Abstract
Exon 20 insertion (Ex20Ins) mutations are the third most prevalent epidermal growth factor receptor (EGFR) activating mutation and the most prevalent HER2 mutation in non-small cell lung cancer (NSCLC). Novel therapeutics for the patients with Ex20Ins mutations are urgently needed, due to their poor responses to the currently approved EGFR and HER2 inhibitors. Here we report the discovery of highly potent and broadly effective EGFR and HER2 Ex20Ins mutant inhibitors. The co-crystal structure of compound 1b in complex with wild type EGFR clearly revealed an additional hydrophobic interaction of 4-fluorobenzene ring within a deep hydrophobic pocket, which has not been widely exploited in the development of EGFR and HER2 inhibitors. As compared with afatinib, compound 1a exhibited superior inhibition of proliferation and signaling pathways in Ba/F3 cells harboring either EGFR or HER2 Ex20Ins mutations, and in the EGFR P772_H773insPNP patient-derived lung cancer cell line DFCI127. Our study identifies promising strategies for development of EGFR and HER2 Ex20Ins mutant inhibitors.
Keywords: carbamates, epidermal growth factor receptor, exon 20 insertion, hydrophobic pocket, medical chemistry
Inhibited cell growth
Highly potent and broadly active EGFR and HER2 exon 20 insertion mutant inhibitors have been developed (EGFR = epidermal growth factor receptor, HER2 = human epidermal growth factor receptor 2). A hydrophobic interaction of a 4-fluorobenzene ring within a deep hydrophobic pocket has led to a strong inhibition of highly refractory EGFR and HER2 exon 20 insertion mutations.
Ex20Ins mutations are the third most common EGFR activating mutations in NSCLC, which collectively account for approximately 4% to 10% of all EGFR mutations.[1] Most of these mutations occur near the end of αC-helix after residue Met766. Patients with NSCLC harboring various Ex20Ins mutations rarely respond to treatment with the approved EGFR inhibitors gefitinib, erlotinib or afatinib [Response rate (RR): 8.7–11%; Progression free survival (PFS): 2.4–2.7 months].[2] Most EGFR Ex20Ins mutant-transfected Ba/F3 cells exhibit resistance to gefitinib and partial responses to second generation EGFR inhibitors including afatinib, dacomitinib and neratinib, whereas EGFR WT transformed cells remain strongly sensitive.[3]
HER2 Ex20Ins mutations make up the majority of HER2 mutations and occur in a similar position as EGFR Ex20Ins mutations after residue Met774. Compared to EGFR Ex20Ins mutations, the spectrum of HER2 Ex20Ins mutations is narrow and HER2 A775_G776insYVMA accounts for approximately 50–80% of HER2 Ex20Ins mutations.[4] Like EGFR Ex20Ins mutations, therapeutic strategies for treating patients harboring NSCLC with HER2 Ex20Ins mutations are limited.[5] In a retrospective study, patients harboring HER2 Ex20Ins mutations showed better response to conventional chemotherapies (with the median duration of 4.3 months) than current HER2 TKIs (with the median duration of only 2.2 months).[4b]
Collectively, the currently approved EGFR and HER2 TKIs are poorly effective in NSCLC patients with EGFR and HER2 Ex20Ins mutations. Thus, a new class of inhibitors that can efficiently inhibit various EGFR and HER2 Ex20Ins mutations is urgently needed.
We describe a highly potent and broadly active EGFR and HER2 Ex20Ins mutant inhibitor which was developed by exploiting a deep hydrophobic pocket, that is not fully occupied by currently approved EGFR and HER2 TKIs. Structure-guided design and biological evaluation of novel 2,4-disubstituted pyrimidines yielded potent Ex20Ins mutant inhibitors of both EGFR and HER2. A co-crystal structure of a representative compound in this series bound to WT EGFR kinase domain revealed a distinct binding mode of the 4-fluorobenzyl carbamate substituent that binds to the deep hydrophobic pocket in EGFR.
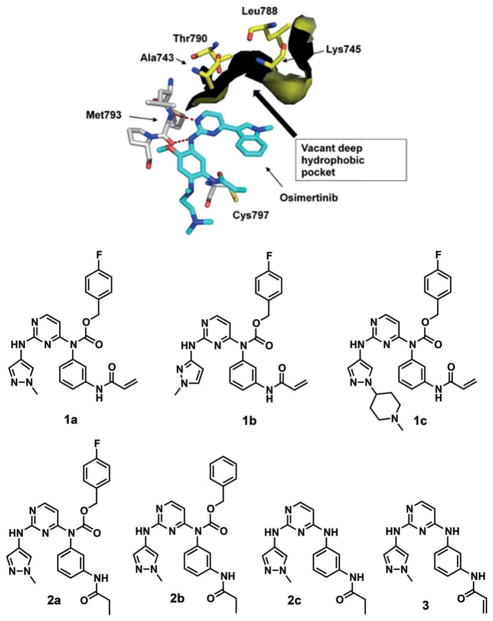
Like the two major activating mutations (L858R and Del), most EGFR Ex20Ins mutations occur away from the ATP binding site, but none of the first line EGFR inhibitors can efficiently inhibit these mutants with selectivity over wild-type EGFR.[6] Treatment of third generation EGFR inhibitor osimertinib, which was recently approved for the first line treatment, against various EGFR Ex20Ins NSCLC xenografts also led to only tumor stasis.[7] We hypothesized that an additional interaction between the small molecule inhibitor and the target protein would improve their inhibitory activity against Ex20Ins mutations. In the crystal structures of WT EGFR in complex with osimertinib (PDB code: 4ZAU, Figure 1A),[8] a pyrimidine-based EGFR inhibitor, and in complex with afatinib (PDB code: 4G5J, see Figure S1A in the Supporting Information),[9] we found a vacant deep hydrophobic pocket flanked by the residues Ala743, Lys745, Leu788 and Thr790. Importantly, this pocket is present in the active conformation of the kinase that is promoted by oncogenic mutants, and is distinct from the void created by the outward displacement of the C-helix that is required by compounds such as lapatinib and neratinib. It is partially occupied by the chlorine atom of afatinib, but completely vacant in the co-crystal structure of osimertinib. The phenethylamine substituent of the pyrrolopyrimidine inhibitor AEE788 extends further into the pocket, but does not fully occupy it (PDB: 2J6M).[10] We also noted a deep hydrophobic pocket in the HER2 crystal structures such as WT HER2 bound to TAK-285 (PDB: 3RCD, Figure S1B), although in these structures the C-helix is displaced outward from its active position.
Figure 1.
A vacant deep hydrophobic pocket in crystal structure of WT EGFR in complex with osimertinib (top, PDB: 4ZAU); a novel pyrimidine-based carbamate series (bottom).
We employed a 2,4-disubstituted pyrimidine core that has been broadly used in the development of EGFR inhibitors such as osimertinib,[11] rociletinib[12] and WZ4002,[13] and installed a 3-acrylamide substituted aniline at the C4 position, inspired by WZ4002, to covalently target EGFR via Cys797 and HER2 via Cys805. We then extended this pyrimidine scaffold by installing a benzyl carbamate moiety, chosen as it provides a suitable trajectory and distance from the pyrimidine core into the deep hydrophobic pocket.
To investigate effects of varying the benzyl carbamate substituents, we established a concise four-step synthetic route that employed 2,4-dichloropyrimidine, 3-nitrophenyl isocyanate and diverse benzylic alcohols and generated 1a–c as lead compounds (Scheme S1). To identify an effect of the additional interaction generated by a benzyl carbamate with the deep hydrophobic pocket independent of covalent bond formation, we first assessed biochemical activities of reversible analogs (2a–c), in which the acrylamide is replaced by a propionamide, against wild-type EGFR [Invitrogen, Z′-LYTE] and EGFR Del746-750 [Invitrogen, LanthaScreen binding] (Table S1). Remarkably, 2b, which contains a benzyl carbamate at C4-position of the pyrimidine showed more than 20-fold higher biochemical inhibitory activities than 2c, which lacks the benzyl carbamate moiety. Additionally, 4-fluorine substitution at the benzyl carbamate (2a) led to an enhancement of potency in both biochemical assays.
This result encouraged us to test the irreversible analogs of the benzyl carbamate series (1a–c) and to investigate their EGFR and HER2 inhibitory activities, especially against Ex20Ins mutations (Table 1). We used proliferation assays to assess the potencies of newly synthesized compounds against Ex20Ins mutants, using Ba/F3 cells transformed by three EGFR Ex20Ins and two HER2 Ex20Ins mutants. Oncogenic kinase transformed Ba/F3 assay is a commonly used model system to assess the potency and selectivity of kinase inhibitors.[14] We primarily focused on EGFR D770_N771insSVD (InsSVD) and EGFR V769_D770in-sASV (InsASV), which are the two most prevalent EGFR Ex20Ins mutants accounting for approximately 40% of EGFR Ex20Ins mutation, as well as HER2 A775_G776in-sYVMA (InsYVMA), the major HER2 Ex20Ins mutation. Three carbamate analogs (1a–c) displayed excellent antiproliferative activities against all tested Ex20Ins mutants in both EGFR and HER2. However, 3, which lacks a carbamate moiety, and 2a, which is a non-covalent inhibitor, were almost inactive in all cell lines. This result demonstrated that both the covalent bond formation and the hydrophobic interaction of the benzyl carbamate are important for inhibition of EGFR and HER2 Ex20Ins mutations in this series. In particular, 1a showed more than 28-fold and 7-fold superior potency against two major EGFR exon 20 insertion mutants (InsSVD and InsASV, respectively), compared to the representative second and third generation EGFR inhibitors except poziotinib which was reported as EGFR and HER2 Ex20Ins inhibitor during the course of our study.[15] It showed strong antiproliferative activities against all Ba/F3 cells tested with EC50 values of lower than 10 nM. Wild-type EGFR inhibition is a well-known cause of target-oriented dose-limiting toxicity. Surprisingly, 1a–c were less potent against WT EGFR than second generation EGFR inhibitors, afatinib, dacomitinib and poziotinib in cellular assays, resulting in an improvement of selectivity between EGFR Ex20Ins mutants and WT EGFR. [InsSVD/WT EGFR: 1.8–2.1 (1a–c) vs. >7.1–240 (afatinib, dacomitinib and poziotinib)]. Particularly, poziotinib displayed much stronger antiproliferative activity against WT EGFR transformed Ba/F3 cells with EC50 value of lower than 1 nM leading to worse InsSVD/WT EGFR ratio than 1a–c. Additionally, 1a displayed a great selectivity over a panel of 468 kinases at a concentration of 1 μM, which was assessed via the KINOMEscan (Figure S3, DiscoverX). At 30% cut-off in this profiling, 1a was active only against the kinases which have a cysteine residue at the same position as the Cys797 of EGFR, such as HER2, HER4, JAK3, BTK, BLK, BMX, TXK, TEC and EGFR mutants, except DDR1. In biochemical and cellular assays, 1a–c displayed excellent inhibitory activities against common EGFR mutants such as L858R, Del, L858R/T790M and Del/T790M. Especially, 1a [EC50 = 2.1 nM (L858R/T790M), 4.6 nM (Del/T790M)] was more potent in Ba/F3 cells harboring the drug resistant mutation T790M than osimertinib [EC50 = 23 nM (L858R/T790M), 12 nM (Del/T790M)] (Table S2). Overall, these data demonstrate that 1a–c are potent and selective EGFR and HER2 Ex20Ins mutant inhibitors.
Table 1.
Biochemical activities and antiproliferative activities against a panel of Ba/F3 cells transformed by EGFR and HER2 wild type and Ex20Ins mutants.
| Compound | Biochemical activity [WT EGFR, 100 μM ATP, nM] | Antiproliferative activity [nM] | InsSVD/WT EGFR ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|
| EGFR Ba/F3 cells | HER2 Ba/F3 cells | ||||||||
| WT | InsSVD | InsASV | InsGY | WT | InsYVMA | InsGSP | |||
| Afatinib | <0.5 | 2.2 ± 0.8 | 520 ± 110 | 240 ± 72 | 85 ± 8.0 | 11 ± 2.9 | 30 ± 9.5 | 31 ± 11 | 240 |
| Poziotinib | <0.5 | <1 | 7.1 ± 0.5 | 5.1 ± 0.2 | 7.3 ± 3.5 | <1 | 3.6 ± 0.2 | 2.2 ± 0.4 | >7.1 |
| Dacomitinib | <0.5 | 3.0 ± 0.4 | 680 ± 160 | 500 ± 200 | 71 ± 27 | 32 ± 7.6 | 77 ± 29 | 46 ± 23 | 230 |
| Osimertinib | 12 ± 2.0 | 60 ± 15 | 420 ± 120 | 520 ± 93 | 690 ± 110 | 31 ± 5.7 | 49 ± 2.9 | 150 ± 17 | 7.0 |
| WZ4002 | 36 ± 3.5 | 760 ± 110 | 910 ± 250 | 1100 ± 96 | 1400 ± 330 | 100 ± 1.1 | 140 ± 15 | 370 ± 13 | 1.2 |
| 2a[d] | 240 ± 38[a] | 7900 ± 940 | NA[b] | NA[b] | NA[b] | NA[b] | NA[b] | NA[b] | ND[c] |
| 3[d] | 1700 ± 1000 | 2400 ± 320 | 3100 ± 370 | 2600 ± 210 | 2200 ± 260 | 760 ± 110 | 970 ± 56 | 1200 ± 110 | 1.3 |
| 1a[d] | <0.5 | 7.6 ± 1.9 | 15 ± 2.2 | 34 ± 2.1 | 11 ± 1.5 | 2.5 ± 0.36 | 13 ± 1.4 | 9.2 ± 0.98 | 1.9 |
| 1b[d] | 0.8 ± 0.10 | 21 ± 4.2 | 37 ± 3.4 | 94 ± 5.3 | 30 ± ND[3] | 3.8 ± 0.74 | 31 ± 3.4 | 19 ± 3.6 | 1.8 |
| 1c[d] | 0.7 ± 0.61 | 19 ± 2.2 | 39 ± 3.8 | 57 ± 8.2 | 35 ± 3.8 | 4.3 ± 0.29 | 15 ± 1.5 | 19 ± 2.1 | 2.1 |
IC50 values were determined at an ATP concentration of Km (11.5 μM).
Not active (EC50 value was higher than 10 μM).
Not determined.
EC50 values were measured from single experiment with six replicates.
Errors are reported as ±95% confidence interval.
We further evaluated effects of lead compound 1a on the phosphorylation of EGFR, HER2 and their downstream signaling effector Erk, in Ba/F3 cells transformed by WT EGFR and EGFR InsSVD as well as WT HER2 and HER2 InsYVMA (Figure 2). Treatment with 1a induced a dose-dependent reduction of EGFR and Erk phosphorylation in EGFR InsSVD transformed Ba/F3 cells with strong inhibition at a concentration of 0.1 μM, which was compatible with the effects observed using a 1.0 μM concentration of afatinib. 1a showed similar level of pEGFR and pErk inhibition in both Ba/F3 cells with WT EGFR or EGFR InsSVD in all tested concentrations. However, afatinib displayed stronger inhibition of pEGFR and pErk in WT EGFR transformed Ba/F3 cells than those in EGFR InsSVD transformed Ba/F3 cells. 1a dose-dependently suppressed HER2 and Erk phosphorylation in Ba/F3 cells harboring either HER2 InsYVMA or WT HER2 more efficiently than afatinib. Particularly, 1a was more potent then afatinib at 0.01 μM in both HER2 transformed Ba/F3 cells. However, 3 was not able to efficiently reduce phosphorylation of both EGFR and HER2 as well as Erk in all cell lines. This outcome was consistent with the results in antiproliferation assays discussed above.
Figure 2.
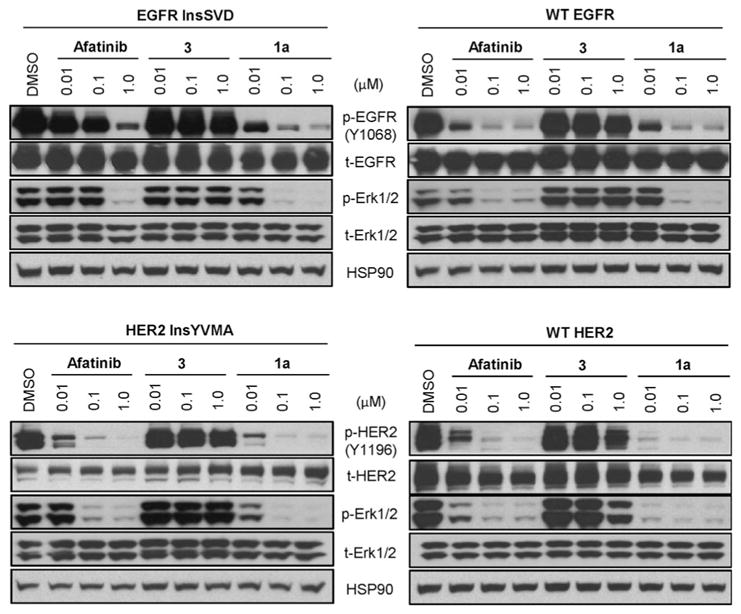
Effects on EGFR, HER2 and its downstream signaling effector, Erk in Ba/F3 cells transformed by EGFR InsSVD, wild type EGFR, HER2 InsYVMA and wild type HER2.
We then assessed the antiproliferative activities of 1a–c, 2a and 3 compared with known EGFR inhibitors in a patient-derived lung cancer cell line DFCI127, which harbors EGFR P772_H773insPNP (Figures 3 and Figure S3).[3] Like the results in Ba/F3 cells, all three carbamate analogs 1a–c achieved excellent antiproliferative activities against DFCI127 cells, but 2a and 3 were inactive. Especially, 1a and 1c showed superior antiproliferative activities relative to known EGFR inhibitors with exception of poziotinib which was consistently more potent than 1a–c. [EC50 = 11.5 nM (1a) and 22.3 nM (1c) vs. 44.0 nM (afatinib), 60.6 nM (dacomitinib) and 4.8 nM (poziotinib)] Moreover, treatment with 1a, the most potent analog, dose-dependently suppressed phosphorylation of EGFR and its downstream signaling effector, Erk. Consistently, afatinib was less effective and poziotinib was more effective at inhibiting EGFR signaling relative to 1a. Treatment of these inhibitors led to up-regulation of Bim that is a marker of EGFR TKI-mediated apoptosis.[16]
Figure 3.
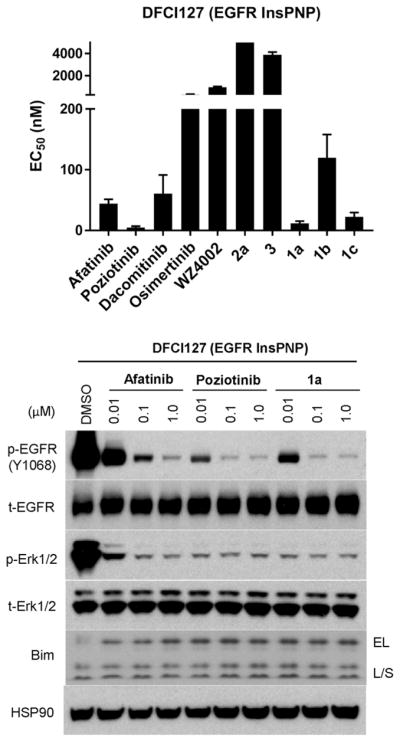
Antiproliferative activity (top) and effects on EGFR signaling pathway and a proapoptotic marker, Bim (bottom) in human lung cancer cell line, DFCI127, harboring EGFR P772 H773insPNP.
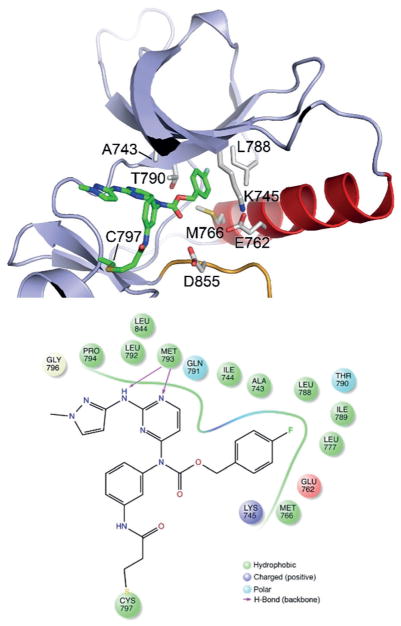
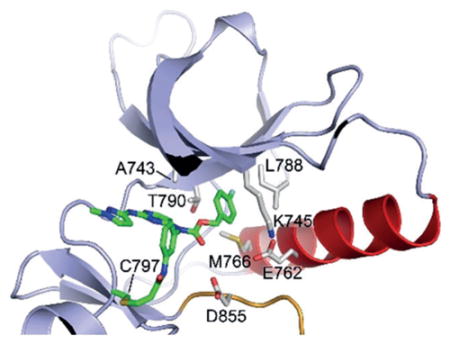
To understand and confirm structural interaction of the newly designed carbamate analogs and the EGFR kinase domain, we determined the crystal structure of the WT EGFR kinase domain in complex with 1b at a resolution of 2.5 Å (Figure 4, Table S3 and Figure S4). Compound 1b occupies the ATP binding pocket with an overall binding mode closely similar to that of WZ4002 and rociletinib, including formation of the expected covalent bond with Cys797 and hydrogen bonds between N1 nitrogen atom of the pyrimidine and the 2-substituted NH group with the hinge residue Met793.[13,17] The aminopyrazole substituent at the C2 position of pyrimidine forms an edge-to-face π-π interaction with the C4-aniline substituent, resembling WZ4002 and rociletinib, which yields a suitable angle of C4-aniline substituent for making the covalent bond with either Cys797 in EGFR or Cys805 in HER2. Consistent with the design strategy, the carbamate linkage positions the 4-fluorobenzene group deep in the hydrophobic pocket at the back of the ATP-site, where it contacts Lys745, Glu762, Leu 777, Leu788, and Thr790. The fluorine substituent makes favorable hydrophobic interactions with the methionine and leucine residues at the back of the pocket. This unique structural feature provides a reasonable explanation for the high and broad potency of this pyrimidine-based carbamate series against EGFR and HER2, especially Ex20Ins mutants.
Figure 4.
Co-crystal structure of wild type EGFR in complex with 1b.
In summary, we have developed a pyrimidine-based benzyl carbamate series which potently and broadly inhibited proliferation and signaling pathways of EGFR and HER2 Ex20Ins mutants, which have been associated with drug resistance to the currently approved EGFR and HER2 inhibitors. The additional hydrophobic interactions of the 4-fluorobenzyl carbamate substituent, together with covalent bond formation with Cys797 in EGFR and Cys805 in HER2, resulted in potent EGFR and HER2 Ex20Ins inhibitory activities in our series of pyrimidine-based EGFR inhibitors. Especially, 1a displayed broad and superior antiproliferative activities against EGFR and HER2 Ex20Ins mutants than currently approved 2nd and 3rd generation irreversible EGFR inhibitors and suppressed phosphorylation of EGFR, HER2 and their downstream signaling component, Erk in both Ba/ F3 cells and in the patient-derived DFCI127 cell line. These compounds displayed superior potency relative to afatinib in all cell lines except Ba/F3 cells harboring WT EGFR. Consequently, 1a has improved mutant selectivity against Ex20Ins over wild type EGFR than currently approved EGFR inhibitors and would be anticipated to exhibit less WT EGFR-dependent toxicology. Crystal structure of 1b in complex with WT EGFR revealed the unique binding mode of this series, in which a 4-fluorobenzene ring occupies a deep hydrophobic pocket that is present in the active conformation of the kinase but not fully occupied by current EGFR or HER2 inhibitors. Discovery of this carbamate series provides a way to inhibit the largely refractory EGFR and HER2 Ex20Ins mutations. Since 1a displayed poor in vitro ADME profile (Table S4), our current efforts focus on improving the pharmacokinetic properties that showed low oral bioavailability (11%), short half-life (0.5 h) and high clearance rate (773.37 mLmin−1kg−1, PO) (Table S5). Future work will report on the pharmacology and efficacy of these inhibitors in murine tumor models.
Supplementary Material
Acknowledgments
This work is supported by American Cancer Society (CRP-17-111-01-CDD (P.A.J.)), the Denise and Kevin Hanlon Family Fund for Lung Cancer Research (P.A.J.), and NIH grants CA116020 (M.J.E.) and CA221830 (E.P.). Diffraction data for the crystal structure described here were recorded using NE-CAT beamlines at the Advanced Photon Source, which are funded by NIGMS(GM103403) and the U.S. Department of Energy (DE-AC02-06CH11357), respectively.
Footnotes
Conflict of interest
J.J., P.A.J. and N.S.G. are inventors on a patent (WO 2017/ 015363) that covers the finding discussed.
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.201805187.
Contributor Information
Dr. Jaebong Jang, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School Boston, MA 02115 (USA)
Dr. Jieun Son, Lowe Center for Thoracic Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA), and Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Medicine, Harvard Medical School, Boston, MA 02115 (USA)
Dr. Eunyoung Park, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School Boston, MA 02115 (USA)
Dr. Takayuki Kosaka, Lowe Center for Thoracic Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA), and Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Medicine, Harvard Medical School, Boston, MA 02115 (USA)
Dr. Jamie A. Saxon, Lowe Center for Thoracic Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA), and Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Medicine, Harvard Medical School, Boston, MA 02115 (USA)
Dr. Dries J. H. De Clercq, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School Boston, MA 02115 (USA)
Dr. Hwan Geun Choi, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School Boston, MA 02115 (USA)
Dr. Junko Tanizaki, Lowe Center for Thoracic Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA), and Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Medicine, Harvard Medical School, Boston, MA 02115 (USA)
Dr. Michael J. Eck, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School Boston, MA 02115 (USA)
Dr. Pasi A. Jänne, Lowe Center for Thoracic Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA), and Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Medicine, Harvard Medical School, Boston, MA 02115 (USA)
Dr. Nathanael S. Gray, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215 (USA) and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School Boston, MA 02115 (USA)
References
- 1.a) Yasuda H, Kobayashi S, Costa DB. Lancet Oncol. 2012;13:e23–31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]; b) Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, Jackman DM, Johnson BE, Jänne PA. J Thorac Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, Zakowski MF, Kris MG, Ladanyi M. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Naidoo J, Sima CS, Rodriguez K, Busby N, Nafa K, Ladanyi M, Riely GJ, Kris MG, Arcila ME, Yu HA. Cancer. 2015;121:3212–3220. doi: 10.1002/cncr.29493. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C, Massey D, Zazulina V, Wu YL. Lancet Oncol. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 3.Kosaka T, Tanizaki J, Paranal RM, Endoh H, Lydon C, Capelletti M, Repellin CE, Choi J, Ogino A, Calles A, Ercan D, Redig AJ, Bahcall M, Oxnard GR, Eck MJ, Janne PA. Cancer Res. 2017;77:2712–2721. doi: 10.1158/0008-5472.CAN-16-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, Paik PK, Zakowski MF, Kris MG, Ladanyi M. Clin Cancer Res. 2012;18:4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Eng J, Hsu M, Chaft JE, Kris MG, Arcila ME, Li BT. Lung Cancer. 2016;99:53–56. doi: 10.1016/j.lungcan.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Buttitta F, Barassi F, Fresu G, Felicioni L, Chella A, Paolizzi D, Lattanzio G, Salvatore S, Camplese PP, Rosini S, Iarussi T, Mucilli F, Sacco R, Mezzetti A, Marchetti A. Int J Cancer. 2006;119:2586–2591. doi: 10.1002/ijc.22143. [DOI] [PubMed] [Google Scholar]; d) Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, Wistuba II, Fong KM, Toyooka S, Shimizu N, Fujisawa T, Minna JD, Gazdar AF. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 5.Herter-Sprie GS, Greulich H, Wong KK. Front Oncol. 2013;3:1–10. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang M, Xu X, Cai J, Ning J, Wery JP, Li QX. Int J Cancer. 2016;139:171–176. doi: 10.1002/ijc.30047. [DOI] [PubMed] [Google Scholar]
- 7.Floc’h N, Martin MJ, Riess JW, Orme JP, Staniszewska AD, Menard L, Cuomo ME, O’Neill DJ, Ward RA, Finlay MRV, McKerrecher D, Cheng M, Vang DP, Burich RA, Keck JG, Gandara DR, Mack PC, Cross DAE. Mol Cancer Ther. 2018;17:885–896. doi: 10.1158/1535-7163.MCT-17-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yosaatmadja Y, Silva S, Dickson JM, Patterson AV, Smaill JB, Flanagan JU, McKeage MJ, Squire CJ. J Struct Biol. 2015;192:539–544. doi: 10.1016/j.jsb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, Kraemer O, Himmelsbach F, Haaksma E, Adolf GR. J Pharmacol Exp Ther. 2012;343:342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 10.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ, Hughes G, Rahi A, Jacobs VN, Red Brewer M, Ichihara E, Sun J, Jin H, Ballard P, Al-Kadhimi K, Rowlinson R, Klinowska T, Richmond GH, Cantarini M, Kim DW, Ranson MR, Pao W. Cancer Discovery. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Finlay MR, Anderton M, Ashton S, Ballard P, Bethel PA, Box MR, Bradbury RH, Brown SJ, Butterworth S, Campbell A, Chorley C, Colclough N, Cross DA, Currie GS, Grist M, Hassall L, Hill GB, James D, James M, Kemmitt P, Klinowska T, Lamont G, Lamont SG, Martin N, McFarland HL, Mellor MJ, Orme JP, Perkins D, Perkins P, Richmond G, Smith P, Ward RA, Waring MJ, Whittaker D, Wells S, Wrigley GL. J Med Chem. 2014;57:8249–8267. doi: 10.1021/jm500973a. [DOI] [PubMed] [Google Scholar]
- 12.Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, Sheets M, St Martin T, Karp R, van Kalken D, Chaturvedi P, Niu D, Nacht M, Petter RC, Westlin W, Lin K, Jaw-Tsai S, Raponi M, Van Dyke T, Etter J, Weaver Z, Pao W, Singh J, Simmons AD, Harding TC, Allen A. Cancer Discovery. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W, Ercan D, Chen L, Yun HC, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Janne PA. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melnick JS, Janes J, Kim S, Chang JY, Sipes DG, Gunderson D, Jarnes L, Matzen JT, Garcia ME, Hood TL, Beigi R, Xia G, Harig RA, Asatryan H, Yan SF, Zhou Y, Gu XJ, Saadat A, Zhou V, King FJ, Shaw CM, Su AI, Downs R, Gray NS, Schultz PG, Warmuth M, Caldwell JS. Proc Natl Acad Sci USA. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, Li S, Chen T, Poteete A, Estrada-Bernal A, Le AT, Truini A, Nilsson BM, Sun H, Roarty E, Goldberg SB, Brahmer JR, Altan M, Lu C, Papadimitrakopoulou V, Politi K, Doebele RC, Wong KK, Heymach JV. Nat Med. 2018;24:638–646. doi: 10.1038/s41591-018-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, Tenen DG, Kobayashi S. PLoS Med. 2007;4:e315. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan XE, Zhu SJ, Liang L, Zhao P, Choi HG, Yun CH. Oncotarget. 2017;8:53508–53517. doi: 10.18632/oncotarget.18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.