to determine the Cas9 target sites present in the coding sequence of p40phox. An oligomer was cloned into the BsmBI site of pLentiCRISPR v2. The constructs were grown in E. coli Stbl3. Lentiviral particles were generated by the transient cotransfection of HEK293T cells with pLentiCRISPRv2, p40phox-KO, psPAX2, and pCMV-VSVg using TransIT-LT1 Transfection Reagent (Mirusbio). The day after transfection, the cells were plated on NB4 medium. The virus-containing supernatant was harvested on days 2 and 3 after transfection and filtered through 0.45-μm pores, and 1 ml was used with 5 × 105 NB4 cells on 2 successive days. Transduced NB4 cells were selected with 1 μg/ml puromycin (InvivoGen). Surviving cells were plated on limiting dilution medium in a 96-well plate at a density of 0.5 cells/150 μl, and growing clones were routinely maintained. The p40phox levels of these clones were assessed by Western blotting. A clone with no p40phox expression was obtained when the 5′-GTCAGCAATGTTGGCCGAGA-3′ target sequence was used.
Statistics.
Statistical analysis was performed with GraphPad Prism, version 7.02 (GraphPad Software). All data sets were considered non-normally distributed, as the numbers were too low for normality tests. A 2-tailed Mann Whitney U test was used for statistical analysis of 2-group comparisons: e.g., controls vs. kindred A, controls vs. kindred B, etc. A P value of less than 0.05 was considered statistically significant. For data sets containing multiple independent variables, the Bonferroni correction for multiple testing was applied, adjusting the P value depending on the number of variables. When the sample number was 3 or more, error bars were added to the graph. Where no error bars are visible, the SEM was smaller than the symbol (for n ≥3).
Study approval.
The study was approved by the IRBs of the INSERM, The Rockefeller University, and Necker Hospital for Sick Children; the Medical Ethics Committee of the Academic Medical Center/Emma Children’s Hospital; and by the bioethics committee for research in humans (CBE-SIU) of the UdeA. All procedures were conducted in accordance with the 1975 Declaration of Helsinki, as revised in 2013.
Author contributions
TWK and JB are the principal investigators and conceived the study. AVDG, ANP, DBK, ATJT, PJHV, AAA, LP, MVH, MG, COQ, CAAF, MEA, NW, CD, EN, RPG, JLVH, and KM performed the experiments. DR, DBK, JIG, JLC, MCD, and TKVDB contributed to the design of the study. ARB, SOB, JV, SM Holland, ET, BB, MO, MMV, SV, IM, JLF, JSL, AC, FCH, PI, AMM, NCB, PDA, AW, CB, DF, CHH, HW, SM Hughes, and HLM contributed to the collection of samples from the patients and provided a clinical overview. CG, MDB, KVL, JFA, FC, VR, VB, MB, and LA were responsible for genetic analysis. All authors revised the manuscript and approved the final manuscript. AVDG, ANP, DR, JLC, JB, and TWK analyzed the results and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank all the patients and their relatives as well as the treatment teams for their participation in this study. We also thank Céline Desvallées, Dominick Papandrea, Cécile Patissier, and Yelena Nemirovskaya at the HGMI laboratory (Rockefeller University branch) and the Institute Imagine branch for administrative assistance. We thank all the members of the Blood Cell Research Department and the HGMI Laboratory for helpful discussions. We acknowledge the use of the biological resources of the Imagine Institute DNA-Biobank (BB-33-00065). AVDG was supported by the Sanquin Blood Supply Product and Process Development Cellular Products Fund (PPOC 1957). ANP was supported by the CONACYT National PhD Fellowship Program. The HGMI Laboratory was funded in part by the NIAID (5R01AI089970 and 5R37AI095983), the National Center for Research Resources, and the National Center for Advancing Sciences of the NIH (8UL1TR000043), The Rockefeller University, the St. Giles Foundation, INSERM, Paris Descartes University, the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), and the French National Research Agency (ANR) under the “Investments for the Future” program (ANR-10IAHU-01, to the Imagine Institute), ANR-IFNPHOX/ANR-13-ISV3-0001-01, and ANR-GENMSMD/ANR-16-CE17-0005-01 (to JB), ECOSNord-C14S01 (to JB and JLF), and COLCIENCIAS (576-2013:111556934990, to JLF). KM was supported by a Japan Foundation for Pediatric Research fellowship grant. AMM was funded by the Canadian Institutes of Health Research (CIHR) and The Leona M. and Harry B. Helmsley Charitable Trust. This work was supported in part by the Division of Intramural Research, NIAID, NIH and was funded in part with federal funds from the National Cancer Institute, NIH (HHSN261200800001E). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Version 1. 07/03/2018
In-Press Preview
Version 2. 08/06/2018
Electronic publication
Version 3. 08/16/2018
PDF formatting corrected.
Version 4. 08/31/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(9):3957–3975.https://doi.org/10.1172/JCI97116.
Contributor Information
Annemarie van de Geer, Email: a.vandegeer@sanquin.nl.
Alejandro Nieto-Patlán, Email: alejandro.nieto@inserm.fr.
Douglas B. Kuhns, Email: dkuhns@mail.nih.gov.
Anton T.J. Tool, Email: a.tool@sanquin.nl.
Andrés A. Arias, Email: aaugusto.arias@udea.edu.co.
Matthieu Bouaziz, Email: matthieu.bouaziz@inserm.fr.
Martin de Boer, Email: m.deboer@sanquin.nl.
José Luis Franco, Email: jose.franco@udea.edu.co.
Roel P. Gazendam, Email: rgazendam@gmail.com.
John L. van Hamme, Email: j.l.vanhamme@amc.uva.nl.
Michel van Houdt, Email: michelvanhoudt@hotmail.com.
Karin van Leeuwen, Email: k.vanleeuwen@sanquin.nl.
Paul J.H. Verkuijlen, Email: P.Verkuijlen@sanquin.nl.
Timo K. van den Berg, Email: t.k.vandenberg@sanquin.nl.
Juan F. Alzate, Email: jfernando.alzate@udea.edu.co.
Carlos A. Arango-Franco, Email: carlos.arango2@udea.edu.co.
Vritika Batura, Email: vritika.batura@sickkids.ca.
Andrea R. Bernasconi, Email: arbernasconi@yahoo.com.ar.
Barbara Boardman, Email: Barbara.Boardman@cmft.nhs.uk.
Claire Booth, Email: c.booth@ich.ucl.ac.uk.
Siobhan O. Burns, Email: siobhan.burns2@nhs.net.
Felipe Cabarcas, Email: felipe.cabarcas@udea.edu.co.
Nadine Cerf Bensussan, Email: nadine.cerf-bensussan@inserm.fr.
Fabienne Charbit-Henrion, Email: fabienne.charbit.henrion@gmail.com.
Anniek Corveleyn, Email: anniek.corveleyn@uzleuven.be.
Caroline Deswarte, Email: caroline.deswarte@inserm.fr.
María Esnaola Azcoiti, Email: mesnaolaazcoiti@gmail.com.
Dirk Foell, Email: dfoell@uni-muenster.de.
John I. Gallin, Email: jgallin@nih.gov.
Carlos Garcés, Email: calichegarces@hotmail.com.
Margarida Guedes, Email: margguedes@gmail.com.
Claas H. Hinze, Email: claas.Hinze@ukmuenster.de.
Steven M. Holland, Email: sholland@niaid.nih.gov.
Stephen M. Hughes, Email: Stephen.Hughes@cmft.nhs.uk.
Patricio Ibañez, Email: patricio.ibanezlazo@gmail.com.
Harry L. Malech, Email: hmalech@nih.gov.
Isabelle Meyts, Email: isabelle.meyts@uzleuven.be.
Marcela Moncada-Velez, Email: marcela.moncada@udea.edu.co.
Kunihiko Moriya, Email: kunihiko.moriya@inserm.fr.
Esmeralda Neves, Email: m.esmeraldaneves@gmail.com.
Matias Oleastro, Email: moleastro@yahoo.com.ar.
Laura Perez, Email: leperez77@gmail.com.
Vimel Rattina, Email: vimel.rattina@gmail.com.
Carmen Oleaga-Quintas, Email: carmen.oleaga@inserm.fr.
Neil Warner, Email: neil.warner@sickkids.ca.
Aleixo M. Muise, Email: aleixo.muise@sickkids.ca.
Eunice Trindade, Email: eunice_trindade@netcabo.pt.
Helmut Wittkowski, Email: Helmut.Wittkowski@ukmuenster.de.
Austen Worth, Email: Austen.worth@gosh.nhs.uk.
Laurent Abel, Email: laurent.abel@inserm.fr.
Mary C. Dinauer, Email: dinauer_m@kids.wustl.edu.
Peter D. Arkwright, Email: peter.arkwright@nhs.net.
Dirk Roos, Email: d.roos@sanquin.nl.
Jean-Laurent Casanova, Email: Jean-Laurent.Casanova@rockefeller.edu.
Taco W. Kuijpers, Email: t.w.kuijpers@amc.uva.nl.
Jacinta Bustamante, Email: jacinta.bustamante@inserm.fr.
References
- 1. Roos D, Tool ATJ, van Leeuwen K, de Boer M. Biochemical and genetic diagnosis of chronic granulomatous disease. In: Seger RA, Roos D, Segal BH, Kuijpers TW, eds. Chronic Granulomatous Disease. Geneticcs, Biology And Clinical Management. New York, New York, USA: Nova Biomedical Press; 2017:231–300. [Google Scholar]
- 2.Janeway CA, Craig J, Davidson M, Downey W, Gitlin D, Sullivan JC. Hypergammaglobulinemia associated with severe, recurrent and chronic non-specific infection. Am J Dis Child. 1954;88:388–392. [Google Scholar]
- 3.Holmes B, Quie PG, Windhorst DB, Good RA. Fatal granulomatous disease of childhood. An inborn abnormality of phagocytic function. Lancet. 1966;1(7449):1225–1228. doi: 10.1016/s0140-6736(66)90238-8. [DOI] [PubMed] [Google Scholar]
- 4.Royer-Pokora B, et al. Cloning the gene for an inherited human disorder — chronic granulomatous disease — on the basis of its chromosomal location. Nature. 1986;322(6074):32–38. doi: 10.1038/322032a0. [DOI] [PubMed] [Google Scholar]
- 5.Dinauer MC, Orkin SH, Brown R, Jesaitis AJ, Parkos CA. The glycoprotein encoded by the X-linked chronic granulomatous disease locus is a component of the neutrophil cytochrome b complex. Nature. 1987;327(6124):717–720. doi: 10.1038/327717a0. [DOI] [PubMed] [Google Scholar]
- 6.Segal AW, et al. Absence of cytochrome b-245 in chronic granulomatous disease. A multicenter European evaluation of its incidence and relevance. N Engl J Med. 1983;308(5):245–251. doi: 10.1056/NEJM198302033080503. [DOI] [PubMed] [Google Scholar]
- 7.Winkelstein JA, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000;79(3):155–169. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 8.de Oliveira-Junior EB, et al. Clinical and Genotypic Spectrum of Chronic Granulomatous Disease in 71 Latin American Patients: First Report from the LASID Registry. Pediatr Blood Cancer. 2015;62(12):2101–2107. doi: 10.1002/pbc.25674. [DOI] [PubMed] [Google Scholar]
- 9.van den Berg JM, et al. Chronic granulomatous disease: the European experience. PLoS One. 2009;4(4):e5234. doi: 10.1371/journal.pone.0005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marciano BE, et al. Common severe infections in chronic granulomatous disease. Clin Infect Dis. 2015;60(8):1176–1183. doi: 10.1093/cid/ciu1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parvaneh N, et al. Visceral leishmaniasis in two patients with IL-12p40 and IL-12Rβ1 deficiencies. Pediatr Blood Cancer. 2017;64(6):e26362. doi: 10.1002/pbc.26362. [DOI] [PubMed] [Google Scholar]
- 12.Magnani A, et al. Inflammatory manifestations in a single-center cohort of patients with chronic granulomatous disease. J Allergy Clin Immunol. 2014;134(3):655–662.e8. doi: 10.1016/j.jaci.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 13. Marciano BE, Segal BH, Holland SM. Inflammatory manifestations in chronic granulomatous disease. In: Seger RA, Roos D, Segal H, Kuijpers TW, eds. Chronic Granulomatous Disease. Geneticcs, Biology And Clinical Management. New York, New York, USA: Nova Biomedical Press; 2017:163–201. [Google Scholar]
- 14.Huang C, et al. Genetic Risk for Inflammatory Bowel Disease Is a Determinant of Crohn’s Disease Development in Chronic Granulomatous Disease. Inflamm Bowel Dis. 2016;22(12):2794–2801. doi: 10.1097/MIB.0000000000000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van de Veerdonk FL, Dinauer MC. Dysfunctional processes regulating IL-1B in chronic granulomatous disease. In: Seger RA, Roos D, Segal H, Kuijpers TW, eds. Chronic Granulomatous Disease. Geneticcs, Biology And Clinical Management. New York, New York, USA: Nova Biomedical Press; 2017:69–87. [Google Scholar]
- 16.Ben Abdallah Chabchoub R, Turki H, Mahfoudh A. [Systemic lupus erythematosus in a boy with chronic granulomatous disease: case report and review of the literature] Arch Pediatr. 2014;21(12):1364–1366. doi: 10.1016/j.arcped.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Kuhns DB, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363(27):2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolach B, et al. Chronic granulomatous disease: Clinical, functional, molecular, and genetic studies. The Israeli experience with 84 patients. Am J Hematol. 2017;92(1):28–36. doi: 10.1002/ajh.24573. [DOI] [PubMed] [Google Scholar]
- 19.Köker MY, et al. Clinical, functional, and genetic characterization of chronic granulomatous disease in 89 Turkish patients. J Allergy Clin Immunol. 2013;132(5):1156–1163.e5. doi: 10.1016/j.jaci.2013.05.039. [DOI] [PubMed] [Google Scholar]
- 20.Kang EM, Marciano BE, DeRavin S, Zarember KA, Holland SM, Malech HL. Chronic granulomatous disease: overview and hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2011;127(6):1319–26;quiz1327. doi: 10.1016/j.jaci.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000;79(3):170–200. doi: 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Roos D. Chronic granulomatous disease. Br Med Bull. 2016;118(1):50–63. doi: 10.1093/bmb/ldw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roos D, de Boer M. Molecular diagnosis of chronic granulomatous disease. Clin Exp Immunol. 2014;175(2):139–149. doi: 10.1111/cei.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roos D, et al. Two CGD families with a hypomorphic mutation in the activation domain of p67phox. J Clin Cell Immunol. 2014;5(3):1000231. [PMC free article] [PubMed] [Google Scholar]
- 25.Bustamante J, et al. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat Immunol. 2011;12(3):213–221. doi: 10.1038/ni.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matute JD, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114(15):3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan S, et al. Genomic structure, chromosomal localization, start of transcription, and tissue expression of the human p40-phox, a new component of the nicotinamide adenine dinucleotide phosphate-oxidase complex. Blood. 1996;88(7):2714–2721. [PubMed] [Google Scholar]
- 28.Matute JD, Arias AA, Dinauer MC, Patiño PJ. p40phox: the last NADPH oxidase subunit. Blood Cells Mol Dis. 2005;35(2):291–302. doi: 10.1016/j.bcmd.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Nunes P, Demaurex N, Dinauer MC. Regulation of the NADPH oxidase and associated ion fluxes during phagocytosis. Traffic. 2013;14(11):1118–1131. doi: 10.1111/tra.12115. [DOI] [PubMed] [Google Scholar]
- 30.Odell EW, Segal AW. Killing of pathogens associated with chronic granulomatous disease by the non-oxidative microbicidal mechanisms of human neutrophils. J Med Microbiol. 1991;34(3):129–135. doi: 10.1099/00222615-34-3-129. [DOI] [PubMed] [Google Scholar]
- 31.Ellson CD, Davidson K, Ferguson GJ, O’Connor R, Stephens LR, Hawkins PT. Neutrophils from p40phox–/– mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing. J Exp Med. 2006;203(8):1927–1937. doi: 10.1084/jem.20052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagaitkar J, Matute JD, Austin A, Arias AA, Dinauer MC. Activation of neutrophil respiratory burst by fungal particles requires phosphatidylinositol 3-phosphate binding to p40phox in humans but not in mice. Blood. 2012;120(16):3385–3387. doi: 10.1182/blood-2012-07-445619. [DOI] [PubMed] [Google Scholar]
- 33.Deffert C, et al. Hyperinflammation of chronic granulomatous disease is abolished by NOX2 reconstitution in macrophages and dendritic cells. J Pathol. 2012;228(3):341–350. doi: 10.1002/path.4061. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues-Sousa T, et al. Deficient production of reactive oxygen species leads to severe chronic DSS-induced colitis in Ncf1/p47phox-mutant mice. PLoS One. 2014;9(5):e97532. doi: 10.1371/journal.pone.0097532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao S, Carr ED, Xu YH, Hunt NH. Gp91(phox) contributes to the development of experimental inflammatory bowel disease. Immunol Cell Biol. 2011;89(8):853–860. doi: 10.1038/icb.2011.4. [DOI] [PubMed] [Google Scholar]
- 36.Bagaitkar J, et al. PI(3)P-p40phox binding regulates NADPH oxidase activation in mouse macrophages and magnitude of inflammatory responses in vivo. J Leukoc Biol. 2017;101(2):449–457. doi: 10.1189/jlb.3AB0316-139R. [DOI] [PubMed] [Google Scholar]
- 37.Conway KL, et al. p40phox expression regulates neutrophil recruitment and function during the resolution phase of intestinal inflammation. J Immunol. 2012;189(7):3631–3640. doi: 10.4049/jimmunol.1103746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prando C, et al. Paternal uniparental isodisomy of chromosome 6 causing a complex syndrome including complete IFN-gamma receptor 1 deficiency. Am J Med Genet A. 2010;152A(3):622–629. doi: 10.1002/ajmg.a.33291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41(6):445–449. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dusi S, et al. Nicotinamide-adenine dinucleotide phosphate oxidase assembly and activation in EBV-transformed B lymphoblastoid cell lines of normal and chronic granulomatous disease patients. J Immunol. 1998;161(9):4968–4974. [PubMed] [Google Scholar]
- 41.Conti F, et al. Phagocyte nicotinamide adenine dinucleotide phosphate oxidase activity in patients with inherited IFN-γR1 or IFN-γR2 deficiency. J Allergy Clin Immunol. 2015;135(5):1393–1395.e1. doi: 10.1016/j.jaci.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crotzer VL, et al. Cutting edge: NADPH oxidase modulates MHC class II antigen presentation by B cells. J Immunol. 2012;189(8):3800–3804. doi: 10.4049/jimmunol.1103080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voetman AA, Weening RS, Hamers MN, Meerhof LJ, Bot AA, Roos D. Phagocytosing human neutrophils inactivate their own granular enzymes. J Clin Invest. 1981;67(5):1541–1549. doi: 10.1172/JCI110185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Repine JE, Clawson CC. Quantitative measurement of the bactericidal capability of neutrophils from patients and carriers of chronic granulomatous disease. J Lab Clin Med. 1977;90(3):522–528. [PubMed] [Google Scholar]
- 45.Rosen H, Michel BR. Redundant contribution of myeloperoxidase-dependent systems to neutrophil-mediated killing of Escherichia coli. Infect Immun. 1997;65(10):4173–4178. doi: 10.1128/iai.65.10.4173-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kagawa R, et al. Alanine-scanning mutagenesis of human signal transducer and activator of transcription 1 to estimate loss- or gain-of-function variants. J Allergy Clin Immunol. 2017;140(1):232–241. doi: 10.1016/j.jaci.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khangura SK, et al. Gastrointestinal features of chronic granulomatous disease found during endoscopy. Clin Gastroenterol Hepatol. 2016;14(3):395–402.e5. doi: 10.1016/j.cgh.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conti F, et al. Mycobacterial disease in patients with chronic granulomatous disease: a retrospective analysis of 71 cases. J Allergy Clin Immunol. 2016;138(1):241–248.e3. doi: 10.1016/j.jaci.2015.11.041. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian Vignesh K, Landero Figueroa JA, Porollo A, Caruso JA, Deepe GS. Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity. 2013;39(4):697–710. doi: 10.1016/j.immuni.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao XW, et al. Defects in neutrophil granule mobilization and bactericidal activity in familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) syndrome caused by STXBP2/Munc18-2 mutations. Blood. 2013;122(1):109–111. doi: 10.1182/blood-2013-03-494039. [DOI] [PubMed] [Google Scholar]
- 52.Gazendam RP, et al. Human neutrophils use different mechanisms to kill Aspergillus fumigatus Conidia and Hyphae: evidence from phagocyte defects. J Immunol. 2016;196(3):1272–1283. doi: 10.4049/jimmunol.1501811. [DOI] [PubMed] [Google Scholar]
- 53.Boyle KB, Stephens LR, Hawkins PT. Activation of the neutrophil NADPH oxidase by Aspergillus fumigatus. Ann N Y Acad Sci. 2012;1273:68–73. doi: 10.1111/j.1749-6632.2012.06821.x. [DOI] [PubMed] [Google Scholar]
- 54.Lehrer RI, Cline MJ. Interaction of Candida albicans with human leukocytes and serum. J Bacteriol. 1969;98(3):996–1004. doi: 10.1128/jb.98.3.996-1004.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hetzel M, et al. Hematopoietic stem cell gene therapy for IFNγR1 deficiency protects mice from mycobacterial infections. Blood. 2018;131(5):533–545. doi: 10.1182/blood-2017-10-812859. [DOI] [PubMed] [Google Scholar]
- 56.Winter S, Hultqvist Hopkins M, Laulund F, Holmdahl R. A reduction in intracellular reactive oxygen species due to a mutation in NCF4 promotes autoimmune arthritis in mice. Antioxid Redox Signal. 2016;25(18):983–996. doi: 10.1089/ars.2016.6675. [DOI] [PubMed] [Google Scholar]
- 57.De Ravin SS, et al. Chronic granulomatous disease as a risk factor for autoimmune disease. J Allergy Clin Immunol. 2008;122(6):1097–1103. doi: 10.1016/j.jaci.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez-Boyanapalli RF, et al. Impaired apoptotic cell clearance in CGD due to altered macrophage programming is reversed by phosphatidylserine-dependent production of IL-4. Blood. 2009;113(9):2047–2055. doi: 10.1182/blood-2008-05-160564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenlee-Wacker MC, Rigby KM, Kobayashi SD, Porter AR, DeLeo FR, Nauseef WM. Phagocytosis of Staphylococcus aureus by human neutrophils prevents macrophage efferocytosis and induces programmed necrosis. J Immunol. 2014;192(10):4709–4717. doi: 10.4049/jimmunol.1302692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenlee-Wacker MC, Nauseef WM. IFN-γ targets macrophage-mediated immune responses toward Staphylococcus aureus. J Leukoc Biol. 2017;101(3):751–758. doi: 10.1189/jlb.4A1215-565RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez-Boyanapalli R, et al. Impaired phagocytosis of apoptotic cells by macrophages in chronic granulomatous disease is reversed by IFN-γ in a nitric oxide-dependent manner. J Immunol. 2010;185(7):4030–4041. doi: 10.4049/jimmunol.1001778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marks DJ, Miyagi K, Rahman FZ, Novelli M, Bloom SL, Segal AW. Inflammatory bowel disease in CGD reproduces the clinicopathological features of Crohn’s disease. Am J Gastroenterol. 2009;104(1):117–124. doi: 10.1038/ajg.2008.72. [DOI] [PubMed] [Google Scholar]
- 63.Frans G, et al. PID in disguise: molecular diagnosis of IRAK-4 deficiency in an adult previously misdiagnosed with autosomal dominant hyper IgE syndrome. J Clin Immunol. 2015;35(8):739–744. doi: 10.1007/s10875-015-0205-x. [DOI] [PubMed] [Google Scholar]
- 64.Charbit-Henrion F, et al. Diagnostic yield of next-generation sequencing in very early-onset inflammatory bowel diseases: a multicenter study. J Crohns Colitis. doi: 10.1093/ecco-jcc/jjy068. [published online ahead of print May 18, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuijpers TW, Alders M, Tool AT, Mellink C, Roos D, Hennekam RC. Hematologic abnormalities in Shwachman Diamond syndrome: lack of genotype-phenotype relationship. Blood. 2005;106(1):356–361. doi: 10.1182/blood-2004-11-4371. [DOI] [PubMed] [Google Scholar]
- 66.van Pelt LJ, van Zwieten R, Weening RS, Roos D, Verhoeven AJ, Bolscher BG. Limitations on the use of dihydrorhodamine 123 for flow cytometric analysis of the neutrophil respiratory burst. J Immunol Methods. 1996;191(2):187–196. doi: 10.1016/0022-1759(96)00024-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.