Abstract
The t-SNARE protein SNAP23 conventionally functions as a component of the cellular machinery required for intracellular transport vesicle fusion with target membranes and has been implicated in the regulation of fasting glucose levels, BMI, and type 2 diabetes. Surprisingly, we observed that adipocyte-specific KO of SNAP23 in mice resulted in a temporal development of severe generalized lipodystrophy associated with adipose tissue inflammation, insulin resistance, hyperglycemia, liver steatosis, and early death. This resulted from adipocyte cell death associated with an inhibition of macroautophagy and lysosomal degradation of the proapoptotic regulator BAX, with increased BAX activation. BAX colocalized with LC3-positive autophagic vacuoles and was increased upon treatment with lysosome inhibitors. Moreover, BAX deficiency suppressed the lipodystrophic phenotype in the adipocyte-specific SNAP23-KO mice and prevented cell death. In addition, ATG9 deficiency phenocopied SNAP23 deficiency, whereas ATG7 deficiency had no effect on BAX protein levels, BAX activation, or apoptotic cell death. These data demonstrate a role for SNAP23 in the control of macroautophagy and programmed cell death through an ATG9-dependent, but ATG7-independent, pathway regulating BAX protein levels and BAX activation.
Keywords: Cell Biology, Metabolism
Keywords: Adipose tissue, Apoptosis, Autophagy
Introduction
SNARE complexes are a large group of membrane-bound proteins that regulate the fusion of intracellular transport vesicles with their target membranes (1–3). All SNAREs contain a characteristic coiled-coil SNARE motif (a cysteine-rich domain) of approximately 70 aa composed of heptad repeats and can function on either target membranes (t-SNAREs) or transport vesicles (v-SNAREs) (4). In mammalian cells, there are approximately 35 SNARE proteins, and the specificity of SNARE protein assembly and function is dependent on the interaction between defined cognate SNAREs, subcellular compartmentalization, and interaction with various accessory factors. Mechanistically, SNARE proteins have been extensively investigated in transport vesicle trafficking and granule secretion, however, recent studies in yeast have demonstrated the involvement of SNARE proteins in autophagy processes. For example, yeast orthologs of mammalian syntaxin 1 (Sso2p), syntaxin 16 (Tlg2), SNAP23 (Sec9), SEC22B (Sec22p), and YKT6 (Ykt6p) have all been implicated in autophagosome biogenesis through the reorganization of the ATG9 tubulovesicular clusters (5). In mammalian cells, the interactions between VAMP7, VTI1B, and syntaxin 7 and -8 regulate the homotypic fusion of preautophagosomal structures necessary for phagophore and autophagosome formation (6). Likewise, VAMP3 was found to be necessary for the maturation of ATG16L1-positive structures required for fusion with ATG9-positive vesicles and autophagosome formation (7).
In our studies of SNARE protein function, we generated whole-body SNAP23-KO mice, and, as recently reported by others, SNAP23 deficiency resulted in early embryonic lethality (8). We therefore generated SNAP23-floxed mice that were crossed with adiponectin Cre (Adipoq-Cre) mice to generate adipocyte-specific (white and brown) SNAP23-KO mice. Surprisingly, we found that these mice displayed a temporal development of severe generalized lipodystrophy associated with adipose tissue inflammation, insulin resistance, hyperglycemia, liver steatosis, and early death. Molecular analyses demonstrated that adipocyte deficiency of SNAP23 resulted in impaired autophagosome function during nutrient-depleted (ND) states (fasting) that in turn prevented the lysosome-mediated degradation of the proapoptotic protein BAX, thus leading to adipocyte-specific apoptotic cell death. The accumulation of BAX protein was selective to impaired ATG9 function, as inhibition of macroautophagy by ATG7 deficiency had no significant effect on BAX protein levels, BAX activation, or cell death.
Results
Adipocyte-specific SNAP23 deficiency induces general lipodystrophy.
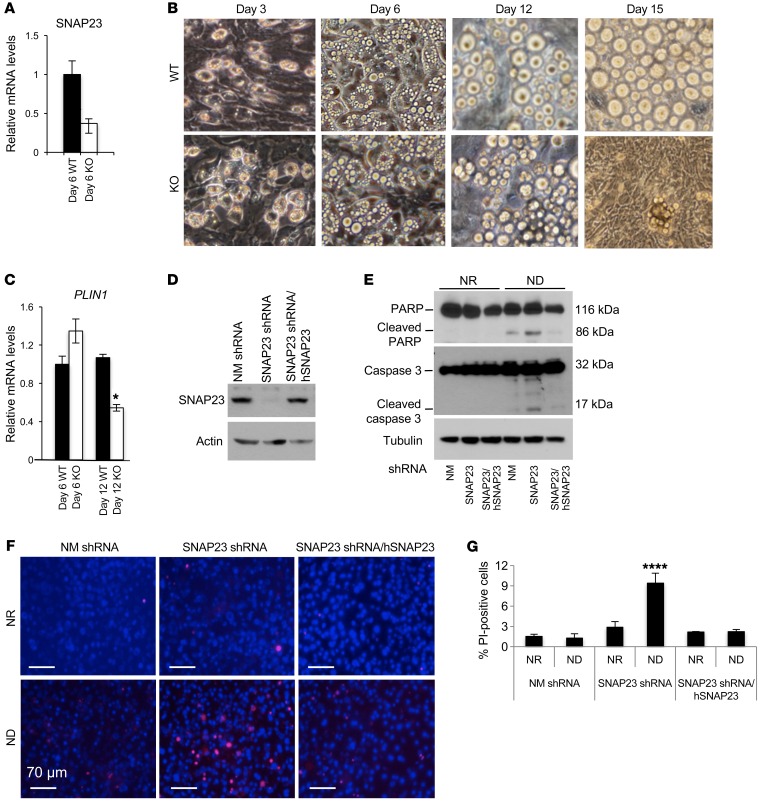
As previously reported by Kaul et al. (8), we also observed that germline deletion of SNAP23 resulted in early embryonic lethality in mice. We therefore generated adipocyte-specific SNAP23-deficient mice by crossing floxed SNAP23 mice with Adipoq-Cre mice (Adipoq-Cre+/– Snap23fl/fl mice, hereafter referred to as KO mice). Quantitative real-time PCR (qRT-PCR) showed no significant decrease in SNAP23 mRNA from the KO mice compared with mRNA levels in control Snap23fl/fl mice (hereafter referred to as WT mice) in total adipose tissue extracts (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI99217DS1). However, we observed a marked increase (~5-fold) in the amount of the macrophage marker F4/80 mRNA in the KO mice. This is consistent with a large increase in adipose tissue inflammation (Figure 1, J and L), and the cross contamination with other cell types probably accounts for the apparent lack of a decrease in SNAP23 transcripts in adipose tissue. We therefore isolated primary adipocytes from 2-week-old mice, and quantitative qRT-PCR analysis revealed a significant decrease in SNAP23 mRNA (approximately 4-fold), with a 2-fold increase in F4/80 mRNA (Supplemental Figure 1B). The residual SNAP23 mRNA in the isolated adipocytes from the KO mice probably reflects the residual contamination by inflammatory cells as indicated by F4/80 mRNA levels. Immunoblotting of the isolated primary adipocytes showed approximately 50% and 80% reductions in SNAP23 protein from the heterozygotic and homozygotic KO mice, respectively (Supplemental Figure 1C). Since the LoxP sites flank exons 3 and 4 and there is an in-frame ATG codon located in exon 5, a potential approximately 18-kDa truncated fragment could be generated. Longer exposure revealed the presence of a nonsignificant trace of this band in the KO adipocytes.
Figure 1. Adipocyte-specific SNAP23-KO mice display severe lipodystrophy associated with liver steatosis and adipose tissue inflammation.
(A) Thirty-two-week-old male KO mice had extended abdomens (first panel on left) with enlarged, pale livers (star in second panel from left), loss of epididymal adipose tissue (triangles in second panel from left), subcutaneous adipose tissue (inside outlined shapes in third panel from left), perirenal adipose tissue (inside outlined shapes in fourth panel from left), and interscapular BAT (circle in last panel on right). (B) Plasma glucose, (C) leptin, (D) adiponectin, and (E) triglyceride levels were determined as described in Methods. (F) Hepatic triglyceride content was normalized to total protein levels (n = 5 WT mice and n = 5 KO mice). (G) Echo-MRI analysis of total fat mass in Snap23fl/fl (WT) and adipocyte-specific SNAP23-deficient (KO) mice at 2, 3, 4, 8, 12, 16, 24, and 32 weeks of age (n = 5–10 mice). (H) Perilipin immunofluorescence (red) and DAPI staining (blue) for nuclei in epididymal adipose tissue from 4-week-old WT and KO mice. Arrows indicate perilipin-depleted cells. Scale bars: 40 μm. (I) Quantification of perilipin-depleted cells was performed on epididymal adipose tissue (epi) from 4-week-old mice and subcutaneous adipose tissue (s.c.) from 1-week-old mice. (J) Epididymal adipose tissue from 4-week-old WT and KO mice was fixed, stained with H&E, and examined by light microscopy. Arrowheads indicate selected areas of inflammation and the presence of crown-like structures. Scale bars: 50 μm. (K) The relative diameter of the morphologically normal–appearing adipocytes from panel J was quantified (n = 500 cells). (L) Epididymal adipose tissue from 4-week-old WT and KO mice was extracted and subjected to qRT-PCR to determine the indicated mRNA levels (n = 5 WT mice and n = 5 KO mice). All data represent the mean ± SEM. *P < 0.05 and ***P < 0.001, by Student’s t test.
The KO mice visually appeared normal before weaning. However, by 32 weeks of age, the male KO mice had visually distended abdomens (Figure 1A, first panel from left). Dissection revealed a greatly enlarged and pale liver with an absence of all fat pads including epididymal fat (Figure 1A, second panel from left), subcutaneous fat (Figure 1A, third panel from left), perirenal fat (Figure 1A, fourth panel from left), and interscapular brown fat (Figure 1A, fifth panel from left) pads. Likewise, KO female mice showed an essentially identical lipodystrophic phenotype (data not shown). Compared with WT mice, the body weights of the KO mice were increased, with increased weights for the liver, seminal vesicles, lung, intestines, pancreas, kidney, and brain (Supplemental Figure 1D). In KO mice, random blood glucose levels were greater than 300 mg/dl compared with 150 mg/dl for the WT littermate mice (Figure 1B). The KO mice also had a near-complete absence of plasma leptin (Figure 1C) and adiponectin (Figure 1D), with increased plasma triglyceride levels (Figure 1E) and hepatic triglyceride levels (Figure 1F).
Consistent with other generalized lipodystrophy mouse models, the KO mice were glucose and insulin intolerant as assessed by intraperitoneal glucose tolerance tests (GTTs) and insulin tolerance tests (ITTs) (Supplemental Figure 1, E and F). Insulin levels were elevated by approximately 10-fold in the KO mice following a 5-hour fast and by approximately 4-fold following a 16-hour fast (Supplemental Figure 1G). The hyperinsulinemia accounts for the similar fasting glucose levels observed during the intraperitoneal GTTs (16-h fast) and ITTs (5-h fast) in the WT and KO mice. The compensating hyperinsulinemia in the fasted state also accounts for the elevated levels of randomly sampled glucose levels observed in Figure 1B, which more closely represent the fed state. The KO mice also manifested increased daily food intake (Supplemental Figure 1H), and indirect calorimetry showed an increased respiratory exchange rate (RER) in the light cycle but not in the dark cycle, indicating metabolic inflexibility (Supplemental Figure 1I). Moreover, the KO mice showed reduced energy expenditure (Supplemental Figure 1J), with no significant change in spontaneous locomotor activity (Supplemental Figure 1K). In addition, these metabolic alterations correlated with a reduced lifespan of approximately 12 months (data not shown). Together, these data indicate that adipocyte-specific deficiency of SNAP23 results in insulin resistance and liver steatosis, most likely as a result of generalized lipodystrophy.
SNAP23 deficiency induces adipocyte-intrinsic cell death in vivo.
To examine the basis of this phenotype, MRI quantification of adipose tissue mass indicated no significant difference in the amount of adipose tissue mass between the WT and KO mice at 2 to 3 weeks of age (Figure 1G). At 4 weeks, we observed a significant decrease in adipose tissue mass in the KO mice that continued to decline and that by 24 weeks was essentially undetectable. To determine whether the loss of adipose tissue was due to adipocyte cell death, we examined the expression of the adipocyte-lipid coating protein perilipin 1 (Figure 1H) (9). Most epididymal adipocytes from 4-week-old WT mice showed well-defined perilipin staining that outlined the lipid droplet. In contrast, epididymal adipocytes from KO mice had numerous regions that were devoid of perilipin labeling, and these regions were surrounded by multi-nuclei (DAPI) staining and typical crown-like structures. Quantification of cell remnants (perilipin negative) indicated that nearly 50% of the epididymal adipocytes from the KO mice were dead at 4 weeks of age (Figure 1I). In subcutaneous adipocytes at 1 week of age, approximately 70% of the KO adipocytes cells were perilipin depleted compared with 5% in the WT mice (Figure 1I).
The presence of crown-like structures was also indicative of macrophages that surround adipocyte cell remnants (Figure 1J), and the presence of inflammation was confirmed by increased gene expression of F4/80, CD11c, and TNFA in the epididymal adipose tissue of the KO mice (Figure 1L). Consistent with the loss of adipocytes, we detected a concomitant increase in the size of the remaining perilipin-positive adipocytes (Figure 1, J and K) and a reduction in expression of the adipogenic transcriptional factors PPARG and PGC1A and the adipocyte markers adiponectin (ADIPOQ), leptin, perilipin 1, and GLUT4 (Figure 1L).
To confirm in vivo that the loss of SNAP23 induced adipocyte cell death rather than prevented adipogenesis, we crossed the floxed SNAP23 mice with tamoxifen-inducible, adipocyte-specific Cre mice (Adipoq-CreERT2+/– Snap23fl/fl). The mice were allowed to age to 16 weeks, when de novo adipogenesis is relatively low (10, 11), and the control Adipoq-CreERT2+/– Snap23+/+ (Ind WT) and Adipoq-CreERT2+/– Snap23fl/fl (Ind-KO) mice were injected daily (for 5 days) with tamoxifen and analyzed 21 days later. Under these conditions, the isolated adipocyte SNAP23 mRNA levels were reduced to approximately 70% of control levels (Supplemental Figure 2A), and the remaining SNAP23 mRNA was again probably due to the large increase in inflammatory cells that contaminated the isolation of primary adipocytes (Supplemental Figure 2B). Nevertheless, the adipocyte deficiency of SNAP23 resulted in a large increase in the number of perilipin-depleted adipocytes (Supplemental Figure 2, C and D).
SNAP23 is necessary for adipocyte cell survival during ND.
Having established that SNAP23 deficiency in adipocytes in vivo induces cell death, we recapitulated these findings in adipocytes differentiated in isolated stromal vascular progenitor cells (SVCs) derived from WT and KO mice. Adipoq-Cre is not expressed in the progenitor cells but is induced following adipocyte differentiation, resulting in the subsequent inactivation of the SNAP23 gene. Progenitor cells grew normally, and the percentage of cells that differentiated into adipocytes was approximately 80%. By day 6 after differentiation, 70% of the SNAP23 mRNA was depleted from the KO cells (Figure 2A). From day 6 to day 20, these adipocytes appeared healthy when the medium was changed every 2 days (Supplemental Figure 2E). However, when the medium was changed every 4 days, we observed no significant effect on the WT cells, but the SNAP23-depleted cells started to die, with fibroblasts replacing the adipocytes by day 15 (Figure 2B). Quantification of perilipin 1 (PLIN1) mRNA on day 6 indicated that adipocyte differentiation was similar between WT and KO mice (Figure 2C). However, by day 12, PLIN1 mRNA levels in WT adipocytes remained unchanged, while PLIN1 mRNA levels in KO adipocytes decreased by 50%.
Figure 2. SNAP23 deficiency induces adipocyte apoptotic cell death.
(A) Subcutaneous adipose tissue SVCs from 3-week-old WT and KO mice were isolated and differentiated into adipocytes, and SNAP23 mRNA levels were determined on day 6. Data represent the mean ± SEM from 3 independent experiments. (B) Cell morphology was examined on days 3, 6, 12, and 15 following differentiation, with medium changes every 2 days for the first 6 days and then every 4 days thereafter. Images are representative of 5 independent SVC isolations. Original magnification, x20. (C) mRNA from WT and KO on days 6 and 12 of differentiation was subjected to qRT-PCR for perilipin expression. Data represent the mean ± SEM from 3 independent experiments. (D) Control (NM shRNA), SNAP23-specific shRNA (SNAP23 shRNA), and shRNA-resistant human SNAP23 cDNA–transfected SNAP23 shRNA (SNAP23 shRNA/hSNAP23) 3T3L1 adipocytes were differentiated for 12 days under NR conditions and immunoblotted for SNAP23 and actin. Immunoblots are representative of 3 independently performed experiments. (E) The adipocytes differentiated for 12 days were maintained under NR conditions, with a medium change every 2 days, or under ND conditions, with a medium change every 4 days after differentiation for 6 days. Cell extracts were then immunoblotted for PARP, caspase 3, and actin. Immunoblots are representative of 3 independent experiments. (F) The adipocytes differentiated for 12 days and maintained under NR or ND conditions for 6 hours were subjected to PI and DAPI labeling. Scale bars: 70 μm. (G) Quantification of PI-positive nuclei was determined by counting approximately 400 cells from 3 independent determinations. Data represent the mean ± SEM from 3 independent experiments. *P < 0.05, by Student’s t test (A and C) and ****P < 0.0001, by ANOVA with Dunnett’s post hoc test (G).
To confirm these data in cultured cell lines, we used lentiviral SNAP23-targeted shRNAs to knock down SNAP23 in 3T3L1 adipocytes and nonmammalan-targeted (NM-targeted) shRNAs as a control. We established 4 lentiviral SNAP23 shRNA–expressing 3T3L1 cell lines (knockdown efficiency ranging from 75% to 90% at the protein level) and 2 control cell lines (1 pair shown in Figure 2D). In addition, to control for potential off-target effects, an shRNA-resistant human SNAP23 cDNA was also expressed in the context of the SNAP23 shRNA cells (SNAP23 shRNA/hSNAP23) (Figure 2D). All the SNAP23-knockdown and SNAP23 cDNA–expressing 3T3L1 fibroblasts grew at the same rate as the control 3T3L1 NM shRNA cells, reaching similar cell densities and extent of differentiation into adipocytes when maintained in nutrient-replete (NR) conditions (data not shown). However, 6 days without changing medium (ND conditions) following adipocyte differentiation resulted in enhanced PARP and caspase 3 cleavages in the SNAP23 shRNA cells (Figure 2E), but not under the NR condition (medium changed every 2 days). Importantly, these markers of apoptosis were prevented in the SNAP23 cDNA–rescued cells. The SNAP23 shRNA cells displayed increased cell death as determined by propidium iodide (PI) labeling compared with NM shRNA adipocytes and compared with the SNAP23 cDNA–rescued cells (Figure 2, F and G). These data indicate that in the context of SNAP23 deficiency, ND enhances the induction of adipocyte programmed cell death in culture.
To assess whether relative changes in nutrient status that occur during the normal diurnal cycle could account for adipocyte cell death and the subsequent lipodystrophic phenotype in vivo, 8-week-old control and KO mice were subjected to ad libitum feeding or starved for 48 hours. As shown in Figure 1H, the KO mice fed ad libitum had reduced perilipin labeling (Supplemental Figure 3A). However, mice starved for 48 hours showed a marked increase in the number of perilipin-depleted adipocytes. Quantification demonstrated that under normal ad libitum conditions, the percentage of perilipin-depleted adipocytes in the control mice was less than 1%, whereas for the KO mice, it was approximately 35% (Supplemental Figure 3B). Following starvation, adipocytes from control mice were unaffected, with approximately 1% remaining perilipin negative, although adipocyte sizes were reduced (Supplemental Figure 3A). In contrast, the number of perilipin-depleted adipocytes in the KO mice had increased to approximately 80% (Supplemental Figure 3B).
SNAP23 deficiency impairs nutrient-deficient activation of macroautophagy.
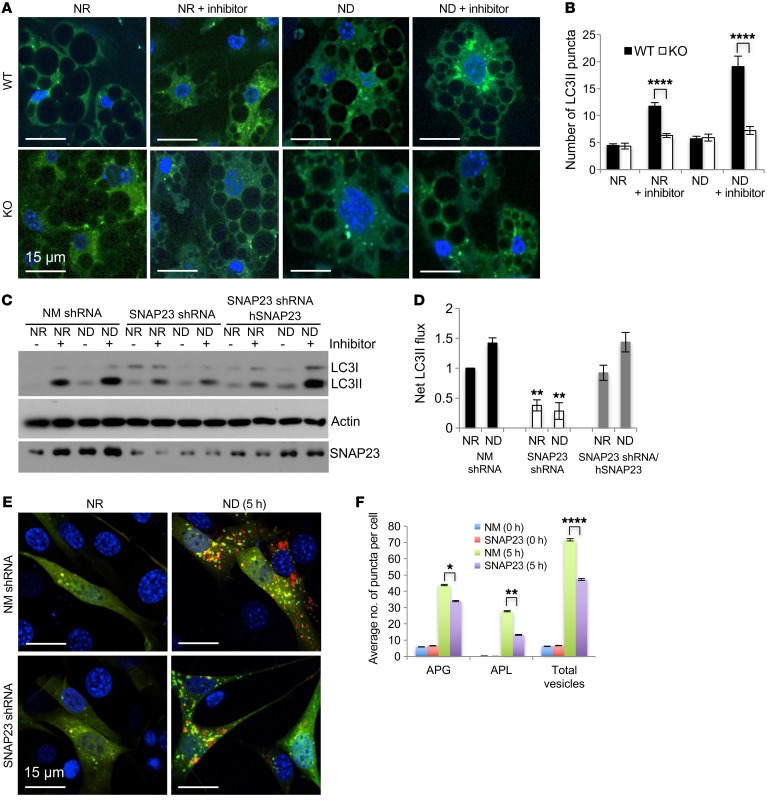
Since ND/starvation enhanced adipocyte cell death in vitro and in vivo, we examined the role of SNAP23 in the regulation of macroautophagy. GFP-LC3–transgenic mice (12) were crossed with WT and KO mice, and adipocyte precursors were isolated and differentiated in culture. As shown in Figure 3A and quantified in Figure 3B, the numbers of GFP-LC3 puncta were similar between WT and KO adipocytes under NR and ND conditions. Following treatment with lysosomal inhibitors (NH4Cl and leupeptin), the number of GFP-LC3 puncta increased in WT adipocytes. By contrast, KO adipocytes failed to show an increase in GFP-LC3 puncta under both NR and ND conditions, indicating the suppression of macroautophagy flux. Immunoblot analysis of LC3 from the SNAP23 shRNA 3T3L1 adipocytes also revealed inhibition of net LC3II flux compared with NM shRNA and SNAP23 shRNA/hSNAP23–rescued cells (Figure 3, C and D). SNAP23 deficiency also suppressed p62 flux, another autophagy marker (Supplemental Figure 3, C and D).
Figure 3. SNAP23 deficiency suppresses adipocyte macroautophagy.
(A) Snap23fl/fl (WT) and Adipoq-Cre Snap23fl/fl (KO) mice were crossed with GFP-LC3–transgenic mice as described in Methods. At 3 weeks of age, subcutaneous SVCs were isolated and differentiated into adipocytes for 6 days under NR conditions. The cells were then maintained in NR (DMEM with 10% FBS) or subjected to ND (RPMI-aa medium) conditions for 2 hours in the absence and presence of the lysosomotropic agents NH4Cl and leupeptin. The cells were then fixed and visualized for GFP (green) and DAPI (blue) by fluorescence microscopy. Scale bars: 15 μm. (B) The average number of GFP-LC3 puncta, with the SEM, was quantified in approximately 400 cells from 3 independent experiments. (C) NM shRNA, SNAP23 shRNA, and rescued SNAP23 shRNA/hSNAP23 3T3L1 adipocytes were differentiated under NR conditions for 8 days. The cells were then maintained in NR conditions or subjected to ND conditions for 4 hours in the absence and presence of the lysosomotropic agents NH4Cl and leupeptin. Cell extracts were immunoblotted for LC3, actin, and SNAP23. Immunoblot is representative of 5 independent experiments. (D) Net LC3II flux was calculated from 3 independent experiments. (E) Mouse NIH3T3 fibroblasts were transfected with NM shRNA or SNAP23 shRNA lentiviruses and subsequently transfected with the dual mCherryGFPLC3 reporter as described in Methods. Cells were then maintained under NR conditions or ND conditions for 5 hours and subjected to fluorescence microscopy. Scale bars: 15 μm. (F) The average number of red puncta per cell, with the SEM, was quantified in 500 individual cells from 3 independent experiments. *P < 0.05, **P < 0.01, and ****P < 0.0001, by ANOVA with Tukey’s post hoc test.
Cold induction through β-adrenergic signaling has been shown to induce triglyceride breakdown through lipophagy in brown adipose tissue (BAT) (13). However, isoproterenol stimulation of NM shRNA, SNAP23 shRNA, and SNAP23 shRNA/hSNAP23–rescued 3T3L1 adipocytes showed the same extent of fatty acid or glycerol release (Supplemental Figure 3, E and F), indicating that the defect in autophagy is not a significant contributor to lipolysis in SNAP23-deficient adipocytes.
To determine whether the inhibition of macroautophagy by SNAP23 deficiency occurred in other cell types, we established control NM shRNA and SNAP23 shRNA NIH3T3 fibroblasts. These cells were then transfected with the tandem mCherry-GFP-LC3 macroautophagy reporter construct (14). With this reporter, LC3-positive autophagosomes are labeled yellow (autophagosome [APG]) because of the overlapping of red and green signals, whereas the acidic environment of the lysosome quenches the GFP signal, resulting in red puncta (autolysosome [APL]) (14–16). In the NR state, APL was not readily apparent in either the control NM shRNA or SNAP23 shRNA cells (Figure 3, E and F). However, following 3 and 5 hours of starvation in ND medium, we observed an increase in the number of APLs in control NM shRNA cells, whereas the increase in APLs was significantly reduced in the SNAP23-knockdown cells. In addition, SNAP23 shRNA NIH3T3 cells also showed a marked reduction of net LC3II flux by immunoblot analysis (Supplemental Figure 4, A and B). In parallel, the SNAP23 shRNA cells had decreased p62 flux (Supplemental Figure 4, C and D). These data demonstrate that depletion of SNAP23 in NIH3T3 cells also impairs macroautophagy flux. Furthermore, like adipocytes in vivo and in vitro, SNAP23 deficiency in NIH3T3 cells also resulted in increased cell death following ND, whether determined by PI labeling or the release of oligonucleosomes (Supplemental Figure 4, E–G).
To directly assess autophagy in vivo, we performed transmission electron microscopy (TEM) of BAT. Multiple TEM images revealed the presence in WT mice of double-membrane autophagosomes that were found adjacent to intact mitochondria (Supplemental Figure 5A). In contrast, BAT from KO mice displayed damaged mitochondria with abnormal cristae and significant lipid droplet (LD) accumulation, indicating altered quality control. More important, we observed a substantial reduction in the number of autophagosomes (Supplemental Figure 5, A and B). Consistent with an early defect in the autophagosome formation process, AMPK activation site phosphorylation at Thr172 was reduced in the SNAP23 shRNA cells in parallel with a reduction in ULK1 activation site phosphorylation at Ser555 (Supplemental Figure 5C).
SNAP23 deficiency leads to cell death through reduced degradation of proapoptotic BAX protein.
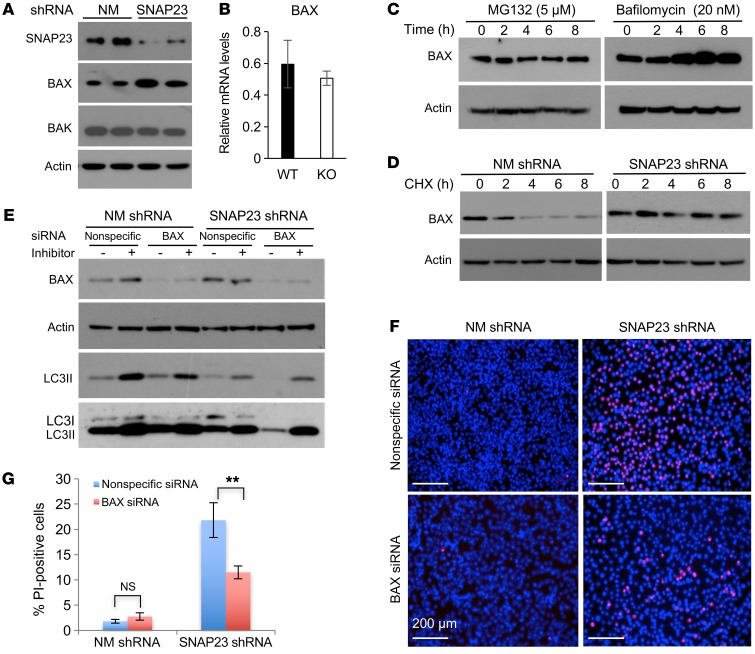
As SNAP23 deficiency–induced cell death was associated with PARP and caspase 3 cleavage, we performed immunoblotting for several of the BH3 domain–containing intrinsic cell death signaling proteins. We found that SNAP23 shRNA cells had elevated levels of BAX, the major proapoptotic BH3 protein, with no significant change in BAX mRNA levels (Figure 4, A and B). In addition, SNAP23 knockdown had no significant effect on the relative amounts of the proapoptotic BAK protein or the antiapoptotic BH3 proteins Bcl2, Bcl-xL, or Mcl1 (Supplemental Figure 6A). BAX protein undergoes both proteasome- and lysosome-mediated degradation (17). Treatment of control adipocytes with the proteasome inhibitor MG132 had no effect on BAX protein levels (Figure 4C). However, treatment with the lysosome H+/K+-ATPase inhibitor bafilomycin A1 resulted in a time-dependent increase in BAX protein levels. Inhibition of protein synthesis in NM shRNA cells with cycloheximide (CHX) resulted in a time-dependent decrease in BAX protein, whereas SNAP23-KO cells showed no significant decline in BAX protein levels (Figure 4D). LC3 immunoblotting demonstrated that BAX knockdown (siRNA) in the context of SNAP23 knockdown had no significant effect on macroautophagy (Figure 4E). However, siRNA knockdown of BAX substantially protected the SNAP23-knockdown cells from the induction of cell death by ND (Figure 4, F and G). Immunoblotting of interscapular brown and epididymal white adipocytes also revealed increased levels of BAX protein in the KO mice (Supplemental Figure 6B). We crossed BAX–/– mice with Adipoq-Cre+/– Snap23fl/+ mice to generate Adipoq-Cre+/– Snap23fl/fl BAX+/– (KO BAX+/–) mice, which resulted in partial rescue from the loss of adipose tissue mass in the KO mice (Supplemental Figure 6, C and D).
Figure 4. SNAP23 deficiency increases BAX protein levels by inhibiting BAX protein degradation.
(A) Duplicate NM shRNA and SNAP23 shRNA 3T3L1 adipocytes 8 days after differentiation were immunoblotted for BAX, BAK, and actin. The actin immunoblot is from the same samples run on parallel gels. Immunoblots shown are representative of 4 independently performed experiments. (B) BAX mRNA levels were quantified by qRT-PCR from 4 independent determinations. Data represent the mean ± SEM. (C) Control NM shRNA differentiated adipocytes were treated with 5 μM MG132 or 20 nM bafilomycin A1 for the indicated durations. Cell extracts were prepared and immunoblotted for BAX protein and actin. Immunoblots are representative of 6 independently performed experiments. (D) NM shRNA and SNAP23 shRNA cells under NR conditions were treated with 20 μg/ml CHX and at the time points indicated were immunoblotted for BAX and actin. The NM shRNA BAX immunoblot is from the same samples run on parallel gels. Immunoblot is representative of 3 independent experiments. (E) In the context of the SNAP23 shRNA, the NIH3T3 cells were transfected with nonspecific siRNA or BAX-specific siRNA. Cells were then subjected to ND conditions for 2 hours in the presence and absence of lysosomotropic agents and immunoblotted for the indicated proteins. A relatively lighter and darker exposure for the LC3 immunoblot is shown. Immunoblot is representative of 3 independent experiments. (F) Nonspecific siRNA and BAX siRNA–knockdown cells in the context of the control NM shRNA and SNAP23 shRNA cells were subjected to ND conditions for 6 hours. The cells were then labeled for PI and DAPI. Scale bars: 200 μm. (G) Quantification of PI-positive nuclei was determined by counting 500 cells from 3 independent determinations per genotype. Data represent the mean ± SEM. **P < 0.01, by ANOVA with Tukey’s post hoc test.
SNAP23 and BAX are causally linked to ATG9 function.
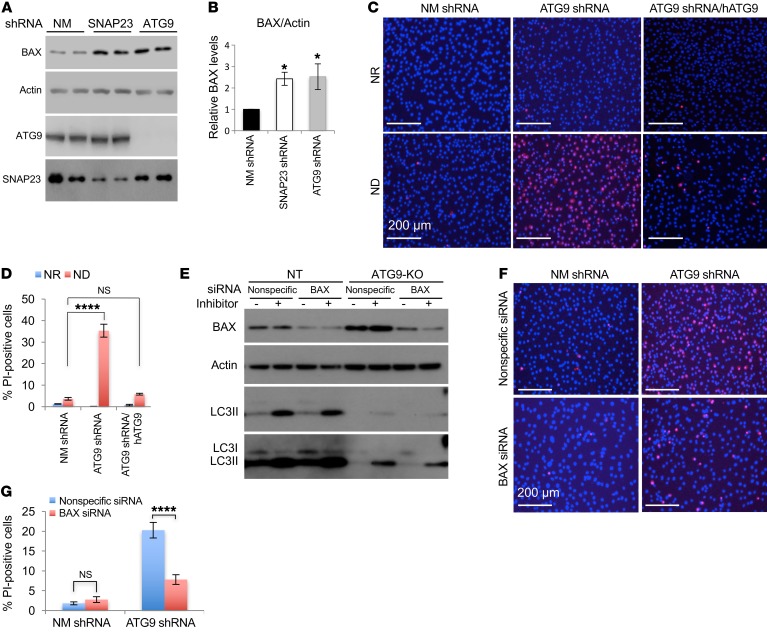
In yeast, Sec9 is the ortholog of SNAP23 and was shown to be necessary for the appropriate trafficking of atg9p required for autophagosome biogenesis (18). We therefore generated ATG9 shRNA as well as CRISPR/Cas9 ATG9 (ATG9-KO) NIH3T3 cells. As with SNAP23 shRNA cells, we observed that ATG9 shRNA cells had a comparable increase in steady-state BAX protein levels (Figure 5, A and B) and showed increased cell death upon ND that was rescued by the reexpression of shRNA-resistant ATG9 cDNA (Figure 5, C and D). ATG9 deficiency also inhibited macroautophagy as determined by LC3II formation in the presence and absence of lysosomotropic agents (Figure 5E). Moreover, we found that BAX knockdown in the context of ATG9 deficiency did not restore macroautophagy but substantially protected against cell death (Figure 5, F and G). As controls, the loss of ATG9 protein also prevented the degradation of BAX protein following CHX addition (Figure 6A), and reexpression of ATG9 in the context of ATG9 deficiency reduced the elevation of BAX protein levels and increased macroautophagy flux (Supplemental Figure 6, E–G). We also found that SNAP23 deficiency or BAX deficiency in the context of SNAP23 knockdown had no significant effect on ATG9 protein levels (Supplemental Figure 6H). In addition, BAX associated with autophagosome structures, as demonstrated by colocalization with LC3II-positive autophagic vacuoles, and this was further increased following treatment with the lysosomotropic agents (Supplemental Figure 7, A and B).
Figure 5. ATG9 deficiency increases BAX protein levels and induces cell death.
(A) Cell extracts from NM shRNA, SNAP23 shRNA, and ATG9 shRNA NIH3T3 cells maintained under NR conditions were immunoblotted for BAX, actin, ATG9, and SNAP23. The immunoblots for ATG9 and SNAP23 are from the same samples run on parallel gels. Immunoblots are representative of 3 independent experiments on 2 cell lines each. (B) BAX protein levels were quantified. Data represent the mean ± SEM. (C) NM shRNA, ATG9 shRNA, and rescued ATG9 shRNA/hATG9 cells were maintained under NR or ND conditions for 6 hours. The cells were labeled with PI and DAPI and visualized by fluorescence microscopy. Scale bars: 200μm. (D) Quantification of the PI-positive nuclei was determined by counting 500 cells from 3 independent determinations per genotype. Data represent the mean ± SEM of 3 independent experiments for each genotype. (E) Independent control and ATG9 single-guide RNA (sgRNA) NIH3T3 cells were generated and transfected with nonspecific or BAX-specific siRNA. Cells were then placed under ND conditions for 2 hours in the presence and absence of lysosomotropic agents. Cell extracts were immunoblotted for the indicated proteins. Relatively lighter and darker exposures for the LC3 immunoblot are shown. Immunoblot is representative of 3 independent experiments. (F) Nonspecific siRNA and BAX siRNA–knockdown cells in the context of the control NM shRNA– and ATG9 shRNA–knockdown cells were placed in ND conditions for 6 hours. Cells were then subjected to PI and DAPI labeling and visualized by fluorescence microscopy. Scale bars: 200 μm. (G) Quantification of PI-positive nuclei was determined by counting 500 cells from 3 independent determinations per genotype. Data represent the mean ± SEM. *P < 0.05 and ****P < 0.0001, by ANOVA with Dunnett’s or Tukey’s post hoc test.
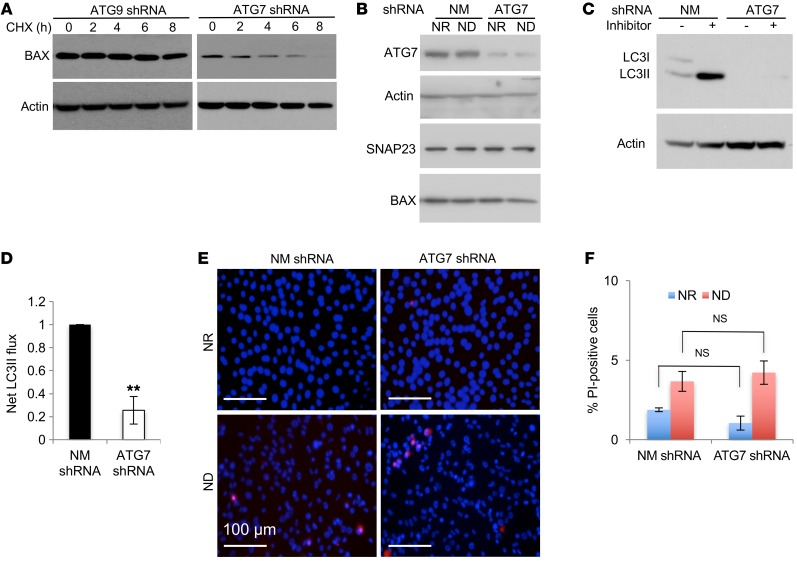
Figure 6. ATG7 deficiency suppresses macroautophagy but does not induce BAX protein levels or cell death.
(A) ATG9 shRNA and ATG7 shRNA NIH3T3 cells under NR conditions were treated with 20 μg/ml CHX, and at the indicated time points, cell extracts were prepared and immunoblotted for BAX and actin. Immunoblot is representative of 3 independent experiments. (B) NM shRNA– and ATG7 shRNA–knockdown NIH3T3 cells were maintained under NR or ND conditions for 2 hours. Cell extracts were prepared and immunoblotted for ATG7, actin, SNAP23, and BAX. Immunoblots for ATG7 and actin are from the same samples run on parallel gels. Immunoblots are representative of 3 independent experiments on 2 cell lines each. (C) NM shRNA– and ATG7 shRNA–knockdown NIH3T3 cells were then maintained under ND conditions for 2 hours in the absence and presence of lysosomotropic agents. Cell extracts were immunoblotted for LC3 and actin. (D) Net LC3II flux was calculated as the difference between LC3II protein levels in the presence and absence of the lysosomotropic agents as described in Methods. Data represent the average, with the SEM determined from 3 independent experiments. (E) Cells were then maintained under NR or ND conditions for 6 hours and subjected to PI and DAPI labeling followed by visualization under fluorescence microscopy. Scale bars: 100 μm. (F) Quantification of the PI-positive nuclei was determined by counting 500 cells. Data represent the mean ± SEM of 3 independent experiments. **P < 0.01, by Student’s t test (D) and NS, by ANOVA with Tukey’s post hoc test (F).
Inhibition of macroautophagy flux per se is not sufficient to induce BAX or to increase cell death.
Previous studies have reported that tissue-specific ATG7 KO in the liver suppresses lipophagy, resulting in liver steatosis, and in adipocytes, resulting in a cell-fate switch from white to brown/beige adipocytes, with no apparent effect on liver or adipocyte cell death (19, 20). We therefore generated ATG7 shRNA–expressing NIH3T3 cells (Figure 6B). ATG7 deficiency did not increase BAX protein levels or inhibit BAX degradation (Figure 6, A and B), nor did ATG7 deficiency result in an inhibition of macroautophagy flux as measured by LC3II protein levels in the absence and presence of lysosomotropic agents (Figure 6, C and D). Despite the inhibition of macroautophagy flux, we observed no significant effect on cell death (Figure 6, E and F). Taken together, these data indicate that inhibition of macroautophagy flux does not by itself augment cell death under ND conditions, but rather that the induction of cell death is a consequence of elevated BAX protein levels. Further, these data also demonstrate that the control of BAX protein levels is SNAP23 and ATG9 dependent, but ATG7 independent.
Since deletion of SNAP23 affects mitochondrial integrity (Supplemental Figure 5), we also assessed the potential role of mitophagy in mediating cell death. Control NM shRNA cells maintained under ND conditions for 16 hours in the presence of lysosome inhibitors displayed an increase in colocalization of the mitochondria marker TOM20 with the lysosome marker LAMP1 (Supplemental Figure 7C), along with increased TOM20 autophagy flux (Supplemental Figure 7, D and E). In the NR state, we observed no discernible colocalization of TOM20 with LAMP1 in either control or SNAP23-knockdown cells (Supplemental Figure 7C) and no increase in TOM20 flux (Supplemental Figure 7, D and E). ND for 3 hours was insufficient to induce mitophagy in control cells, as determined by either colocalization of TOM20 with LAMP1 or by TOM20 flux (Supplemental Figure 7C; Supplemental Figure 7, D and E). Likewise, we detected no significant induction of mitophagy in the SNAP23-knockdown cells. Although it was not possible to measure longer durations of ND in the SNAP23-knockdown cells, as these cells began to undergo BAX-dependent cell death, these data demonstrate that altered mitophagy did not account for the differential sensitivity to cell death by ND.
SNAP23 and ATG9 deficiency sensitizes BAX activation to ND.
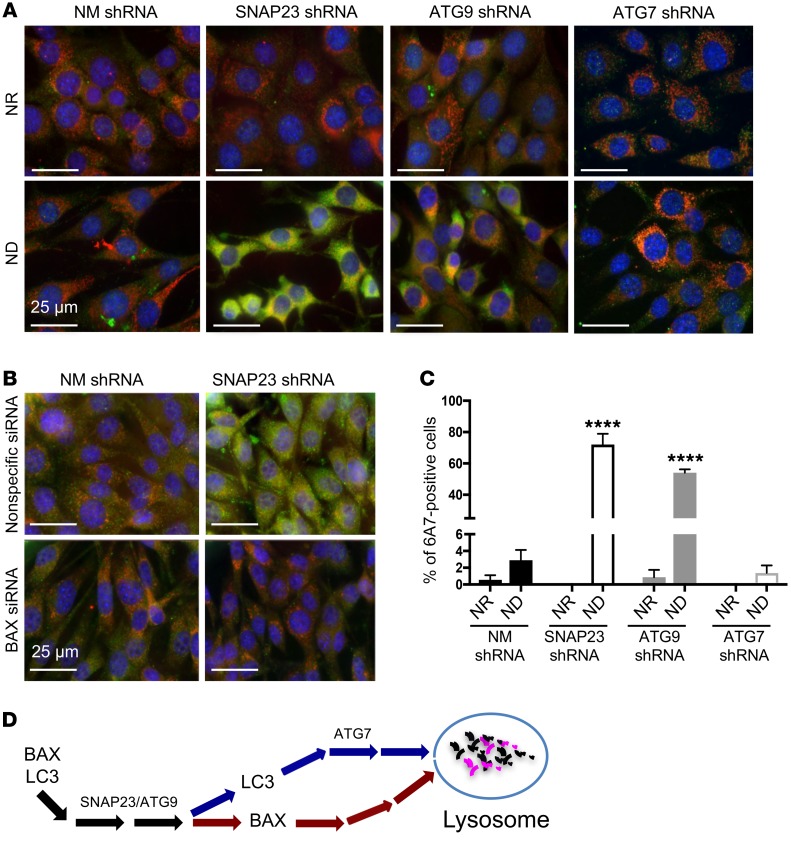
Although BAX protein levels were elevated in SNAP23- and ATG9-knockdown cells, we detected no significant increase in cell death when cells were maintained under NR conditions. This suggests that under NR conditions BAX, remains inactive but only becomes activated upon ND. To examine BAX activation, we took advantage of the 6A7 monoclonal antibody that recognizes aa 13–19 of BAX, which is only accessible in the conformational active state (21). In the NR state, control NM shRNA, SNAP23 shRNA, and ATG9 shRNA cells had relatively low levels of 6A7 immunofluorescence (Figure 7A, top 3 panels from left). However, following ND, we observed a marked increase in 6A7 immunofluorescence in the SNAP23 shRNA and ATG9 shRNA cells (Figure 7A, bottom 3 panels from left). In addition, we detected substantial colocalization of 6A7 immunofluorescence with the mitochondria marker ATP5α. In contrast, we detected relatively low 6A7 immunoreactivity in the ATG7 shRNA cells in the NR state, and ND had little effect on BAX activation in the ATG7 shRNA cells (Figure 7A, top and bottom far right panels). The percentage of 6A7 immunofluorescence–positive cells is quantified in Figure 7C, and the specificity of the 6A7 antibody is confirmed in Figure 7B. Together, these data demonstrate that SNAP23 and ATG9 deficiency not only increases BAX protein levels but also activates a pathway that is more permissive for BAX activation by ND. Moreover, this ND activation of BAX is independent of ATG7-dependent macroautophagy.
Figure 7. SNAP23 and ATG9 deficiency enhances BAX activation by ND.
(A) NM shRNA, SNAP23 shRNA, ATG9 shRNA, and ATG7 shRNA NIH3T3 cells were maintained under NR or ND conditions for 1 hour. Cells were fixed and subjected to immunofluorescence microscopy using the BAX activation–specific monoclonal antibody 6A7 (green), the mitochondria-specific antibody ATP5α (red), and DAPI (blue). Images are representative of 4 independent determinations. Scale bars: 25 μm. (B) Nonspecific siRNA and BAX siRNA–knockdown cells in the context of control NM shRNA and SNAP23 shRNA cells were maintained under ND conditions for 1 hour. Cells were fixed and subjected to immunofluorescence microscopy using the BAX activation–specific monoclonal antibody 6A7 (green), the mitochondria-specific antibody ATP5α (red), and DAPI (blue). Images are representative of 3 independent determinations. Scale bars: 25 μm. (C) Quantification of 6A7-positive cells from A was determined by counting 500 cells for each condition from 4 independent experiments. ****P < 0.0001, by ANOVA with Tukey’s post hoc test. Data represent the mean ± SEM. (D) Schematic model depicting a selective SNAP23/ATG9-dependent, but ATG7-independent, pathway mediating lysosomal degradation of BAX.
SNAP23 deficiency does not suppress nutrient-dependent calcium regulation or plasma membrane trafficking events.
SNAP23 primarily resides at the plasma membrane and in various endosomal compartments, including late endosomes, the trans-Golgi network, and lysosomes, and is responsible for various membrane fusion events (22–24). ATG9 is a trafficking membrane protein that has been reported to traffic through the plasma membrane, the trans-Golgi network, and early, late, and recycling endosomes (7, 25–28). Several studies have reported that ATG9 trafficking from intracellular sites and plasma membrane endocytosis are necessary for autophagy initiation (7, 29–31). To assess whether SNAP23 deficiency alters plasma membrane trafficking events that impinge on autophagosome formation, we examined transferrin receptor endocytosis that occurs in an AP2-, clathrin-, and dynamin-dependent manner (32, 33). Labeled transferrin underwent efficient endocytosis, as detected by intracellular Alexa Fluor 594–transferrin labeling in the control, SNAP23-, and ATG9-deficient NIH3T3 cells, and this was selectively blocked by the dynamin GTPase inhibitor dynasore (Supplemental Figure 8A) (34).
Similarly, Alexa Fluor 594–transferrin underwent a similar extent of plasma membrane endocytosis in adipocytes differentiated from WT and KO SVCs that was also prevented by dynasore (Supplemental Figure 8B). Together, these data indicate that neither SNAP23 nor ATG9 deficiency alters the classical pathway of plasma membrane endocytosis exemplified by the transferrin receptor.
SNAP23 has also been proposed to be required for adipocyte GLUT4 translocation to the plasma membrane (35–37). However, in contrast to these previous reports, we did not observe any significant reduction in glucose uptake between the control NM shRNA and SNAP23 shRNA 3T3L1 adipocytes (Supplemental Figure 8C). In addition, using the HA-GLUT4-GFP reporter, we detected no significant reduction in insulin-stimulated GLUT4 plasma membrane translocation (Supplemental Figure 8, D and E).
Since many SNARE-dependent trafficking events are dependent on intracellular calcium levels (38, 39), control NM shRNA and SNAP23 shRNA cells were preloaded with the calcium indicator Fluro-8. As shown in Supplemental Figure 8F, there was no difference in basal levels of cytosolic calcium in the NM shRNA and SNAP23 shRNA cells. Similarly, following 1 or 3 hours of ND, there was no significant change in cytosolic calcium levels.
Discussion
Intracellular membrane transport is essential for the movement of cargo between intracellular organelles and the secretion of various factors into the circulation. The large family of SNARE proteins provides the specificity and energetics necessary to drive the fusion of these numerous transport events (40). Of particular interest, SNAP23 has been implicated as a SNARE protein required for insulin-stimulated glucose uptake, GLUT4 translocation to the plasma membrane, triglyceride lipid droplet expansion, and association of lipid droplets with the mitochondria necessary for β-oxidation (35, 37, 41–43). To examine the physiologic role of SNAP23 in vivo, we generated adipocyte-specific SNAP23-KO mice. As detailed in this study, these mice displayed a surprisingly severe lipodystrophic phenotype, with essentially no adipose tissue present by 4 months of age. Consistent with the absence of adipose tissue, these mice phenocopied other lipodystrophic mouse models characterized by insulin resistance, glucose intolerance, hyperlipidemia, and hepatomegaly due to ectopic lipid accumulation in the liver.
The loss of adipose tissue specifically resulted from an induction of the death of mature adipocytes and with no significant alteration in adipocyte differentiation. This conclusion was based on the presence of normal adipose tissue in newborn mice, the loss of adipose tissue in adult mice upon induction of SNAP23 deficiency, and on analyses of in vitro differentiation and cell death markers for primary isolated adipocyte precursor cells and SNAP23-knockdown cultured 3T3L1 adipocytes. Although whole-body SNAP23 KO results in early embryonic lethality (44), and adipocyte-specific SNAP23-KO mice undergo adipocyte cell death, this is not apparently true for all cell types or tissues. Consistent with a previous study (45), we have also generated pancreatic β cell SNAP23-KO mice, and although they displayed altered insulin secretion properties, there was no significant effect on the number of β cells, or number or size of islets (unpublished observations). Interestingly, β cells express the SNAP25 isoform that is functionally interchangeable with SNAP23 in terms of insulin secretion (46), suggesting that SNAP25 may also play a role similar to that of SNAP23 in the regulation of cell death.
It should also be noted that during the preparation of the revised manuscript, we became aware that in platelets, Cre recombination results in the generation of a fragment of approximately 18 kDa that is likely initiated from an internal in-frame ATG in exon 5 (Roberto Adachi, Institute of Biosciences and Technology, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA; personal communication). However, only a nonsignificant trace of this band was detectable in adipocytes, suggesting that either the truncated message or the protein is not expressed and/or is unstable in adipocytes.
In any case, previous studies showed that a carboxyl terminal 8-aa deletion of SNAP23, but not a 49-aa carboxyl terminal deletion, in 3T3-L1 adipocytes suppressed insulin-stimulated glucose uptake by approximately 40% (35). Also, 2 other studies using polyclonal antibodies or adenovirus-mediated siRNA knockdown of SNAP23 reported an approximate 45% reduction in insulin-stimulated glucose uptake (36, 37). Since these studies have implicated SNAP23 in the control of insulin-stimulated regulation of glucose uptake, we examined both transient siRNA and stable shRNA SNAP23–knockdown adipocytes. Surprisingly, however, we found that SNAP23 was completely dispensable, with no significant effect on basal or insulin-stimulated glucose uptake or plasma membrane GLUT4 translocation. Although we do not know the basis for the differences between the findings from these previous studies and those reported here, our data clearly demonstrate that SNAP23 deficiency does not have any significant effect on either basal or insulin-stimulated glucose uptake or GLUT4 translocation to the plasma membrane. Further studies will also be necessary to determine whether another SNARE protein can compensate for the absence of SNAP23.
We also found that SNAP23 heterodeficiency had no significant effect on adipose tissue cell mass or glucose metabolism (unpublished observations), suggesting that a reduction of more than 50% in SNAP23 protein is necessary to observe this phenotype. Although several genes have been associated with lipodystrophic phenotypes (47–53), to our knowledge this has not yet been observed for SNAP23. However, since homozygotic disruption of SNAP23 resulted in early embryonic lethality in mice, it is likely that complete SNAP23 deficiency in humans would not result in any surviving offspring. Nevertheless, several GWAS studies have identified multiple SNAP23 SNPs associated with type 2 diabetes, fasting hyperglycemia, and BMI (Type 2 Diabetes Knowledge Portal; http://www.type2diabetesgenetics.org/).
One important clue to the mechanistic role of SNAP23 in the control of cell death came from the observation that SNAP23 knockdown did not impair the growth of NIH3T3 fibroblasts or the growth or in vitro differentiation of 3T3L1 fibroblasts into adipocytes, as long as they were maintained under NR conditions. Moreover, starving young SNAP23-deficient mice had a markedly accelerated rate of adipocyte cell death in vivo. These data suggest that SNAP23 is linked to macroautophagy and are consistent with previous reports showing that the yeast ortholog of SNAP23, -25 (sec9) plays an important role in the control of autophagy through the trafficking of atg9p (5).
In general, autophagy is considered an important cell-survival process, as it removes damaged proteins and organelles to reduce cell stress and provide energy under ND cellular states (54). Thus, both defective or prolonged autophagy activation can result in cell death, in the former by accumulation of defective organelles (i.e., mitochondria) and/or increases in endoplasmic reticulum stress, and in the latter by the degradation of cellular machinery, energy production, and substrates for macromolecular synthesis (55). Our data suggest that there are also specific molecular signaling pathways linking selective components of the macroautophagy machinery to intrinsic programmed cell death through control of the proapoptotic protein BAX, as schematically illustrated in Figure 7D.
SNAP23 deficiency in culture or in KO mice showed increased levels of BAX protein, without any significant effect on other proapoptotic, antiapoptotic, or sensitizer BH3 protein family members. Importantly, BAX knockdown in the context of SNAP23 deficiency in vitro protected against the induction of cell death, and BAX haploinsufficiency in vivo substantially, though not totally, protected against the decrease in adipose tissue mass in SNAP23-KO mice. Likewise, in culture, ATG9 deficiency also resulted in the elevation of BAX protein levels, and BAX knockdown protected against ATG9 deficiency–induced cell death. BAX has been reported to have a relatively short half-life and undergoes both proteasome- and lysosome-mediated degradation (17, 56). Given the use of proteasome and lysosome inhibitors in WT adipocytes, the majority of BAX degradation is lysosome mediated, with an apparent half-life of 4 to 6 hours. In our cell culture systems, the inhibition of macroautophagy by SNAP23 and ATG9 deficiency or with lysosomotropic agents resulted in an approximately 2-fold elevation of steady-state BAX protein levels. Importantly, however, inhibition of macroautophagy through other means does not necessarily result in an increase in BAX protein levels or enhancement of cell death. For example, adipocyte-specific ATG7-KO mice have defective macroautophagy, however, in this case, white adipocytes appear to undergo an alteration in developmental pattern to become beige adipocytes, but with no evidence of cell death (19, 20). Our data also demonstrate that ATG7 knockdown in cultured adipocytes also display defective macroautophagy, but without any evidence of cell death or elevation in BAX protein levels. Thus, it appears that the lysosomal component of BAX protein degradation is selective for specific functions of SNAP23 and ATG9, but independent of other parts of the macroautophagy pathway, at least for ATG7.
Our data indicate that neither SNAP23 nor ATG9 deficiency perturbs plasma membrane endocytosis, at least as assessed by transferrin uptake by the transferrin receptor, a classical clathrin-dependent endocytosis process, or plasma membrane exocytosis, as assessed by insulin-stimulated GLUT4 translocation. Although we do not yet know the specific point of intersection between SNAP23 and ATG9, our data showing SNAP23- and ATG9-dependent, but ATG7-independent, regulation of BAX protein levels are similar to those reported in other studies indicating alternative noncanonical pathways that control macroautophagy. For example, etoposide-induced, stress-activated autophagic vesicle formation and macroautophagy-dependent protein degradation formation occurred normally in Atg5–/– mouse embryonic fibroblasts, despite inhibition of LC3II flux (57). At the onset of puparium formation, the Drosophila intestine undergoes a reduction in midgut length through a macroautophagic process (58, 59). In this system, ATG3 and ATG7, but not other ATG genes such ATG1, -2, -8 and -18, are dispensable for midgut size reduction. In contrast, ATG7 is required for nutrient deprivation–induced autophagy activation in the larval fat body (58). These data suggest that alternative autophagy pathways are used in a signal- and cell context–dependent manner.
Our data also beg the question that if steady-state BAX levels are elevated by SNAP23 and ATG9 deficiency, why did adipocyte death only occur under ND conditions, and why was it not apparent under NR conditions? BAX is predominantly localized in the cytosol, where it exists as a quiescent monomer or in an inactive dimer conformation (60). Following a proapoptotic trigger, the autoinhibited BAX dimer dissociates to BAX monomers, and these monomers undergo a multistep activation process that includes conformational changes exposing the amino and carboxyl terminal domains, insertion into the mitochondria outer membrane, and homo- and heterotypic oligomerization (61). The relatively weak immunoreactivity with the conformation-specific BAX antibody 6A7 demonstrated that, despite the increased levels of BAX protein in the SNAP23- and ATG9-deficient cells, BAX remained in an inactive state under NR conditions. In contrast, upon ND, BAX protein became activated, resulting in mitochondrial outer membrane permeabilization and a consequent sensitization of the cells to apoptotic cell death. Importantly, enhanced activation of BAX was also independent of generalized macroautophagy, as defined by LC3II flux, since the ATG7-knockdown cells also had suppressed macroautophagy but did not result in enhanced BAX activation upon ND. Further studies are now needed to determine the specific mechanism(s) responsible for BAX activation under ND conditions and the unique role of SNAP23 and ATG9, distinct from ATG7-dependent macroautophagy, in mediating the lysosomal trafficking and degradation of BAX.
Methods
Generation of KO and Adipoq-CreERT2+/–Snap23fl/fl mice. The Snap23fl/fl mice were generated by Ozgene by introducing a LoxP site flanking exons 3 and 4 of SNAP23. Adipocyte-specific SNAP23-KO mice (Adipoq Snap23fl/fl) were obtained by crossing Snap23fl/fl mice with Adipoq-Cre mice, which were provided by Evan Rosen (Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA). Likewise, the tamoxifen-inducible, adipocyte-specific KO (Adipoq-CreERT2+/– Snap23fl/fl) mice were obtained by crossing Snap23fl/fl mice with Adipoq-CreERT2 mice, which were provided by Stefan Offermanns (Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany). BAX–/– mice were purchased from The Jackson Laboratory.
Tamoxifen preparation and administration.
Tamoxifen (T5648, MilliporeSigma) was dissolved in 100% ethanol at 10 mg/ml. Immediately before injection, 200 μl of 10 mg/ml tamoxifen was mixed with 300 μl corn oil (C8267, MilliporeSigma) and then vacuum centrifuged to remove the ethanol. The Adipoq-CreERT2+/– Snap23fl/fl and control Adipoq-CreERT2+/– Snap23+/+ mice were then treated with daily intraperitoneal tamoxifen injections over a 5-day period. The mice were analyzed 21 days later.
Isolation of SVCs and adipocyte differentiation.
SVCs were isolated and differentiated as previously described (9). Briefly, epididymal fat pads were dissected and then minced in HEPES-buffered DMEM supplemented with 10 mg/ml fatty acid–free BSA. The tissue fragments were centrifuged at 1,000 g for 10 minutes at 4°C to pellet erythrocytes and other blood cells, and the tissue suspension was incubated with 0.05 mg/ml Liberase (Roche) and 50 units/ml DNase I (MilliporeSigma) at 37°C in an orbital shaker (150 Hz) for 25 to 30 minutes. The digested samples were filtered through a sterile 250-μm nylon mesh, and the suspension was centrifuged at 1,000 g for 5 minutes to pellet the SVCs. The isolated SVCs were cultured in high-glucose DMEM (11965, Gibco, Thermo Fisher Scientific) supplemented with 20% FBS and penicillin-streptomycin and maintained until confluent (for the typical 2-day period) prior to initiation of the differentiation protocol. Adipocyte differentiation was induced by treating confluent cells for 48 hours in medium containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 μM dexamethasone, 850 nM insulin, 1 nM T3, and 1 μM rosiglitazone. Two days after induction, the cells are switched to maintenance medium containing 10% FBS, 850 nM insulin, 1 nM T3, and 1 μM rosiglitazone for another 2 days. Cells are maintained in regular medium, and the medium was changed every 2 or 4 days.
TEM.
BAT from 2-week-old WT and KO mice was subjected to TEM as previously described (62). Briefly, the tissue was fixed in 2.5% glutaraldehyde in 100 mM sodium cacodylate, pH 7.4, and postfixed in 1% osmium tetroxide in sodium cacodylate followed by 1% uranyl acetate. After ethanol dehydration and embedment in LX112 resin (LADD Research Industries), ultrathin sections were stained with uranyl acetate followed by lead citrate. Autophagic vacuoles (vesicles <0.5 μm) were classified as autophagosomes when they met 2 or more of the following criteria: double membranes (complete or at least partially visible), absence of ribosomes attached to the cytosolic side of the membrane, luminal density similar to that of cytosol, and identifiable organelles or regions of organelles in their lumen.
Cell culture.
Murine 3T3L1 preadipocyte culture and adipocyte differentiation were prepared as previously described (63). Human embryonic kidney 293T (HEK293T) cells and mouse fibroblast NIH3T3 cells were cultured in NR medium containing DMEM supplemented with 10% FBS and 1× penicillin-streptomycin. To induce ND conditions, cells were placed in RPMI medium without aa, glucose, or glutamine (R9010-01, USBiological Life Sciences) for 1 hour for BAX activation and 6 hours for PI staining experiments. For autophagy flux experiments, cells were placed in RPMI medium without aa (R8999, USBiological Life Sciences) for 4 hours, with or without lysosome inhibitors. Cell lines were maintained in a 5% CO2 incubator at 37°C.
Catecholamine-stimulated lipolysis.
3T3L1 adipocytes were differentiated in NR medium and on day 8 were placed in KRH buffer (136 mM NaCl, 4.7 mM KCl, 1.25 mM MgSO4, 1.25 mM CaCl2, 20 mM HEPES, pH 7.4, and 1% BSA) and then stimulated with or without isoproterenol (1 μM) for 120 minutes. Free fatty acids and glycerol, which were released from adipocytes into KRH buffer, were analyzed using a nonesterified fatty acid (NEFA) kit (Wako) and free glycerol reagent (MilliporeSigma) and normalized to total adipocyte protein levels.
Intracellular calcium levels.
NIH3T3 cells were cultured under NR and ND conditions as described above for 0, 1, or 3 hours, washed once with HBSS buffer, and then incubated with 100 μl Fluo-8 solution (Abcam) for 30 minutes at 37°C, followed by 1 wash with HBSS and an additional 30-minute incubation in their own medium. Calcium intensity was analyzed at excitation/emission (Ex/Em) = 490:525 nm.
Cell death determination.
Plasma membrane permeability was determined by incubating cells with 250 ng/ml PI and 5 μg/ml Hoechst 33342 for 10 minutes and then analyzing them under a microscope. Cell death was also assessed by the internucleosomal degradation of genomic DNA using the Cell Death Detection ELISA Plus Kit (Roche Molecular Biochemicals). Background values (incubation with buffer alone) were subtracted, and optical density values representing nucleosomal DNA fragments in treated samples were compared with those values obtained from the untreated control cells and expressed as the fold increase of oligonucleosomes. Lentiviral shRNA knockdown and cDNA overexpression MISSION lentiviral shRNA bacterial glycerol stocks for SNAP23, ATG9a, ATG7, ATG5, ATG16, and NM control plasmids were obtained from MilliporeSigma. The lentiviral human SNAP23 and ATG9a cDNA plasmids were obtained from GeneCopoeia. The plasmid DNAs were purified using the HiSpeed Plasmid Maxi Kit (QIAGEN) and transfected into HEK293T cells along with Lentiviral Packaging Mix (MilliporeSigma) to produce lentivirus packed with shRNA and cDNA according to the manufacturer’s instructions. 3T3-L1 preadipocytes (80% confluence) were lentivirus infected, selected with puromycin (2.5 μg/ml), and subjected to standard adipocyte differentiation. NIH3T3 cells were infected with the lentivirus and also selected with puromycin (2.5 μg/ml) for 3 days.
Perilipin and H&E staining of adipose tissue.
Perilipin staining was performed as previously described (9). Briefly, adipose tissue was fixed for 24 to 36 hours at room temperature in zinc-formalin fixative and embedded in paraffin. Paraffin-embedded adipose tissue was sectioned, deparaffinized, and heated in antigen unmasking solution for antigen retrieval, and then cooled, washed, and blocked with 10% goat serum in Tris-buffered saline with Tween (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20). The sections were incubated overnight with F4/80 (70076, Cell Signaling Technology) and perilipin 1 (3470, Cell Signaling Technology) primary antibodies followed by fluorescence-conjugated secondary antibodies. The sections were counterstained in ProLong Gold Antifade Reagent (Invitrogen, Thermo Fisher Scientific) with DAPI and visualized by confocal fluorescence microscopy. Zinc-fixed, paraffin-embedded fragments of adipose tissues were sectioned (5-μm-thick) and subjected to standard H&E staining at the Albert Einstein College of Medicine Histology Core facility.
Total RNA extraction and qRT-PCR analysis.
Cellular total RNA was extracted using QIAzol Lysis Reagent and an RNeasy Mini Kit (QIAGEN). First-strand cDNA was synthesized using SuperScript VILO cDNA Synthesis Kit (Invitrogen, Thermo Fisher Scientific). Gene expression levels were determined by SYBR green (QIAGEN) or by TaqMan (Applied Biosystems) according to the manufacturer’s instructions.
Macroautophagy and mitophagy measurements.
Macroautophagy was calculated by counting the number of GFP-LC3 puncta under NR and ND conditions with and without treatment for 2 hours with the lysosomotropic agents leupeptin (100 μM) and NH4Cl (20 mM). Autophagic flux was determined by transient transfection with the mCherry-GFP-LC3 reporter plasmid (16). Quantification of yellow fluorescent cytosolic particles (APG) and red fluorescent cytosolic particles (APL) per cell was determined using the Analyze Particles function of ImageJ (NIH) after applying a fixed threshold for puncta in all images (64). Net LC3II and p62 flux was calculated as the difference in levels of LC3II or p62 protein normalized to loading-control actin protein levels, with and without lysosomotropic agent treatment for 2 hours. Mitophagy was calculated by colocalization of TOM20 with Lamp1 under long-term starvation (DMEM without serum for 16 h) or short-term starvation (RPMI without aa, glucose, glutamine, or serum) for 1 hour or 3 hours. TOM20 flux was determined as the difference in TOM20 protein levels (Western blotting) in the presence and absence of a lysosomotropic agent.
Energy balance and physiologic characterization.
Body composition parameters that included total body weight, total water, and lean and fat mass were determined by quantitative nuclear magnetic resonance noninvasive imaging as described previously (65). Spontaneous locomotor activity, energy expenditure, oxygen consumption (VO2), and carbon dioxide production (VCO2) were determined by indirect calorimetry (Oxymax CLAMS System, Columbus Instruments) as described previously (65).
GTTs and ITTs.
GTTs were performed on mice fasted for 16 hours and then given an intraperitoneal injection of glucose (1 g/kg body weight). ITTs were performed on mice fasted for 5 hours and then given an intraperitoneal injection of insulin (1 U insulin/kg body weight). Blood was collected at various time points, and glucose concentration was determined using the OneTouch Glucose Monitoring System (LifeScan Inc.). Insulin levels were analyzed using an Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem Inc.)
Immunoblot analysis.
Cultured cells were washed with cold PBS and scraped and homogenized using Ceria Stabilized Zirconium Oxide Beads (MidSci) in RIPA lysis buffer (sc-24948, Santa Cruz Biotechnology) containing Protease and Phosphatase Inhibitor Cocktail (78442, Thermo Fisher Scientific). The cell homogenates were centrifuged for 15 minutes at 21,000 g at 4°C, and supernatants were collected for protein assay. Protein samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane using the iBlot Blotting System (Thermo Fisher Scientific). The immunoblot membrane was blocked with SuperBlock (TBS) Blocking Buffer (37535, Life Technologies, Thermo Fisher Scientific) and incubated with a primary antibody in the blocking buffer. The immunoblot membranes were washed in Tris-buffered saline with Tween-20 (TBST) and incubated with HRP-conjugated secondary antibody in TBST with 3% BSA. The Membrane was then washed with TBST and visualized through ECL (Pierce, Thermo Fisher Scientific). ImageJ software was used to compare relative protein levels.
Detection of BAX activation by 6A7 immunofluorescence.
NIH3T3 cells were grown on coverslips and treated for 1 hour with NR or ND medium, as described above. Cells were washed once with PBS and fixed with 4% paraformaldehyde at room temperature for 15 minutes and permeabilized with PBS containing 0.1% Triton X-100 for 10 minutes. Cells were blocked with PBS containing 3% w/v Bovine Serum Albumin Fraction V (Roche) for 1 hour and then incubated overnight with a 1:100 dilution of the activation-specific BAX antibody 6A7 (556467, BD Biosciences, 1:100 dilution) and a 1:100 dilution of the inner mitochondrial protein ATP5α (Abcam) at 4°C. After washing with PBS, slides were incubated with Alexa Fluor secondary antibodies (T13343, Thermo Fisher Scientific) in blocking solution at room temperature for 1 hour, mounted with VECTASHIELD HardSet Mounting Medium and stained with DAPI (Vector Laboratories). Fluorescent signals were visualized with an Axio Observer Z1 microscope (Carl Zeiss), and signal intensity was quantitated using ImageJ software to integrate the 6A7 fluorescence intensity per cell.
Transferrin receptor endocytosis.
NIH3T3 and primary isolated adipocyte SVCs differentiated into adipocytes were maintained under NR conditions. The cells were placed in ND medium for 1 hour and then incubated with vehicle (DMSO) or 80 μM dynasore for 30 minutes. Alexa Fluor 594–conjugated transferrin (Life technologies, Thermo Fisher Scientific) was added to cells for 10 minutes followed by acid wash (1-min wash with 0.5 M glycine, pH 2.8) and then PBS (pH 7.0) prior to fixation in 4% paraformaldehyde. Cells were imaged by confocal fluorescence microscopy.
Insulin-stimulated glucose uptake and GLUT4 translocation.
3T3L1 adipocytes were differentiated in NR media for 8 days and then incubated with 50 μM 2-deoxyglucose containing 0.5 μCi of 2-[3H] deoxyglucose in the absence or presence of 100 nM insulin. The cells were washed, and incorporated radioactivity was extracted by lysing cells with 1.0 ml of 0.2N NaOH for 30 minutes. The extracts were subjected to scintillation counting, and the are data presented as 2-deoxyglucose uptake per gram protein per minute (66). GLUT4 translocation was determined as previously described (67). Briefly, 3T3L1 cells 4 days after initiation of the differentiation protocol were transfected with HA-GLUT4-GFP cDNA and siRNAs. Forty-eight hours later, the cells were either left untreated or stimulated with insulin for 15 minutes. Cells were fixed and incubated with a mouse HA monoclonal antibody (MMS-101R, Covance) in the absence of detergent. Cells were then incubated with Cy3 goat anti-mouse, and the ratio of Cy3 to GFP fluorescence signals was quantified using MetaMorph processing software (Molecular Devices).
Statistics.
Group data are presented as the mean ± SEM. Statistical significance was calculated using a paired or unpaired 2-tailed Student’s t test and ANOVA with Tukey’s or Dunnett’s post hoc test for multiple comparisons where appropriate. A P value of less than 0.05 was considered statistically significant.
Study approval.
The IACUC of the Albert Einstein College of Medicine approved all animal experiments performed in this study.
Author contributions
DF performed all the experiments presented in Figures 1–6 and Supplemental Figures 1–3, 5, and 6, analyzed the data, prepared figures, and participated in the drafting and editing of the manuscript. DA performed and analyzed the data presented in Figure 7 and edited the manuscript. TPW and J Wei performed the glucose uptake assays (Supplemental Figure 8) and edited the manuscript. J Wen and TEM performed the GLUT4 translocation assays (Supplemental Figure 8) and edited the manuscript. RS performed and quantified the TEM data presented in Supplemental Figure 5 and edited the manuscript. RNK assisted in the experimental design and interpretation of the data and participated in the drafting and editing of the manuscript. JEP supervised the project, assisted in the experimental design and interpretation of the data, and helped draft and edit the manuscript.
Supplementary Material
Acknowledgments
We wish to thank Evan Rosen (Beth Israel Deaconess Medical Center, Boston, USA) and Stefan Offermanns (Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany) for their gifts of the Adipoq-Cre and Adipoq-CreERT2 mice. This study was support in part by grants from the NIH (DK033823, DK020541, HL13086, and HL128071), the American Heart Association (15CSA26240000HL130861), and the Foundation Leducq Transatlantic Networks of Excellence.
Version 1. 08/13/2018
Electronic publication
Version 2. 08/15/2018
Error corrected in Figure 3A
Version 3. 08/31/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(9):3941–3956.https://doi.org/10.1172/JCI99217.
Contributor Information
Daorong Feng, Email: daorong.feng@einstein.yu.edu.
Dulguun Amgalan, Email: Dulguun.amgalan@phd.einstein.yu.edu.
Rajat Singh, Email: rajat.singh@einstein.yu.edu.
Jianwen Wei, Email: jiw2018@med.cornell.edu.
Jennifer Wen, Email: jew2019@med.cornell.edu.
Tszki Peter Wei, Email: wei.pet2020@gmail.com.
Timothy E. McGraw, Email: temcgraw@med.cornell.edu.
Jeffrey E. Pessin, Email: jeffrey.pessin@einstein.yu.edu.
References
- 1.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362(6418):318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92(6):759–772. doi: 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- 3.Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2(2):98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 5.Nair U, Klionsky DJ. Autophagosome biogenesis requires SNAREs. Autophagy. 2011;7(12):1570–1572. doi: 10.4161/auto.7.12.18001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146(2):303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154(6):1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaul S, Mittal SK, Feigenbaum L, Kruhlak MJ, Roche PA. Expression of the SNARE protein SNAP-23 is essential for cell survival. PLoS ONE. 2015;10(2):e0118311. doi: 10.1371/journal.pone.0118311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng D, et al. High-fat diet-induced adipocyte cell death occurs through a cyclophilin D intrinsic signaling pathway independent of adipose tissue inflammation. Diabetes. 2011;60(8):2134–2143. doi: 10.2337/db10-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SM, et al. Loss of white adipose hyperplastic potential is associated with enhanced susceptibility to insulin resistance. Cell Metab. 2014;20(6):1049–1058. doi: 10.1016/j.cmet.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenwood MR, Hirsch J. Postnatal development of adipocyte cellularity in the normal rat. J Lipid Res. 1974;15(5):474–483. [PubMed] [Google Scholar]
- 12.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Lopez N, et al. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2016;23(1):113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 15.Nyfeler B, Bergman P, Wilson CJ, Murphy LO. Quantitative visualization of autophagy induction by mTOR inhibitors. Methods Mol Biol. 2012;821:239–250. doi: 10.1007/978-1-61779-430-8_14. [DOI] [PubMed] [Google Scholar]
- 16.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 17.Guan JJ, Zhang XD, Sun W, Qi L, Wu JC, Qin ZH. DRAM1 regulates apoptosis through increasing protein levels and lysosomal localization of BAX. Cell Death Dis. 2015;6:e1624. doi: 10.1038/cddis.2014.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119(11):3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106(47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peyerl FW, et al. Elucidation of some Bax conformational changes through crystallization of an antibody-peptide complex. Cell Death Differ. 2007;14(3):447–452. doi: 10.1038/sj.cdd.4402025. [DOI] [PubMed] [Google Scholar]
- 22.Valdez AC, Cabaniols JP, Brown MJ, Roche PA. Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-Golgi network. J Cell Sci. 1999;112(Pt 6):845–854. doi: 10.1242/jcs.112.6.845. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Whiteheart SW. Intracellular localization of SNAP-23 to endosomal compartments. Biochem Biophys Res Commun. 1999;255(2):340–346. doi: 10.1006/bbrc.1999.0173. [DOI] [PubMed] [Google Scholar]
- 24.Leung SM, Chen D, DasGupta BR, Whiteheart SW, Apodaca G. SNAP-23 requirement for transferrin recycling in Streptolysin-O-permeabilized Madin-Darby canine kidney cells. J Biol Chem. 1998;273(28):17732–17741. doi: 10.1074/jbc.273.28.17732. [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W. AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci. 2012;125(Pt 7):1706–1715. doi: 10.1242/jcs.093203. [DOI] [PubMed] [Google Scholar]
- 26.Mari M, Reggiori F. Atg9 reservoirs, a new organelle of the yeast endomembrane system? Autophagy. 2010;6(8):1221–1223. doi: 10.4161/auto.6.8.13792. [DOI] [PubMed] [Google Scholar]
- 27.Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119(Pt 18):3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 28.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21(22):3998–4008. doi: 10.1091/mbc.e10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou C, et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27(2):184–201. doi: 10.1038/cr.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popovic D, Dikic I. TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep. 2014;15(4):392–401. doi: 10.1002/embr.201337995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. ATG16L1 meets ATG9 in recycling endosomes: additional roles for the plasma membrane and endocytosis in autophagosome biogenesis. Autophagy. 2014;10(1):182–184. doi: 10.4161/auto.27174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Renswoude J, Bridges KR, Harford JB, Klausner RD. Receptor-mediated endocytosis of transferrin and the uptake of fe in K562 cells: identification of a nonlysosomal acidic compartment. Proc Natl Acad Sci U S A. 1982;79(20):6186–6190. doi: 10.1073/pnas.79.20.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dam EM, Stoorvogel W. Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol Biol Cell. 2002;13(1):169–182. doi: 10.1091/mbc.01-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10(6):839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Kawanishi M, Tamori Y, Okazawa H, Araki S, Shinoda H, Kasuga M. Role of SNAP23 in insulin-induced translocation of GLUT4 in 3T3-L1 adipocytes. Mediation of complex formation between syntaxin4 and VAMP2. J Biol Chem. 2000;275(11):8240–8247. doi: 10.1074/jbc.275.11.8240. [DOI] [PubMed] [Google Scholar]
- 36.Foster LJ, Yaworsky K, Trimble WS, Klip A. SNAP23 promotes insulin-dependent glucose uptake in 3T3-L1 adipocytes: possible interaction with cytoskeleton. Am J Physiol. 1999;276(5 Pt 1):C1108–C1114. doi: 10.1152/ajpcell.1999.276.5.C1108. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi T, et al. The t-SNAREs syntaxin4 and SNAP23 but not v-SNARE VAMP2 are indispensable to tether GLUT4 vesicles at the plasma membrane in adipocyte. Biochem Biophys Res Commun. 2010;391(3):1336–1341. doi: 10.1016/j.bbrc.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 38.Peters C, Mayer A. Ca2+/calmodulin signals the completion of docking and triggers a late step of vacuole fusion. Nature. 1998;396(6711):575–580. doi: 10.1038/25133. [DOI] [PubMed] [Google Scholar]
- 39.Hay JC. Calcium: a fundamental regulator of intracellular membrane fusion? EMBO Rep. 2007;8(3):236–240. doi: 10.1038/sj.embor.7400921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24(1):35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Min J, et al. Synip: a novel insulin-regulated syntaxin 4-binding protein mediating GLUT4 translocation in adipocytes. Mol Cell. 1999;3(6):751–760. doi: 10.1016/S1097-2765(01)80007-1. [DOI] [PubMed] [Google Scholar]
- 42.Jägerström S, et al. Lipid droplets interact with mitochondria using SNAP23. Cell Biol Int. 2009;33(9):934–940. doi: 10.1016/j.cellbi.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Boström P, et al. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9(11):1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 44.Suh YH, et al. Deletion of SNAP-23 results in pre-implantation embryonic lethality in mice. PLoS ONE. 2011;6(3):e18444. doi: 10.1371/journal.pone.0018444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunii M, et al. Opposing roles for SNAP23 in secretion in exocrine and endocrine pancreatic cells. J Cell Biol. 2016;215(1):121–138. doi: 10.1083/jcb.201604030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadoul K, et al. SNAP-23 is not cleaved by botulinum neurotoxin E and can replace SNAP-25 in the process of insulin secretion. J Biol Chem. 1997;272(52):33023–33027. doi: 10.1074/jbc.272.52.33023. [DOI] [PubMed] [Google Scholar]
- 47.Cao H, Alston L, Ruschman J, Hegele RA. Heterozygous CAV1 frameshift mutations (MIM 601047) in patients with atypical partial lipodystrophy and hypertriglyceridemia. Lipids Health Dis. 2008;7:3. doi: 10.1186/1476-511X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim CA, et al. Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy. J Clin Endocrinol Metab. 2008;93(4):1129–1134. doi: 10.1210/jc.2007-1328. [DOI] [PubMed] [Google Scholar]
- 49.Hayashi YK, et al. Human PTRF mutations cause secondary deficiency of caveolins resulting in muscular dystrophy with generalized lipodystrophy. J Clin Invest. 2009;119(9):2623–2633. doi: 10.1172/JCI38660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Lay S, et al. The lipoatrophic caveolin-1 deficient mouse model reveals autophagy in mature adipocytes. Autophagy. 2010;6(6):754–763. doi: 10.4161/auto.6.6.12574. [DOI] [PubMed] [Google Scholar]
- 51.Shastry S, Delgado MR, Dirik E, Turkmen M, Agarwal AK, Garg A. Congenital generalized lipodystrophy, type 4 (CGL4) associated with myopathy due to novel PTRF mutations. Am J Med Genet A. 2010;152A(9):2245–2253. doi: 10.1002/ajmg.a.33578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, et al. BSCL2/seipin regulates adipogenesis through actin cytoskeleton remodelling. Hum Mol Genet. 2014;23(2):502–513. doi: 10.1093/hmg/ddt444. [DOI] [PubMed] [Google Scholar]
- 53.Salle-Teyssières L, et al. Maladaptative autophagy impairs adipose function in congenital generalized lipodystrophy due to Cavin-1 deficiency. J Clin Endocrinol Metab. 2016;101(7):2892–2904. doi: 10.1210/jc.2016-1086. [DOI] [PubMed] [Google Scholar]
- 54.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci U S A. 2000;97(8):3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461(7264):654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 58.Chang TK, et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013;15(9):1067–1078. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee CY, Cooksey BA, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250(1):101–111. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- 60.Garner TP, et al. An Autoinhibited Dimeric Form of BAX Regulates the BAX Activation Pathway. Mol Cell. 2016;63(3):485–497. doi: 10.1016/j.molcel.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westphal D, Kluck RM, Dewson G. Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 2014;21(2):196–205. doi: 10.1038/cdd.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams D, Pessin JE. Mapping of R-SNARE function at distinct intracellular GLUT4 trafficking steps in adipocytes. J Cell Biol. 2008;180(2):375–387. doi: 10.1083/jcb.200709108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bejarano E, Yuste A, Patel B, Stout RF, Spray DC, Cuervo AM. Connexins modulate autophagosome biogenesis. Nat Cell Biol. 2014;16(5):401–414. doi: 10.1038/ncb2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zong H, Armoni M, Harel C, Karnieli E, Pessin JE. Cytochrome P-450 CYP2E1 knockout mice are protected against high-fat diet-induced obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2012;302(5):E532–E539. doi: 10.1152/ajpendo.00258.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanzaki M, Pessin JE. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J Biol Chem. 2001;276(45):42436–42444. doi: 10.1074/jbc.M108297200. [DOI] [PubMed] [Google Scholar]
- 67.Lampson MA, Racz A, Cushman SW, McGraw TE. Demonstration of insulin-responsive trafficking of GLUT4 and vpTR in fibroblasts. J Cell Sci. 2000;113(Pt 22):4065–4076. doi: 10.1242/jcs.113.22.4065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.