Abstract
Strains of the filamentous fungus Cochliobolus carbonum that produce the host-selective compound HC-toxin, a cyclic tetrapeptide, are highly virulent on certain genotypes of maize (Zea mays L.). Production of HC-toxin is under the control of a complex locus, TOX2, which is composed of at least seven linked and duplicated genes that are present only in toxin-producing strains of C. carbonum. One of these genes, TOXE, was earlier shown to be required for the expression of the other TOX2 genes. TOXE has four ankyrin repeats and a basic region similar to those found in basic leucine zipper (bZIP) proteins, but lacks any apparent leucine zipper. Here we show that TOXE is a DNA-binding protein that recognizes a ten-base motif (the “tox-box”) without dyad symmetry that is present in the promoters of all of the known TOX2 genes. Both the basic region and the ankyrin repeats are involved in DNA binding. A region of TOXE that includes the first ankyrin repeat is necessary and sufficient for transcriptional activation in yeast. The data indicate that TOXE is the prototype of a new family of transcription factor, so far found only in plant-pathogenic fungi. TOXE plays a specific regulatory role in HC-toxin production and, therefore, pathogenicity by C. carbonum.
Interactions between plants and their pathogens frequently show a high degree of specificity, in which single genes in the host and the pathogen control whether the interaction will result in susceptibility or resistance (1). Among the known agents of specificity are the host-selective toxins, low-molecular-weight compounds produced by certain fungal pathogens, notably species of Cochliobolus and Alternaria (2, 3). In the interaction between the filamentous fungus Cochliobolus carbonum and its host, maize, specificity is controlled by HC-toxin, a cyclic peptide host-selective toxin with the structure cyclo-(d-Pro-l-Ala-d-Ala-l-Aeo), where Aeo stands for 2-amino-9,10-epoxy-8-oxodecanoic acid. Disease occurs only when HC-toxin-producing (Tox2+) isolates of C. carbonum infect maize plants that are homozygous recessive at the Hm1 and Hm2 loci. These genes encode NADPH-dependent reductases that confer insensitivity to HC-toxin, and hence resistance to C. carbonum, by reducing the 8-carbonyl group of Aeo (4–6).
HC-toxin, like other Aeo-containing cyclic tetrapeptides such as trapoxin (7), is a potent inhibitor of histone deacetylases from maize and other organisms (8, 9). By a mechanism that remains to be elucidated, inhibition of histone deacetylase during the early stages of infection permits C. carbonum to infect and colonize maize plants (10).
HC-toxin production by C. carbonum is controlled by a complex genetic locus called TOX2 (3). TOX2 contains at least seven genes with an established or putative role in HC-toxin biosynthesis. HTS1 encodes a 570-kDa nonribosomal peptide synthetase (11, 12). HTS1 is tightly clustered with TOXA, which encodes a putative HC-toxin efflux carrier (13). TOXC encodes a fatty acid synthase beta subunit that is proposed to be involved in the biosynthesis of the side chain of Aeo (14). TOXD is predicted to encode a dehydrogenase, and although its role in HC-toxin biosynthesis has not been established, it amino acid sequence is most closely related to that of lovC, which is required for biosynthesis of the polyketide lovastatin by Aspergillus terreus (15). TOXF encodes a putative branched-chain amino acid transaminase and is closely clustered with TOXG, which encodes an alanine racemase (16, 17). All of the known TOX2 genes are present in multiple, linked copies in all Tox2+ isolates that have been studied, and are completely absent from toxin nonproducing (Tox2−) isolates (17, 18).
The seventh known gene of the TOX2 cluster is TOXE. TOXE is required for HC-toxin biosynthesis and for mRNA expression of the other genes of TOX2. TOXE mutants display no phenotype other than loss of HC-toxin production and virulence, and thus TOXE appears to be a specific regulator of HC-toxin biosynthesis (19). The product of TOXE (TOXE) contains two motifs that are individually widespread in nature but which had not previously been found together in the same protein (Fig. 1). An amino acid sequence at its N terminus matches the consensus sequence of the basic DNA binding domain of the leucine zipper (bZIP) family of DNA-binding transcriptional regulators (20). However, TOXE does not contain a leucine zipper, a feature essential to the function of true bZIP proteins. At its C terminus TOXE is predicted to contain four repeats of the ankyrin motif (Fig. 1). This motif is found in a number of different types of proteins and has been shown to mediate protein–protein interactions (21, 22).
Figure 1.
Structure of TOXE. TOXE has a molecular mass of 49 kDa and 441 aa. The bZIP-like basic domain is between residues 19 and 34 and the four ankyrin repeats are located between residues 290 and 315, 323 and 350, 357 and 384, and 415 and 441 (19).
From its requirement for expression of other TOX2 genes and its possession of a predicted DNA binding motif, it seemed possible that TOXE might be a transcription factor that coordinates expression of the TOX2 genes. However, its overall unusual structure compared with other known proteins, including known transcription factors, left its precise function unclear. In this study, we demonstrate that TOXE is a transcription factor that binds as a monomer or homomultimer to a specific nonpalindromic DNA sequence that is present in one or two copies within the promoters of the known TOX2 genes. In addition to its ability to bind DNA, TOXE has intrinsic ability to activate transcription in yeast.
Materials and Methods
Expression of TOXE in Escherichia coli.
The plasmid pAJ39 was used as a source of a TOXE cDNA (GenBank accession no. AF038874; ref. 19). The 5′ end of TOXE was amplified by PCR using the primers JDW623 (5′-CACGGATCCGGCACGACTTCCCCGAATAGC-3′) and JDW624 (5′-CCTTACGCTGGCTAGTTCACGAAGC-3′) to introduce a BamHI site (italicized). The 237-bp product was digested with BamHI and StyI and used to replace the 5′ end of the TOXE cDNA insert in pAJ39, creating plasmid pKP9. The TOXE cDNA insert was removed from pKP9 with BamHI and KpnI and cloned into the BamHI and KpnI sites of the bacterial expression vector pQE30 (Qiagen, Chatsworth, CA) to create the TOXE expression vector pKP10. Plasmids pKP36 and pKP37 were constructed for expressing C-terminal deletions of TOXE in E. coli by using either a BamHI–EcoRV or BamHI–EcoRI fragment from pKP10 cloned into pQE30, respectively. For expression, the E. coli strain M15 containing pREP4 (Qiagen) was transformed with pKP10. A sample of an overnight culture (250 μl) was used to inoculate 5 ml of 2 × YT medium (16 g/l tryptone/10 g/l yeast extract/5 g/l NaCl, pH 7.5) containing ampicillin and kanamycin. Isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1 mM) was added after 2.5 h of growth, and the cells were grown for an additional 30 min before harvest.
Determination of Transcriptional Start Sites.
Random amplification of cDNA ends (RACE), using a kit from Life Technologies (Rockville, MD), was used to determine the length of the 5′ untranslated regions (5′-UTRs) of TOXC and TOXD (17, 23).
Southwestern Blot Analysis.
E. coli cell pellets were resuspended in sample buffer [0.25 M Tris⋅HCl, pH 6.8/20% 2-mercaptoethanol (vol/vol)/8% SDS (wt/vol)/30% sucrose (wt/vol)/0.01% bromophenol blue] and boiled for 5 min. The denatured protein extracts were fractionated by SDS/PAGE (10% acrylamide). After equilibration with blotting buffer [25 mM Tris/192 mM glycine/20% methanol (vol/vol)] the proteins were transferred to nitrocellulose (Schleicher & Schuell) by using a Mini Trans-Blot cell (Bio-Rad). The membrane was incubated in renaturation buffer (100 mM Hepes, pH 7.5/100 mM KCl/1 mM DTT/0.1 mM EDTA/10 mM MgCl2/5% nonfat milk) at 4°C for 18 h. After rinsing with TNE-50 buffer (10 mM Tris, pH 7.5/50 mM NaCl/1 mM EDTA/1 mM DTT), the blot was incubated in the same buffer containing a [32P]-labeled DNA probe (106 cpm/ml) and nonspecific competitor DNA (10 mg/ml sheared salmon sperm DNA) for 6 h at 25°C. The blot was then washed two times with TNE-50 at 25°C for 15 min followed by autoradiography.
Construction and Labeling of Probes for Southwestern Blotting.
Gene promoter fragments were amplified by PCR, using specific primers for each of the promoters. Primers were 18–26 nt in length (sequences available on request). PCR conditions were 35 cycles (94°C for 1 min; 55°C for 2 min; 72°C for 1 min) after an initial denaturation for 2 min at 94°C, followed by a final extension of 5 min at 72°C. Reaction products were purified by gel filtration chromatography and labeled with α-[32P]dCTP by random priming (24). To make double-stranded probes, complementary oligonucleotides JDW-831 (5′-AACACAATCTCACGTAAGTCTGCAG-3′) and JDW-832 (5′-CCTGCAGACTTACGTGAGATTGTGT-3′) representing a TOXE recognition element (tox-box) from the TOXA promoter, and JDW-833 (5′-AACACAGCAGGACGTAAGTCTGCAG-3′) and JDW-834 (5′-CCTGCAGACTTACGTCCTGCTGTGT-3′), representing a mutant version of this site, were annealed before use. Double-stranded oligonucleotides were end-labeled with γ-[32P]ATP by using polynucleotide kinase (24).
Yeast Strains and Methods.
Standard methods were used (25). Yeast strain MG106 (MATa ade2–1 can1–100 his 3–11 15 leu 2–3 112 trp 1–1 ura 3–1; ref. 26) was used for testing DNA binding, and Y190 (MATa, ura3–52, his3–200, lys2–801, ade2–101, trp1–901, leu2–3, 112, gal4Δ, gal80Δ, cyhr2, LYS2∷GAL1UAS-HIS3TATA-HIS3, URA∷GAL1UAS-GAL1TATA-lacZ; CLONTECH) for testing TOXE transcriptional activation function.
Analysis of DNA Binding in Yeast.
Complementary oligonucleotides containing either four tandem copies of the wild-type TOXE recognition sequence (tox-box) (JDW-825, 5′-tcgaATCTCACGTAATCTCACGTAATCTCACGTAATCTCACGTA-3′, and JDW-823, tcgaTACGTGAGATTACGTGAGATTACGTGAGATTACGTG-3′; lowercase letters indicate XhoI overhangs used for cloning) or mutant versions (JDW-836, 5′-tcgaGCAGGACGTAGCAGGACGTAGCAGGACGTAGCAGGACGTA-3′, and JDW-837, 5′-tcgaTACGTCCTGCTACGTCCTGCTACGTCCTGCTACGTCCTGC-3′) were annealed and ligated into the XhoI site of the lacZ reporter vector pBgl-lacZ (27). The resulting reporter constructs were integrated into the ura3 locus of strain MG106 by transformation and selection for uracil prototrophy.
TOXE was expressed in yeast by using the pG-1 expression vector (28) containing the TOXE cDNA from pAJ39 modified by PCR to create restriction sites for cloning. The resulting plasmid pKP40 was transformed into the reporter strains by selecting for tryptophan prototrophy.
Analysis of TOXE Activation Activity.
To test the activation activity of fragments of TOXE fused with the GAL4 DNA binding domain (GAL4DBD), NcoI sites (underlined) were introduced into TOXE at sites corresponding to amino acids 1, 167, and 254 by using PCR primers JDW-768 (5′-GCTGGATCCACACCATGGGCACGACTTCCCCG-3′), JDW-868 (5′-CGTGGATCCCTGGCCATGGACTTGCGTTCTGGT-3′), and JDW-1059 (5′-GGAGGGATCCATGGATTCAGTTATAGTAACCTC-3′), respectively. The carboxy termini of the fragments tested for activation activity were created by using native restriction sites or reverse primer JDW-1061 (5′-GGGGGTCGACTAGTCTTCCTTTGGGCCATG-3′) corresponding to amino acid 289. The resulting products were cloned into pAS2–1 (CLONTECH) to express GALDBD:TOXE fusion proteins.
Site-Directed Mutagenesis.
Amino acid substitutions were introduced into the TOXE basic region by using PCR primers containing the desired mutations (sequences available on request). The products were cloned into pKP9 by using restriction sites present in the TOXE sequence. All changes were confirmed by DNA sequencing. Amino acid changes in the activation region of TOXE were made by using the “megaprimer” mutagenesis protocol (29). All changes were confirmed by DNA sequencing. Mutated copies of TOXE were cloned into pAS2–1 and used to express GAL4DBD:TOXE fusions in yeast strain Y190.
β-Galactosidase Assay.
Yeast transformants expressing β-galactosidase were assayed in two ways. For a qualitative assay, yeast cells were grown on SD plates for 2–3 days at 30°C and overlaid with a nitrocellulose membrane filter (Schleicher & Schuell) for 5 min. The nitrocellulose filter was frozen in liquid nitrogen and placed on a piece of filter paper saturated with 50 nM 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) dissolved in Z-buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/38 mM β-mercaptoethanol, pH 7.0). The filters were monitored for the appearance of blue color after incubation at 30°C for 1 h. For a quantitative assay, yeast cells from a 2-ml overnight culture in SD medium were used to inoculate 8 ml of fresh YPD medium. The cultures were grown at 28°C with shaking (230–250 rpm) until the cells reached a mid-log phase (OD600 0.3–0.5). Cells (1.5 ml) were harvested by centrifugation at 14,000 rpm for 30 sec and washed in Z-buffer. Cells were resuspended in 300 μl of Z-buffer and disrupted with three freeze–thaw cycles. Aliquots of the disrupted cells (100 μl) were diluted with 700 μl of Z-buffer and mixed with 160 μl of 4 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) dissolved in Z-buffer. After incubation at 30°C for 10 min, 400 μl of 1 M Na2CO3 was added and the A410 measured. One unit of βgalactosidase is defined as the amount which hydrolyzes 1 μmol of ONPG per min per cell (30).
Results
Before analysis of the interaction between TOXE and the TOX2 promoters, it was essential to know the transcriptional start sites of the TOX2 genes. HTS1 and TOXA are clustered and divergently transcribed. Their 5′-UTRs are 162 and 145 bp, respectively (13). The 5′-UTR of TOXD was determined by random amplification of cDNA ends (RACE) to be 17 bp (Y. Cheng, K.F.P., and J.D.W., unpublished results). TOXF and TOXG are also clustered and divergently transcribed, and have 5′-UTRs of 59 and 46 bp, respectively (17). The 5′-UTR of TOXC was determined by RACE to be 167 bp (data not shown).
TOXE Binds to a Specific Sequence in the Promoters of the TOX2 Genes.
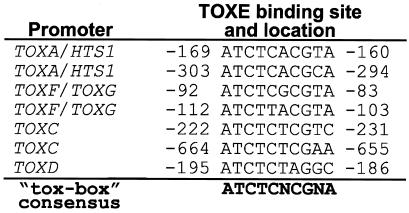
It was not possible to express TOXE in soluble form in E. coli, thereby precluding the analysis of DNA binding by gel mobility shift assays. TOXE was instead expressed in E. coli and analyzed by southwestern blotting (i.e., separation of proteins by SDS/PAGE followed by transfer to nitrocellulose, renaturation, and probing with double-stranded radiolabeled DNA fragments). For each TOX2 gene promoter tested, one or more DNA fragments were found to bind strongly to a protein of the correct size in TOXE-expressing E. coli extracts (Fig. 2). Analysis of the sequences of the DNA fragments that bound to TOXE revealed the presence of a conserved nucleotide sequence, which was therefore a candidate for the TOXE recognition site. This 10-bp motif, which we refer to as the tox-box, is highly conserved at eight positions. Nucleotides at five of the positions are absolutely conserved, including the first four, ATCT (Fig. 3). Examination of the DNA sequences upstream and downstream of this element gave no indication that the tox-box sequence has dyad symmetry. The tox-box was present in all DNA fragments that bound TOXE and was absent from all fragments that did not bind TOXE (Fig. 2). The promoters of HTS1/TOXA, TOXC, and TOXF/TOXG contain two copies of the tox-box and TOXD contains one copy (Fig. 2). No evidence was found for a tox-box in the promoter of TOXE, suggesting that TOXE does not regulate itself. As further evidence for the importance of the tox-box for TOXE binding, changing the first five oligonucleotides of the tox-box eliminated binding of TOXE (Fig. 4).
Figure 2.
TOXE binds to the promoters of the TOX2 genes. Total protein extracts from E. coli expressing (+) or not expressing (−) TOXE were separated by SDS/PAGE and transferred to nitrocellulose membranes. After renaturing the bound proteins, the membranes were probed with the indicated 32P-labeled DNA fragments. Hatched boxes indicate tox-box sequences (see Fig. 3) relative to the transcriptional start sites and transcriptional directions of the TOX2 genes, indicated by the arrows. Note that A–D are not drawn to scale.
Figure 3.
Sequences of the TOXE-binding sites from the promoters of the TOX2 genes, as deduced from the results shown in Fig. 2 and from comparative sequence analysis. Sequence locations are relative to the transcriptional start sites. For the HTS1/TOXA and the TOXF/TOXG promoters, the locations are relative to TOXA and TOXF, respectively (see Fig. 2). Note that the first tox-box of TOXC is in the opposite orientation to the others.
Figure 4.
Mutation of the conserved residues in the tox-box eliminate TOXE binding. (A) Representation of the TOXA/HTS1 promoter. Hatched boxes indicate the two tox-boxes. (B) Double-stranded oligonucleotides containing the wild-type (Wt) tox-box sequence (underlined) and surrounding nucleotides, and a mutant version (Mut) in which all five of the highly conserved nucleotides were changed (indicated in bold lettering), were used as 32P-labeled probes against southwestern blots. “−” Indicates total protein extracts from E. coli not expressing TOXE (control), and “+” indicates E. coli expressing TOXE.
TOXE Functions as a Transcription Factor in Yeast.
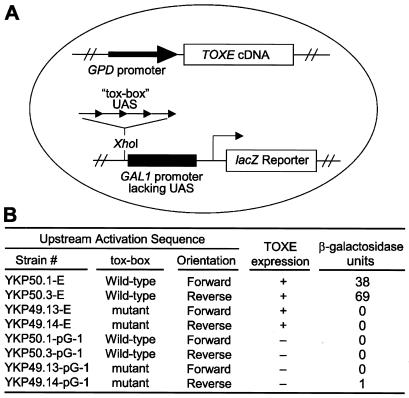
To test the ability of TOXE to bind specifically to the tox-box in vivo as well as in vitro, a yeast expression system was used. Four copies of wild-type or mutant versions of the tox-box sequence, in both forward and reverse orientations, were cloned upstream of a GAL1 promoter driving lacZ expression. This promoter is inactive by itself because of a lack of upstream activation sequences (ref. 27; Fig. 5A). Yeast strains containing the various tox-box constructs were then transformed with an expression plasmid containing TOXE driven by the constitutive yeast GPD promoter. Only strains containing both the wild-type tox-box and expressing TOXE had detectable levels of β-galactosidase activity (Fig. 5B). Both orientations of the wild-type tox-box were active (Fig. 5B).
Figure 5.
TOXE acts as a sequence-specific DNA binding protein and transcriptional activator in yeast. (A) Four tandem copies of a double-stranded oligonucleotide containing either the wild-type or the mutant tox-box (see Fig. 4) were fused in both orientations upstream of GAL1 without upstream activating sequences (UAS) fused to lacZ (27). TOXE driven by the constitutive yeast GPD promoter was expressed from a plasmid. (B) Resulting β-galactosidase activities of the various yeast strains. “TOXE expression” indicates whether the yeast cells contained the plasmid expressing TOXE.
Identification of Regions of TOXE Important for DNA Binding.
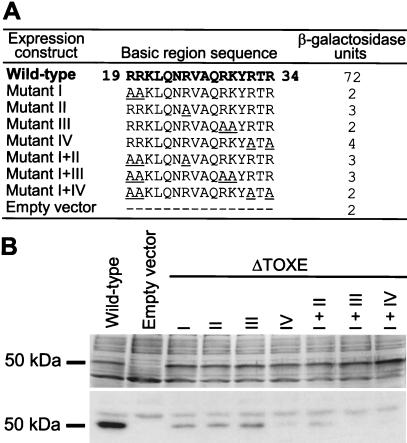
In the bZIP family of proteins the basic region determines DNA binding-site specificity through direct contacts with DNA (20). To test whether this was also true for the basic region of TOXE, we analyzed this region by alanine scanning (31). Seven basic amino acids between residues 19 and 34 were changed to alanines and expressed in yeast strain KP50.1 (see Fig. 5). Only strains expressing wild-type TOXE had detectable levels of β-galactosidase activity (Fig. 6A). Because failure to act in vivo could have been due to poor expression, reduced protein stability, or failure to localize to the nucleus, and not necessarily due to reduced DNA binding, all of the mutant constructs were also tested for their ability to bind to the tox-box by southwestern blotting. All of the mutant proteins were expressed in E. coli at levels comparable to the wild-type (Fig. 6B), but all had reduced ability to bind to the tox-box (Fig. 6B). Mutation of four of the highly conserved basic residues completely eliminated DNA binding (Fig. 6B).
Figure 6.
Mutations in the TOXE basic region reduce in vivo transcriptional activity and in vitro DNA binding. (A) Underlined amino acids indicate the changes made in TOXE in each mutant. β-Galactosidase activities were measured in yeast strain YKP50.1 (see Fig. 5) expressing mutant TOXE constructs. (B) Southwestern blotting. Mutant TOXE constructs were expressed in E. coli. (Upper) The SDS/PAGE gel stained with Coomassie blue; (Lower) the autoradiograph of the blot probed with a 32P-labeled DNA fragment from the TOXA/HTS1 promoter containing a single tox-box.
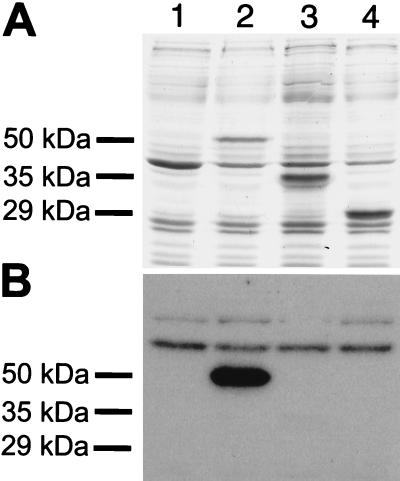
The results of the mutational analysis of the TOXE basic region indicated that the basic region near the N terminus was essential for DNA binding but not necessarily sufficient. To test this, two carboxy-terminal deletion mutants, which eliminated three or four of the four ankyrin repeats, respectively, were tested for in vitro DNA binding. Both of the truncated versions of TOXE were expressed in E. coli at levels comparable to wild-type TOXE (Fig. 7A), but neither was able to bind DNA (Fig. 7B).
Figure 7.
TOXE lacking the carboxy-terminus does not bind DNA. (A) SDS/PAGE (stained with Coomassie blue) of total extracts of E. coli expressing different TOXE constructs. Lane 1, empty plasmid (negative control); lane 2, full-length TOXE (amino acids 1–441) (positive control); lane 3, TOXE amino acids 1–317; lane 4, TOXE amino acids 1–254. The sizes of the expressed proteins are given on the left. (B) Southwestern blotting of the gel shown in A probed with a DNA fragment from the TOXA/HTS1 promoter containing a single tox-box.
Identification of the Transcriptional Activation Region of TOXE.
When fused to the GAL4DBD and expressed in yeast strain Y190, TOXE by itself (i.e., without coexpressing the activation domain of GAL4) induced expression of lacZ, indicating that native TOXE has the capacity not only to bind DNA but also to activate transcription. To characterize the region or regions of TOXE responsible for activation, a series of amino- and carboxy-terminal deletions of TOXE were expressed as fusion proteins to GAL4DBD and tested for their ability to induce expression of lacZ. Of the TOXE fragments tested, the smallest one that could act as an activation domain encompassed amino acids 254–317 (Fig. 8). All active fragments contained this region, which extends from 35 aa in front of the first ankyrin repeat to one amino acid after. These results indicate that TOXE has a discrete activation domain and that, whereas activation possibly involves at least one of the ankyrin repeats, it does not require all four repeats.
Figure 8.
Mapping of the TOXE activation region. The indicated fragments of TOXE were fused to the GAL4DBD, expressed in yeast strain Y190, and the yeast transformants assayed for β-galactosidase activity.
The activation domain composed of amino acids 254–317 was further analyzed by alanine scanning. This region is not particularly rich in any particular single amino acid or class of amino acid, although ≈30% are hydrophobic (Fig. 9A). Hydrophobic residues have been implicated as critical in various transcription activation domains, even in those that are classified on the basis of their predominant amino acids (32–34). All mutations of hydrophobic residues in this region, except one, strongly reduced activation function in yeast (Fig. 9B). The exception was a mutant domain with two changes, I298A and L300A, which actually led to increased activation activity (Fig. 9B).
Figure 9.
Mutational analysis of the activation domain of TOXE. (A) Sequence of the activation domain. Hydrophobic residues that were mutated are boxed, and the first ankyrin repeat is underlined. (B) Full-length TOXE proteins containing the indicated mutations were fused to the GAL4DBD, expressed in yeast strain Y190, and the yeast transformants assayed for β-galactosidase activity.
Discussion
TOXE was earlier shown to be required for the expression of the genes involved in the biosynthesis of HC-toxin, but the mechanism of regulation was not clear because of the unusual structure of TOXE (19). Here we show that TOXE is a DNA binding protein that recognizes a specific sequence, the tox-box, that is present in one or two copies in the promoters of all of the known genes involved in HC-toxin biosynthesis, except TOXE itself. The tox-box is functional in yeast in both orientations, which is biologically significant because two pairs of the TOX2 genes (HTS1/TOXA and TOXF/TOXG) are tightly and divergently clustered and therefore share overlapping promoters (13, 18).
The N-terminal bZIP-type basic region is required for DNA binding. The southwestern blotting results indicate that TOXE binds as a monomer or homomultimer. Because the tox-box does not contain dyad symmetry, TOXE probably binds as a monomer. In this regard, TOXE more closely resembles transcription factors that are known to bind as monomers—e.g., Skn-1 from Caenorhabditis elegans—rather than most true bZIP proteins, which typically bind as homo- or heterodimers to DNA sites with dyad symmetry. Skn-1 lacks a leucine zipper but binds to an asymmetrical DNA binding site as a monomer via its bZIP-like basic region (35). However, Skn-1 differs significantly from TOXE in containing homeodomain elements and lacking ankyrin repeats (35, 36).
Two versions of TOXE that were truncated at the carboxy terminus failed to bind to the tox-box, suggesting that the C-terminal ankyrin repeats contribute either directly or indirectly to DNA binding. The four-and-a-half N-terminal ankyrin repeats of the β subunit of the transcriptional regulator GABP stabilize the interaction between the GABPα/β complex and DNA (37). Therefore it is possible that the ankyrin repeats of TOXE contribute to the stability of TOXE binding to DNA or are critical for the correct positioning of the basic region over the tox-box.
In addition to DNA binding activity, TOXE also has the capacity to activate transcription in a heterologous system, indicating that TOXE contains an activation domain. The six ankyrin repeats of the C. elegans GLP-1 receptor are alone sufficient to activate transcription in yeast (38). However, the activation domain of TOXE defined in our experiments resides in a 63-aa portion of the protein just before and including the first ankyrin repeat. All four ankyrin repeats are not necessary for TOXE to function as a transcriptional activator, although it cannot be excluded from our experiments that the four ankyrin domains might constitute a second activation domain in TOXE.
The only known phenotype of TOXE mutants is inhibition of mRNA expression of the TOX2 genes dedicated to HC-toxin biosynthesis, with concomitant loss of specific pathogenicity of C. carbonum on maize that is homozygous recessive at the Hm1 and Hm2 loci (19). Like the other genes of TOX2, TOXE is present only in Tox2+ isolates of C. carbonum isolates, and at least one of its two copies is clustered with the other TOX2 genes (ref. 19; J.-H. Ahn and J.D.W., unpublished data). Therefore, TOXE appears to be a pathway-specific transcription factor whose sole function is to coordinate the expression of the genes involved in HC-toxin biosynthesis.
A gene whose product has the same structure as TOXE, namely a bZIP basic region, ankyrin repeats, and no leucine zipper, was recently reported in Cladosporium fulvum, a fungal pathogen of tomato (39). Thus, TOXE might be the prototype of a new class of transcription factors, so far found only in plant pathogenic filamentous fungi. Bussink et al. (39) have proposed the name bANK proteins for this class of transcription factor.
Pathway-specific transcription factors have also been found to regulate secondary metabolite biosynthesis in other filamentous fungi. For example, TRI6, a zinc-finger transcription factor, regulates the expression of the trichothecene biosynthetic genes of Fusarium sporotrichiodes (40, 41). In species of Aspergillus, the aflatoxin biosynthetic genes are regulated by AFLR, a zinc cluster transcription factor (42–45). Another zinc cluster protein, ORFR, regulates the ACR-toxin gene cluster of Alternaria alternata (46). In all of these cases, the transcriptional regulators are clustered with the biosynthetic genes, as is the case in TOX2. Because trans-acting transcription factors, such as TOXE, TRI6, AFLR, and ORFR, should be capable of activating genes anywhere in their respective genomes, the biological rationale for the physical linkage of regulatory and regulated genes in fungal secondary metabolite gene clusters is not clear. It may be a result of their evolutionary origins and mechanisms of transmission (47–49).
Acknowledgments
We thank John Pitkin for advice in the early stages of this work and Steve Triezenberg for valuable suggestions throughout. This work was supported by the U.S. Department of Energy, Division of Energy Biosciences.
Abbreviations
- GAL4DBD
GAL4 DNA binding domain
- UTR
untranslated region
- bZIP
basic leucine zipper
References
- 1.Hammond-Kosack K, Jones J D G. In: Biochemistry and Molecular Biology of Plants. Buchanan B B, Gruissem W, Jones R L, editors. Rockville, MD: Am. Soc. Plant Biologists; 2000. pp. 1102–1156. [Google Scholar]
- 2.Kohmoto K, Otani H. Experientia. 1991;47:755–764. doi: 10.1007/BF01922454. [DOI] [PubMed] [Google Scholar]
- 3.Walton J D. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meeley R M, Johal G, Briggs S P, Walton J D. Plant Cell. 1992;4:71–77. doi: 10.1105/tpc.4.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johal G S, Briggs S P. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 6.Multani D S, Meeley R B, Paterson A H, Gray J, Briggs S P, Johal G S. Proc Natl Acad Sci USA. 1998;95:1686–1691. doi: 10.1073/pnas.95.4.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 8.Brosch G, Ransom R, Lechner T, Walton J D, Loidl P. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darkin-Rattray S J, Gurnett A M, Myers R M, Dulski P M, Crumley T M, Alloco J, Cannova C, Meinke P T, Colletti S L, Bednarek M A, et al. Proc Natl Acad Sci USA. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ransom R F, Walton J D. Plant Physiol. 1997;115:1021–1027. doi: 10.1104/pp.115.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panaccione D G, Scott-Craig J S, Pocard J-A, Walton J D. Proc Natl Acad Sci USA. 1992;89:6590–6594. doi: 10.1073/pnas.89.14.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott-Craig J S, Panaccione D G, Pocard J-A, Walton J D. J Biol Chem. 1992;267:26044–26049. [PubMed] [Google Scholar]
- 13.Pitkin J W, Panaccione D G, Walton J D. Microbiology. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- 14.Ahn J-H, Walton J D. Mol Plant–Microbe Interact. 1997;10:207–214. doi: 10.1094/MPMI.1997.10.2.207. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy J, Auclair K, Kendrew S G, Park C, Vederas J C, Hutchinson C R. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y-Q, Ahn J-H, Walton J D. Microbiology. 1999;145:3539–3546. doi: 10.1099/00221287-145-12-3539. [DOI] [PubMed] [Google Scholar]
- 17.Ahn J-H, Walton J D. Plant Cell. 1996;8:887–897. doi: 10.1105/tpc.8.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y-Q, Walton J D. J Biol Chem. 2000;275:4906–4911. doi: 10.1074/jbc.275.7.4906. [DOI] [PubMed] [Google Scholar]
- 19.Ahn J-H, Walton J D. Mol Gen Genet. 1998;260:462–469. doi: 10.1007/pl00008632. [DOI] [PubMed] [Google Scholar]
- 20.Ellenberger T E. Curr Opin Struct Biol. 1994;4:12–21. [Google Scholar]
- 21.Bork P. Proteins. 1991;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 22.Sedgwick S G, Smerdon S J. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 23.Frohmann M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T A. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Guthrie, C. & Fink, G. R., eds. (1991) Methods Enzymol.194.
- 26.Myers L C, Gustafsson C M, Hayashibara K C, Brown P O, Kornberg R D. Proc Natl Acad Sci USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 28.Schena M, Picard D, Yamamoto K R. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar G, Sommer S S. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 31.Cunningham B C, Wells J A. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 32.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 33.Triezenberg S J. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan S M, Horn P J, Olson V A, Koop A H, Niu W, Ebright R H, Triezenberg S J. Nucleic Acids Res. 1998;26:4487–4496. doi: 10.1093/nar/26.19.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackwell T K, Bowerman B, Priess J R, Weintraub H. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- 36.Kophengnavong T, Carroll A S, Blackwell T K. Mol Cell Biol. 1999;19:3039–3050. doi: 10.1128/mcb.19.4.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batchelor A H, Piper D E, de la Brousse F C, McKnight S L, Wolberger C. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 38.Roehl H, Bosenberg M, Blelloch R, Kimble J. EMBO J. 1996;15:7002–7012. [PMC free article] [PubMed] [Google Scholar]
- 39.Bussink H-J, Clark A, Oliver R. Eur J Plant Pathol. 2001;6:655–659. [Google Scholar]
- 40.Proctor R H, Hohn T M, McCormick S P, Desjardins A E. Appl Environ Microbiol. 1995;61:1923–1930. doi: 10.1128/aem.61.5.1923-1930.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hohn T M, Krishna R, Proctor R H. Fungal Genet Biol. 1999;26:224–235. doi: 10.1006/fgbi.1999.1122. [DOI] [PubMed] [Google Scholar]
- 42.Payne G A, Nystrom G J, Bhatnagar D, Cleveland T E, Woloshuk C P. Appl Environ Microbiol. 1993;59:156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J H, Butchko R A E, Fernandes M, Keller N P, Leonard T J, Adams T H. Curr Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes M, Keller N P, Adams T H. Mol Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 45.Todd R B, Andrianopoulos A. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka A, Tsuge T. Mol Plant-Microbe Interact. 2000;13:975–986. doi: 10.1094/MPMI.2000.13.9.975. [DOI] [PubMed] [Google Scholar]
- 47.Keller N P, Hohn T M. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- 48.Walton J D. Fungal Genet Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- 49.Rosewich U L, Kistler H C. Annu Rev Phytopathol. 2000;38:325–363. doi: 10.1146/annurev.phyto.38.1.325. [DOI] [PubMed] [Google Scholar]