Abstract
Nanoparticles (NPs) tend to adsorb matrix molecules like proteins and lipids incubated with biological fluids, forming a biological corona. While the formation and functions of protein corona have been studied extensively, little attention has been paid to lipid adsorption on NPs. However, lipids are also abundantly present in biological fluids and play important roles in processes like cell signaling and angiogenesis. Therefore, in this study, we established the analytical procedure for study of lipid adsorption on three different types of NPs in two matrices: human serum and heavy cream, using nanoflow liquid chromatography-mass spectrometry (nanoflowLC-MS). Serum was chosen to represent the common environment the NPs would be present once entering human body; and heavy cream was the representative food matrix NPs may be added to improve the color or taste. Steps of liquid-liquid extraction were established and optimized to achieve maximum recovery of the adsorbed, standard lipids from the NPs. Then, the LC-MS/MS method was developed to attain base-line separation of the standard lipids that represent the major lipid classes. At last, the lipid adsorption profiles of the three NPs were compared. We found that the lipid adsorption profile on the same type of NP was significantly different between the two matrices. The established method will help us investigate lipid adsorption on additional NPs and reveal how it could be affected by the physiochemical properties of NPs and the presence of proteins and other components in the biological matrix.
Keywords: TiO2 nanoparticle, polystyrene nanoparticle, cellulose nanofibrils, lipid adsorption, serum, heavy cream, LC-MS/MS
Introduction
Since nanoparticles (NPs) carry distinct and superior properties to their bulk counterparts, their industrial manufacture and applications have been augmented dramatically, calling for thorough evaluations of the impacts of NPs on biological systems and environments [1, 2]. Such evaluations are not only focused on the intrinsic physicochemical properties of the NPs, but also on how the NPs would transform under different circumstances. Their high specific surface area and activity prompt the NPs to adsorb many substances in the complex matrix they find themselves in. For example, in biofluids, it has been well studied that the NPs could disguise their surface by adsorption of a layer of proteins, forming the so-called protein corona [3,4]. The NP properties like size, shape, surface coating, core composition, are strong determinants to the formation of protein corona [3,4,5,6,7]. This is further confirmed by studies that use protein adsorption profiles to differentiate NPs varied in size, core structure, and coating [8,9]. The corona formed by protein adsorption changes the surface and size properties of the NPs, and its composition would also vary with the surrounding environment [10]. Thus, for the NPs being found to be able to move through the intestinal barrier, permeate epithelial barrier, or enter regional lymph nodes, etc. [5–11], it is believed that such transportation and other behaviors of NPs in biological systems could be influenced by the protein corona [12,13,14].
While a lot of attention has been paid to protein corona, lipid adsorption has not been studied as frequently [15], although lipids play an equally important role in organisms as proteins. For instance, phospholipids (PLs) are important metabolites connecting with other materials organically and key regulators for cell proliferation [16–17]; lipid rafts constituted of cholesterols and sphingolipids mediate cell signaling [17]; and lysophospholipids (LPLs), even with their relatively low abundance, are deeply involved in development of the central nervous and cardiovascular system [18]. Lipids are also associated with various diseases. Graessler et al. identified the close correlation of hypertension with the contents of PC and phosphatidylinositol [19]. Cholesteryl ester, triacylglycerol (TAG), LPLs and diacylglycerol (DAG) are indicated as key markers for metabolic syndrome including obesity and insulin resistance [20,21,22,23]. Oxidation stress on lipids can affect cell signaling, and even lead to cell apoptosis [24,25]. The diverse roles of lipids and their close correlations support the necessity of studying lipid-NP interaction to reveal their potential influence on the biological behaviors of NPs.
Gas chromatography (GC) has been widely employed for analysis of lipids owing to its high separation resolution, but most lipid analytes require derivatization to become volatile [26,27]. Modern liquid chromatography (LC) technologies have achieved comparable resolution to GC and become more and more popular in lipid profiling when coupled with the highly sensitive nanospray ionization-mass spectrometry (MS) [28, 29], which is further improved with the advancement in ionization modifier [30] and in instrumentation [31]. Development of analytical methods for simultaneous detection of LPL and PL [32], and combination of both top-down and bottom-up analysis [33] also greatly expand the scope of lipids being identified.
In the present study, we adopted the typical analytical procedure developed for analysis of lipids using nanoflowLC-MS, and optimized it for study of the lipids adsorbed on NPs. We chose three NP classes in this study. They were cellulose nanofibrils (CNF), polystyrene (PS) NPs, and TiO2 NPs. Both CNF and TiO2 can be added to food products to adjust nutrition contents and/or change the color and/or taste of the food. Due to their presence in food commonly ingested by humans, these particles represent the classes of NP’s that are likely to interact with lipids in serum or food matrices. The PS NPs were chosen as another group of organic materials to the CNF in contrast to the inorganic metal oxide NPs, but with higher hydrophobicity compared to CNF. Standard lipids were employed for method optimization, and the optimized method was applied to analyze the relative composition of LPLs, PLs, DAGs, TAGs adsorbed on the three selected NPs after incubation in heavy cream and serum. The study focused on the lipid classes of LPLs, PLs and TAGs, because these are the more abundant lipid components in living organisms compared other lipid classes such as sphingolipids, steroids, and cholesterols. These lipids also have very diverse hydrophobicity and it is possible to reveal the correlation between lipid hydrophobicity and adsorption on NPs.
Materials and Methods
Materials and samples
The CNF (mean number of branches per node determined by TEM = 3.33± 0.60 nm; mean fiber diameter = 64 nm± 29 nm and the mean fiber length = 6.71 ± 5.61 μm as determined by SEM) were synthesized and characterized by the authors as reported previously. [34, 35] Commercially available food grade TiO2 E171 was purchased from an online retailer. The particles were characterized in terms of size, shape, crystallinity, density, surface area, porosity, purity, surface chemistry, endotoxins and biological contaminants according to the protocols described by Beltran et al. [34]. The results are summarized in the Electronic Supplementary Material (ESM) (Tables S1–S3, Figs. S1 and S2). The PS NPs with the average diameter of 200 nm were purchased from Duke Scientific Corp. (Palo Alto, CA, USA). Powdered TiO2 was weighed and dissolved in water to make a solution of 1 μg/μL, then sonicated following the protocol described in a previous publication by the authors [36]. The stock solutions of CNF (2.5% w/w in water) and PS NPs (1% v/v) were diluted 10× with water before use. All of NPs were consumed completely within one day.
Seven lipid standards including 16:0-lysophosphatidylchoine (LPC), 14:0/14:0-phosphatidylethanolamine (PE), 16:0/14:0- PC, 16:0/16:0-PE, 18:0/18:1-PC, 16:0/16:0/14:1-TAG, 16:0/18:1-DAG and 18:0/18:0/18:1-TAG were purchased from Avanti Polar lipid Inc. (Alabaster, AL, USA). For optimization of LC condition, each lipid standard was prepared at the concentration of 3 pmol/μL in the same solvent mixture of chloroform/MeOH/water (20:75:5 v/v/v), and 3 μL of the mixture was injected into nanosprayLC-MS for each run. Horizon™ organic heavy whipping cream (WhiteWave Food Company, Denver, CO, USA) was purchased from a supermarket. Human serum was obtained from Innovative Research (Novi, MI, USA), which was donated by a healthy African-American male at the age of 32. All solvents used in the work were acquired from Fisher Scientific (Hampton, NH, USA).
Preparation of lipid vesicles
Lipid vesicles prepared from standard lipids dissolved in ultrapure water or buffer were employed as the simple sample matrix for evaluation of lipid adsorption and recovery from NPs. Formation of lipid vesicles was conducted by a published method [37]. In brief, 10 μg of 16:0-LPC, 16:0-PE, 16:0/14:0-PC, 18:0/18:1-PC, 18:0/18:1/18:1-TAG, 16:0/16:0/14:1-TAG, 16:018:1-DAG, 16:0/16:0-PA (totally, 8 species of lipid standards) were dissolved in the organic solvent mixture of CH3Cl and MeOH (8:2, v/v). To mimic biological matrices, 1×PBS was chosen as the background electrolyte, in which the cationic LPC, PC and PE were easily dissolved. PA was then added to decrease the electrical repulsion between the positive charges on the lipid molecules and also reduce the steric hindrance of the bulky head groups compared to their fatty acid chains, facilitating the formation of lipid vesicles. After transferring the mixture to a 10-mL round flask, the solution was dried by rotary evaporator to form a lipid film with their hydrophilic heads toward on the inner surface of the flask. The film was hydrated with 3 mL of 1× PBS buffer (pH 7.5) (Bioland Scientific LLC, Paramount, CA, USA) under a mild vortexing condition. Unilamellar vesicles with uniform sizes were finally formed by extruding the hydrated lipid through the membrane with an average pore size of 100 nm (Avanti Polar lipid Inc.). The whole process was repeated 2 more times and then all of the obtained vesicles were combined to ensure sufficient amounts of samples for method optimization.
Optimization of lipid extraction methods
Six versions of the modified “Folch method” [38] were tested to identify the optimal extraction conditions for lipid recovery from the NPs after incubation with the lipid vesicle solution. In this part, CNF was used as the representative NPs. Seven vials (6 extraction protocols and one control without NP) containing 800 μL of the lipid vesicle solution and 100 μL of the CNF solution were vortexed gently for 30 min. Afterwards, each vial was centrifuged for 5 min under 10,000×g to precipitate the CNF, and then the supernatant containing the free lipids was removed. The CNF was washed with 300 μL of water, using centrifugation for precipitation and supernatant removal, and a total of two washing steps were conducted.
The typical extraction procedure started with evaporation of the lipid solution completely by using a SpeedVac concentrator (Savant, Irvine, CA USA). Then, 300 μL CHCl3 was added and the solution was vortexed for 15 min before the addition of 900 μL water. A brief vortex was followed with 5 min of centrifugation at 5,000× g, after which the solution divided into two layers: the upper water layer containing the NPs and the bottom organic layer containing the lipids released from CNF. The organic layer was collected and dried by SpeedVac. Several variations of this method were tested in the present work to obtain optimal extractions, either by omitting the SpeedVac before mixing with CHCl3, or employing an additional extraction step. Details of the varied extraction methods were listed in Table 1. The collected dried power was dissolved in 300 μL of organic solvent consisting of chloroform, MeOH, and water (20:75:5, v/v/v). Additionally, 1.5 nmol of 14:0/14:0-PE was added to each sample at 5 pmol/μL to serve as the internal standards (IS) for correction of ionization efficiency.
Table 1.
Summary of the conditions in the six varied extraction approaches tested. (“√” – performed; “×” not performed)
| No | SpeedVac | 1st extraction | 2nd extraction |
|---|---|---|---|
| 1 | √ | CHCl3 300 μL, then H2O 900 μL added | × |
| 2 | × | CHCl3 300 μL | × |
| 3 | × | CHCl3 300 μL | CHCl3 300 μL |
| 4 | × | CHCl3 300 μL | CHCl3 250 μL, MeOH 50 μL |
| 5 | × | CHCl3 300 μL | CHCl3 200 μL, MeOH 100 μL |
| 6 | × | CHCl3 300 μL | CHCl3 150 μL, MeOH 150 μL |
Condition of liquid chromatography-Mass spectrometry
Finnigan LTQ MS (Thermo Fisher Scientific Inc., Chino, CA, USA) connected to a CapLC pump equipped with an autosampler (Waters, Milford, MA, USA) was employed for lipid analysis in the present study. A split flow was generated to provide rapid introduction of the solvent gradient to the separation column and reduce the separation time. A capillary tubing (I.D. 50 μm, O.D. 356 μm; Polymicro technologies, Phoenix, AZ, USA) was used to connect the autosampler, HPLC pump and valve installed on the MS. All fittings such as union, ferrules, adaptors and microcross were attained from IDEX Corporation (Lake Forest, IL, USA). A capillary column was prepared in-house by filling the vacant capillary (I.D. 75 μm, 6 cm length) with the Watchers ODS-P C18 beads (particle diameter of 3 μm, pore diameter of 100 Ȧ, Isu Industry, Seoul, South Korea). One end of the capillary was pulled with Model P-2000 (Sutter instrument Co., Novato, CA, USA) to form a 10 μm I.D. tip. The four ends of a microcross located between LC and MS were coupled with the capillary tubing from the autosampler and capillary column, the Pt wire for ESI, and the flow-regulator capillary (I.D. 20 μm) that controlled the spray flowrate at the tip of the column. At the sample injection step, the flowrate was set as 800 nL/min., with 100% of solvent A and the pressure capillary closed by the valve installed on the MS, thus 100% of the sample was injected to the separation column. Following the 13-min injection step, the flowrate and ratio of solvent B were raised as well as the valve to control the flow-regulator capillary was opened to generate a split flow. An initial flowrate coming from the capillary HPLC pump was 15 μL/min., with the majority of the flow split to the flow-regulator capillary, and the flow rate passing through the separation column was only 300 nL/min. to improve ionization efficiency. Solvent A and B were composed as water:acetonitrile at 9:1(v/v) and isopropanol: chloroform: acetonitrile at 6: 2: 2 (v/v/v), respectively. To increase lipid ionization, 5 mM of ammonium formate and 0.05% of ammonium hydroxide were added [30] and the solvents were degassed prior to use. The full scan and data dependent CID mode of MS was utilized to perform simultaneous quantification and qualification analysis of lipids. The MS was set at a scan range of m/z 470 to 1050 and worked under a positive ion mode. Data dependent CID was applied for lipid fragmentation; and the isolation width of the parent ion and normalized collision energy were set at 2.0 and 50.0, respectively.
Results and Discussion
Scheme of lipid profiling from lipid corona on NPs
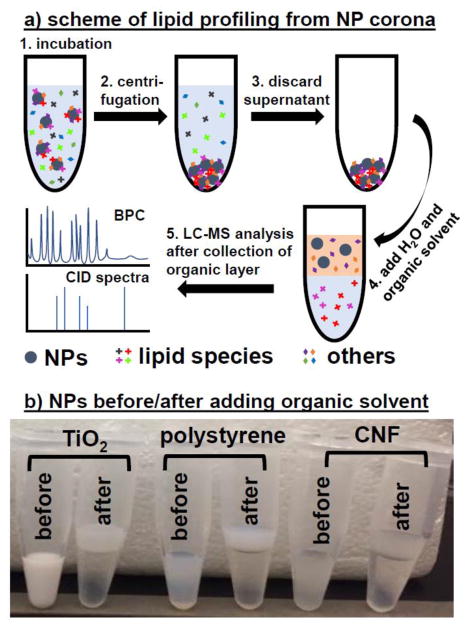
Fig. 1a illustrates the process of analyzing lipid adsorption on NPs incubated with a target matrix. After incubation, the NPs are settled down by centrifugation to eliminate the unbound biomolecules, which are further removed during several rounds of washing. Lipids are co-precipitated with the NPs at bottom of vial, to which the extraction solution is added. The polar molecules including proteins, ions, carbohydrates and the NPs are extracted to the water layer, while the hydrophobic molecules like lipids participate in the organic solvent layer (chloroform has a higher density than water, thus forms the lower layer). The two layers can be well separated using gentle centrifugation that is not strong enough to precipitate the NPs. The collected lipids from the organic layer are identified by LC-MS system.
Figure 1.
(A) Schematic of lipid extraction and analysis process, including collection of NPs incubated with the biological sample, extraction of lipids binding to the NP surface, and analysis of the extracted lipids by LC-MS/MS. (B) Appearance change of TiO2, PS and CNF NPs before and after being mixed with chloroform.
One concern is that, if the NPs aggregate severely during extraction, they would quickly sink down during the centrifugation step and be not separable from the lipid-dissolved organic layer. As shown in Fig. 1b, the cloudy extraction solution containing the NPs of TiO2, PS and CNF separated into two layers after centrifugation, and majority of the NPs seemed to distribute in the water layer, leading to a much more turbid top layer than the transparent chloroform layer. We also evaluated the size of the NPs before and after extraction by Nanosight (Malvern Panalytical Ltd., UK). The water layer was collected and dried by SpeedVac. The pellet was re-hydrated in the same volume of dH2O. For the extraction of lipids from all three types of NPs tested, the particle concentration recovered in the water layer was slightly lower than that added to the extraction solution (ESM, Fig. S3). This indicates that a small proportion of the NPs may have moved to the organic layer by centrifugation or aggregated slightly during extraction. The CNF and TiO2 had negligible change in their mean hydrodynamic size after treatment with chloroform and centrifugation, while the NPs exhibited 20% size increase after extraction. We also detected a low degree of aggregation, which further shows that the extraction was successful and NP loss into the organic layer was insignificant.
Optimization of LC-MS for lipid analysis
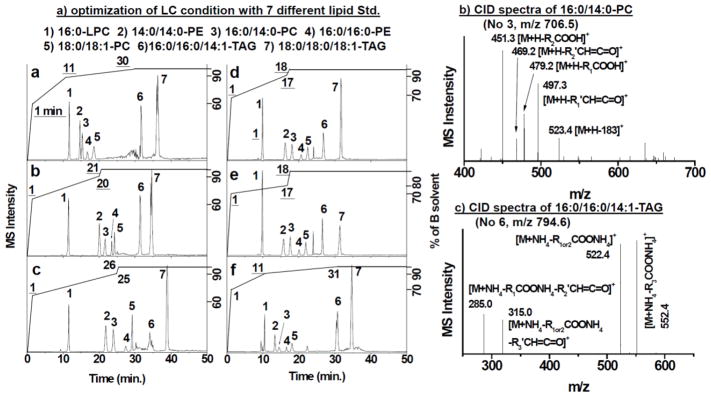
We used 7 lipid standards for the optimization of the gradient condition for HPLC separation. They are 1 LPC, 1 PEs, 1 DAG, 2 PCs and 2 TAGs, covering a wide range of hydrophilicities among lipids. The chromatograms collected at varied elution conditions are shown in Fig. 2a. At first, we ramped up the proportion of solvent B to 60 % within the first minute and increased it to 90% within the next 10 minutes to elute relatively polar lipids the lysophospholipids (LPLs) and phospholipids (PLs). Then, %B went up until 100% at t = 30 min and remained for the rest of the elution period, within which period the rest of the lipid species except TAG were eluted between 10 and 20 min. Considering that most of the fatty acids commonly found in mammalian biomatrix are 16:0, 18:0, 18:1, 18:2, 18:3, 20:4, 22:5 and 22:6 (the first number represents the carbon number of fatty acid and the second number shows the number of double bonds) [28–33], but the lipid standards tested are relatively short fatty acids, we adjusted the gradient to enlarge the elution window to accommodate more PLs from the real samples. We either 1) elongated the time window of ramping up %B from 60% at t = 1 min. to 90% %B at t = 18, 20, and 26 min.; or 2) kept the step of increasing from 60% to 90% between t = 1 and t = 11 min., but elongated the later period (from 90% – 100%) to 31 min. However, under these condition, the retention time difference between LPC and 14:0/14:0-PE leaves a large gap of time where no sample is measured, thus lowering efficiency. From Fig. 2a, we can see that, Condition B and C wasted too much time to elute 14:0/14:0-PE (Lipid #2) after the elution of 16:0-LPC; and Condition D and E exhibited little difference, both giving an elution gap too narrow to profile PL species from real samples. Condition F, which had a long, 20-min ramping window for the high %B of 90%–100%, displayed an ideal elution situation: the short chain lipids were eluted within the first 20 minutes and there was a gap between lipids #4 and #6, which could accommodate the long chain lipids that preceded the elution of TAG. Thus, the gradient Condition F was chosen in the following study.
Figure 2.
(A) Base peak chromatograms (BPC) of 7 lipid species under 6 (a – f) different solvent gradients. The Y-axis on the right side represents the percentage of solvent B (%B) in the gradient elution program; the underlined numbers on the BPC are the time point at which %B reached to the value on X-axis; and the numbers without underlines depict each lipid species. (B) and (C) are the specific CID fragment patterns of PC and TAG, respectively.
CID is an effective fragmentation route to obtain structural information of lipids [28,29,31]. Two representative CID spectra collected from this study were shown in Fig. 2b and 2c, for 16:0/14:0-PC (m/z 706.5) and 16:0/16:0/14:1-TAG, respectively. The first specific fragment produced from PC was the ion at m/z = 523.4 tagged as [M+H-183]+ in Fig. 2b, produced by cleavage of the choline head from the parent ion. Inside the collision cell, specific fragment ions were produced due to the loss of two acyl chains in the form of ketene (m/z 469.2 [M+H-R2′CH=C=O]+ and m/z 497.3 [M+H-R1′CH=C=O]+) or carboxylic acid (m/z 451.3 [M+H-R2COOH]+ and m/z 479.2 [M+H-R1COOH]+). The exact 18 differences between the m/z of the carboxylic acid and ketone forms and the intensities of these fragments further support the location of the fatty acids, which are commonly labeled as R1 and R2 in lipid nomenclature. Similarly, the structure of TAG can be revealed by the CID spectra (Fig. 2c). Compared to the intrinsic molar mass of TAG, since its ionization was assisted by an ammonium ion instead of a hydrogen ion, many of fragmentations occurred with the addition of one ammonium ion. Three different fragments were produced from the cleavages of three fatty acids, but only two large fragments were detected in Fig. 2c owing to the same structure of R1 and R2. Cleavage of fatty acids from the parent ion of 16:0/16:0/14:1-TAG produced the species of [M+NH4-R1or2COONH4]+ (m/z 522.4) and [M+NH4-R3COONH4]+ (m/z 552.4), with the two daughter species having the exact m/z difference of 30 due to an additional 2 carbons and 6 hydrogens from 16:0 fatty acid compared with 14:1 fatty acid. The fragments produced by losing acyl chain in the form of ketene or carboxylic acid showed similar intensity, whereas the intensities of the secondary fragments relatively lower than the first two primary fragments. With CID, the structures of all standard lipids were correctly identified.
Development of methods for lipid extraction from NPs
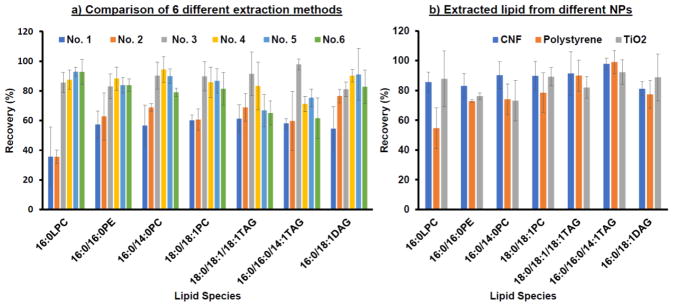
Six different extraction protocols were examined for recovery of lipids from CNF, and the results are shown in Fig. 3a. We calculated the recovery by normalizing the peak area in base peak chromatogram (BPC) to that of the IS, 14:0/14:0-PE. The varied extraction protocols were all based on the widely applied Folch method but with adjustments in whether the SpeedVac-based solvent evaporation was applied before extraction, whether a second round of extractions was conducted, or in the composition of the extraction solutions used in the second extraction round. We can see from Figure 3a that, extraction protocol No. 1 and No. 2 yielded the lowest recovery compared to others probably because they only went through one extraction round. Small differences in recovery were observed between No. 1 and No. 2, indicating that the initial evaporation step to remove the sample solvent was not necessary. This step may even induce more NP aggregation that could conceal lipids inside of aggregated structures and prevent high recovery. Organic solvents with higher hydrophobicity may not be favorable to dissolve the highly polar lipids like LPLs; however, they should be good for releasing the lipids from the hydrophobic surface of NPs. Thus, we employed pure chloroform to release all lipids from NPs, and then adjusted the polarity of chloroform by increasing the percentage of MeOH from 0% to 50% in the second extraction round, expecting to improve solubility of the more polar lipids and enhance recovery. As expected, the recovery of LPC was higher with increasing %MeOH, generally ~ 80%, but the recovery of TAG was reduced to ~ 60% in these protocols (No. 4–6). On the contrary, the recovery of LPC in protocol No. 3, the protocol that utilized the highly hydrophobic chloroform in both rounds of extraction, was not significantly low and still maintained ~80% (Fig. 3a). Therefore, we chose No. 3 to extract a wide range of lipids with different hydrophobicity from NPs.
Figure 3.
(A) Recovery of 7 lipid standards obtained by the 6 different extraction methods. (B) Recovery comparison of 7 lipid standards from CNF, PS and TiO2 NPs using Method No. 3. All recovery values were calculated from triplicated detections.
Figure 3b compared the recovery of the standard lipids from the three NPs chosen in this study: 100 μL of CNF, PS and TiO2 NPs. It is noticeable that while CNF and TiO2 NPs gave out >80% for all of the standard lipids, PS NPs resulted in a relatively lower recovery for the more hydrophilic LPLs, including LPC, PC, and PE. In particular, the recovery of 16:0 LPC dropped to ~ 50%.
Comparative analysis of lipid composition from real sample
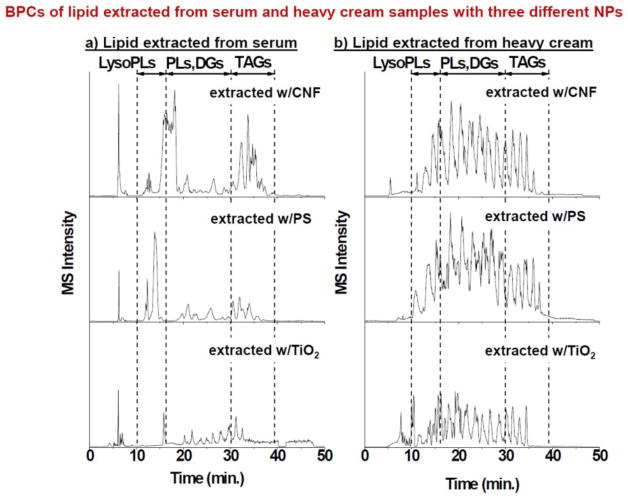
With the optimized extraction protocol and LC-MS/MS method, we analyzed the lipid corona formed on these three NPs when incubated with serum and heavy cream. One hundred μL of the NP solution was mixed with 200 μL of serum or heavy cream and incubated under mild vortexing condition for an hour. Both of two real samples were diluted 4 times with 1× PBS buffer before incubation with NPs. After incubation, NPs were collected and washed to remove the free matrix molecules. Then, lipid extraction was performed on the NPs. The lipids that were pelleted during the last extraction round were dissolved in 1 mL of the organic solvent mixture (Chloroform 20: MeOH 75: water 5, v/v/v); and the internal standard of 14:0/14:0-PE was added to a final concentration of 5 pmol/μL for LC-MS/MS analysis, with the injection volume fixed at 5 μL. The BPCs of lipids obtained from the NPs after incubation with serum or heavy cream are displayed in Figure 4a and 4b, respectively. According to the elution order of the lipid standards observed in Fig. 2a, the whole retention time could be divided into three ranges: 10 ~ 15 min for elution of LPLs, 15~30 min for PLs and DAGs, 30~40 min for TAGs. The general pattern from each NP from the same sample, i.e. serum or heavy cream, looked similar, with fine differences observed among the BPCs from different NPs. However, the patterns between serum and heavy cream were substantially different, which reflect the large difference in lipid composition between these two samples. These results hint that NPs could preserve the main lipid profile originated from the sample, but each NP could differentially adsorb the lipids depending on the properties of NPs and lipids.
Figure 4.
Base peak chromatograms of lipids extracted from three NP coronas after incubation in (A) serum and (B) heavy cream.
To gain more understanding of the lipid adsorption profile, we quantified the concentration of each identified lipid recovered from each type of NP. Since the ionization efficiency of each lipid is different, affected by the structure and fatty acid length, the absolute amount of each lipid species could not be obtained. We calculated the relative amount by comparing the peak area of each lipid with that of the IS, which should be suitable for getting the lipid composition profile in each sample. Except for the lipids with low abundancies, most lipids species show reasonable standard deviation when three repeated extractions were performed. According to their relative amounts, areas of several lipids revealed more than 70 times larger than the peak area of IS, especially some TAGs. Lipids present at low abundancies with the relative amount < 0.05 showed relatively higher standard deviations because their detection was not as consistent as the more abundant lipids.
The relative quantification results are displayed in Table 2. In this study, each lipid was represented by the total carbon number found on the two fatty acids of PC and PE, or the three fatty acids from TAG, followed by the number of double bond on each lipid. Like mentioned above, although number of carbon and double bonds from two fatty acid summed, each fatty acid could be speculated based on the characteristic fatty acids observed from mammalian lipids as mentioned in the Introduction section. For example, 34:2-PC should be from fatty acids 16:0 and 18:2, and 38:6-PC should be from either the combination of 18:2/20:4 or 16:0/22:6. As for TAG species, 50:2-TAG is sum of 16:0,16:0,18:2 and 54:4-TAG may be acquired from the combination of 18:0/18:2/18:2 or 18:1/18:1/18:2 or 16:0/18:0/20:4. A brief overview of the numbers shown in Table 2 indicates that in general, more and higher quantities of lipids were recovered from heavy cream than from serum, which holds true for all three types of NPs tested. This is understandable because heavy cream is much richer than serum in lipid composition. One exception is the LPC with the fatty acids containing a sum of 18 carbon number and various number of double bonds, i.e. 18:0/18:1/18:2, most of which were not recovered at detectible levels by the NPs from heavy cream but low relative amounts were extracted from serum. Among the lipids recovered from serum, very few PC and DAGs were identified, in particular with CNF and PS. TiO2 NPs were able to recover some PC and DAG from serum, but the relative amounts were about 2–10 times lower than those recovered by the same NPs from heavy cream.
Table 2.
List of all lipid species identified on CNF, PS and TiO2 incubated with serum and heavy cream, and their relative amounts obtained by dividing the peak area of each lipid with that of the IS. Each sample was quantified in triplicate. N.D. = Not Detected.
| Head | FA | m/z | serum | cream | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| CNF | PS | TiO2 | CNF | PS | TiO2 | |||||
| LPC | 16:0 | 496.3 | 0.06±0.01 | 0.14±0.02 | 0.02±0.01 | 0.22±0.01 | 0.12±0.01 | 16.58±0.73 | ||
| 18:0 | 524.4 | 0.06±0.04 | 0.06±0.04 | 0.02±0.01 | N.D. | N.D. | 3.03±2.16 | |||
| 18:1 | 522.4 | 0.01±0.01 | 0.04±0.01 | 0.03±0.01 | N.D. | N.D. | N.D. | |||
| 18:2 | 520.3 | 0.01±0.01 | 0.13±0.09 | 0.13±0.03 | N.D. | N.D. | N.D. | |||
| 22:5 | 570.4 | N.D. | 0.17±0.03 | 0.06±0.04 | 27.19±3.41 | 18.75±1.54 | 5.06±0.34 | |||
| 22:6 | 568.4 | N.D. | N.D. | N.D. | 5.37±0.51 | 4.09±0.49 | 8.06±1.46 | |||
|
| ||||||||||
| PC | 32:0 | 734.6 | N.D. | 0.41±0.09 | 0.32±0.02 | 8.64±0.57 | 7.45±1.00 | 3.28±0.09 | ||
| 34:0 | 762.4 | N.D. | N.D. | 0.84±0.16 | 7.02±0.94 | 7.78±0.72 | 1.93±0.12 | |||
| 34:1 | 760.4 | N.D. | N.D. | N.D. | 2.14±0.08 | 1.9±0.25 | N.D. | |||
| 34:2 | 758.5 | N.D. | 0.65±0.06 | N.D. | N.D. | N.D. | 11.37±1.48 | |||
| 36:0 | 790.5 | 0.62±0.05 | 0.50±0.03 | 0.67±0.01 | 11.18±1.26 | 11.52±0.72 | 3.40±0.27 | |||
| 36:1 | 788.5 | N.D. | N.D. | 0.19±0.06 | 2.90±0.52 | 3.89±0.05 | N.D. | |||
| 36:3 | 784.6 | N.D. | N.D. | 0.12±0.02 | 1.70±0.23 | 1.43±0.16 | 0.41±0.09 | |||
| 36:4 | 782.5 | N.D. | N.D. | 3.58±0.27 | 11.85±0.6 | 10.81±1.16 | 3.61±0.50 | |||
| 38:4 | 810.5 | N.D. | N.D. | 0.63±0.11 | 5.19±0.81 | 5.47±1.07 | 1.81±0.40 | |||
| 38:5 | 808.6 | N.D. | N.D. | 0.43±0.03 | 14.83±0.92 | 14.51±1.16 | 4.55±0.70 | |||
| 38:6 | 806.5 | N.D. | N.D. | 0.65±0.15 | 6.59±0.58 | 6.05±0.10 | 1.26±0.18 | |||
| 40:6 | 834.5 | 0.75±1.06 | 0.16±0.22 | 0.49±0.08 | 10.01±1.03 | 9.22±1.02 | 2.86±0.15 | |||
|
| ||||||||||
| LPE | 18:1 | 480.3 | 0.12±0.03 | 0.58±0.11 | 0.17±0.03 | N.D. | 0.21±0.01 | 0.54±0.15 | ||
| 22:6 | 526.3 | N.D. | 0.03±0.01 | N.D. | N.D. | N.D. | 1.26±0.22 | |||
|
| ||||||||||
| PE | 36:2 | 744.5 | 0.23±0.02 | 0.16±0.02 | N.D. | 0.77±0.08 | 0.48±0.04 | 0.26±0.06 | ||
| 38:4 | 768.5 | 3.87±0.64 | 3.17±0.64 | 3.42±0.22 | 47.14±4.34 | 41.87±1.90 | 14.27±1.65 | |||
| 38:5 | 766.5 | 3.12±0.66 | 2.49±0.27 | 3.95±0.19 | 41.55±4.35 | 37.34±2.24 | 14.11±3.18 | |||
| 38:6 | 764.5 | 1.45±0.31 | 0.91±0.05 | 1.38±0.09 | 18.38±2.41 | 18.93±1.24 | 4.52±0.91 | |||
| 40:5 | 794.6 | 4.21±0.43 | 3.01±0.29 | 9.51±1.49 | 50.44±2.81 | 40.26±0.34 | 17.54±2.69 | |||
| 40:6 | 792.5 | 1.51±0.17 | 1.46±0.30 | 2.54±0.38 | 25.46±3.07 | 25.70±3.62 | 7.92±1.51 | |||
|
| ||||||||||
| DAG | 14:0,18:1 | 584.5 | N.D. | N.D. | N.D. | 5.49±0.95 | 5.2±0.22 | 1.31±0.15 | ||
| 14:0,18:2 | 582.6 | N.D. | N.D. | N.D. | 1.05±0.06 | 1.21±0.14 | 0.44±0.09 | |||
| 16:0,16:0 | 586.5 | N.D. | N.D. | 0.93±0.06 | 9.58±1.78 | 12.41±0.31 | 2.47±0.30 | |||
| 16:0,18:0 | 614.5 | N.D. | 0.73±0.10 | N.D. | 18.87±2.06 | 20.01±0.65 | 6.28±0.42 | |||
| 16:0,18:1 | 612.5 | 1.20±0.14 | 1.18±0.10 | 0.35±0.06 | 6.10±0.75 | 10.24±1.10 | 7.15±1.05 | |||
| 16:0,20:0 | 642.6 | N.D. | N.D. | 0.62±0.12 | 12.83±1.64 | 11.38±0.74 | 2.49±0.31 | |||
| 16:0,20:1 | 640.5 | N.D. | N.D. | 0.42±0.14 | 7.19±1.22 | 6.18±0.74 | 1.83±0.25 | |||
| 16:0,24:1 | 696.6 | N.D. | N.D. | 0.55±0.10 | 13.74±2.24 | 10.54±0.53 | 2.81±0.83 | |||
| 16:1,20:1 | 638.5 | 0.35±0.04 | 0.53±0.09 | 0.27±0.09 | 2.46±0.20 | 3.46±0.61 | 0.69±0.05 | |||
| 16:1,22:1 | 666.5 | N.D. | N.D. | N.D. | 2.65±0.27 | 2.12±0.56 | 0.65±0.05 | |||
| 18:1,20:1 | 666.5 | 6.66±0.77 | 7.19±1.23 | 5.92±0.37 | N.D. | N.D. | N.D. | |||
| 18:1,22:1 | 694.6 | N.D. | N.D. | N.D. | 2.44±0.11 | 2.55±0.38 | 1.22±0.11 | |||
| 18:2,22:6 | 682.6 | N.D. | N.D. | 2.85±0.11 | 46.88±2.48 | 50.59±4.28 | 19.94±6.07 | |||
|
| ||||||||||
| TAG | 48:0 | 824.7 | 2.17±0.49 | 2.15±0.24 | 8.10±1.10 | 19.57±1.71 | 25.86±3.65 | 7.57±0.86 | ||
| 48:1 | 822.7 | 4.27±0.64 | 3.59±0.47 | 10.05±1.32 | 55.28±2.98 | 53.62±2.08 | 17.25±1.28 | |||
| 50:0 | 852.8 | 1.70±0.43 | 2.19±0.32 | 5.73±0.49 | 17.59±1.91 | 15.62±2.44 | 4.99±0.22 | |||
| 50:1 | 850.7 | 3.59±0.35 | 4.20±1.01 | 16.08±2.09 | 58.61±0.40 | 61.46±4.25 | 18.77±3.29 | |||
| 50:2 | 848.8 | 5.02±0.73 | 3.95±0.22 | 9.55±0.12 | 41.65±4.20 | 37.82±4.86 | 14.96±1.59 | |||
| 50:3 | 846.8 | 2.94±0.25 | 2.72±0.62 | 4.63±0.22 | 26.54±2.45 | 27.29±1.28 | 7.65±0.69 | |||
| 50:4 | 844.7 | 1.28±0.23 | 1.05±0.15 | 1.70±0.16 | 9.65±0.55 | 10.42±0.28 | 2.79±0.39 | |||
| 52:0 | 880.7 | 2.09±0.13 | 1.62±0.23 | 2.38±0.32 | 14.88±0.61 | 11.86±2.15 | 2.37±0.46 | |||
| 52:1 | 878.8 | 3.05±0.33 | 3.03±0.21 | 9.48±0.09 | 72.47±7.65 | 54.34±2.67 | 15.44±2.29 | |||
| 52:2 | 876.7 | 5.63±0.79 | 6.26±0.36 | 9.55±0.35 | 46.36±2.91 | 54.20±3.00 | 18.48±1.38 | |||
| 52:3 | 874.8 | 10.84±0.99 | 13.39±2.26 | 8.34±0.46 | 25.43±1.22 | 24.98±1.54 | 10.32±1.20 | |||
| 52:4 | 872.8 | 11.59±1.07 | 5.73±0.60 | 4.49±0.51 | 17.21±1.39 | 15.92±0.84 | 4.14±0.61 | |||
| 52:6 | 870.7 | 3.06±0.37 | 2.35±0.40 | 1.82±0.17 | 3.76±0.23 | 3.48±0.37 | 1.49±0.22 | |||
| 54:1 | 906.7 | 1.59±0.07 | 1.34±0.22 | 2.82±0.11 | 39.86±1.21 | 25.15±1.35 | 12.93±1.32 | |||
| 54:2 | 904.7 | 1.74±0.11 | 1.50±0.09 | 4.50±0.19 | 38.34±5.04 | 29.60±1.50 | 9.71±1.75 | |||
| 54:3 | 902.8 | 2.49±0.30 | 2.33±0.30 | 4.32±0.56 | 20.45±1.24 | 20.70±2.22 | 8.18±0.78 | |||
| 54:4 | 900.7 | 4.49±0.15 | 3.32±0.18 | 2.81±0.08 | 14.28±0.36 | 13.85±1.04 | 5.17±0.56 | |||
| 54:5 | 878.8 | 3.05±0.33 | 3.36±0.35 | 9.73±0.32 | 66.37±3.33 | 54.34±2.67 | 15.83±2.21 | |||
| 54:6 | 896.6 | 2.44±0.30 | 1.96±0.06 | 2.40±0.24 | 4.51±0.57 | 3.39±0.34 | 1.21±0.06 | |||
| 56:2 | 932.8 | N.D. | N.D. | 1.34±0.12 | 3.93±0.50 | 3.72±0.11 | 0.49±0.03 | |||
| 56:3 | 930.8 | 0.19±0.01 | 0.19±0.01 | N.D. | 2.23±0.07 | 1.87±0.16 | N.D. | |||
| 56:4 | 928.8 | N.D. | N.D. | N.D. | 1.68±0.04 | 1.78±0.06 | N.D. | |||
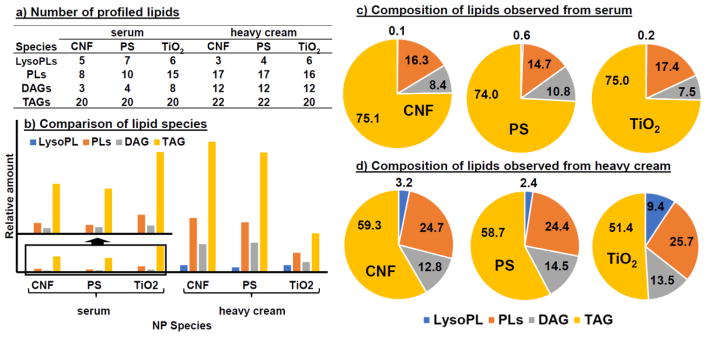
These conclusions can be seen more clearly in Figure 5, which compared the total number (Fig. 5a) and relative contents (Fig. 5b) of each lipid category in a table and a bar plot. The lipids are grouped into LysoPL (LPC + LPE), PL (PC and PE), DAG, and TAG. We can see that higher number of PL and DAG species were found from heavy cream than serum. However, the total numbers of TAG and LysoPL species from both matrices were quite comparable. Although very few LysoPL species were found in both serum and heavy cream, their relative contents in heavy cream were higher than in serum, owing to the higher overall lipid content in heavy cream than in serum. TAG seems to be the main lipid category bound to the NPs. Even though the ionization efficiency of TAG is lower than other lipids owing to their high m/z values, relatively higher amounts of TAG were found in both matrices by all three types of NPs. Interestingly, while in serum, TiO2 NPs adsorbed a higher relative content of TAG than the other two NPs, but in heavy cream they bound to a significantly lower amount of TAG.
Figure 5.
Comparison of the lipid adsorption profiles among NPs when incubated with serum and heavy cream. The profile on each NP in the matrix is represented in three ways: (A) numbers of identified lipids within each lipid category (LysoPLs, PLs, DAG and TAG); (B) the sum relative amounts of all lipid species belonging to each lipid category; and (C) the percentage of each lipid category among all lipids adsorbed by the NPs, calculated by dividing the sum relative amount of each lipid category by the sum relative amount of all four lipid categories considered in the present work.
The pie charts in Fig. 5c&d compare the lipid profiles adsorbed by the three NPs tested. We can see that the lipid profile formed in serum was highly similar among the NPs. More than 70% of the adsorbed lipids were TAG and ~15% were PLs. While the profile did not change much for CNF and PS when incubated in heavy cream, a noticeable difference was found for the lipid adsorption profiles of TiO2 between serum and heavy cream: a lower proportion of TAG content was found in TiO2 than in the other two NPs. This agrees with what was observed in Fig. 5b: TAG adsorption on TiO2 was reduced in heavy cream compared to what occurred in serum. On the contrary, adsorption of LysoPL by TiO2 increased. These results indicate that, the higher relative content of TAG found in the lipid corona formed on the TiO2 NPs when incubated in serum and heavy cream may be due to its higher amount in the matrix than other lipid categories. However, the NPs prefer to bind to LysoPL than TAG, probably because of its lower hydrophobicity as well as the single acryl chain that makes the phosphate group more easily to coordinate with the Ti atom on the surface of TiO2. Thus, when the matrix contains elevated amounts of LysoPL to compete with TAG for the binding sites on the NPs, TAG adsorption would be reduced and the relative content of LysoPL in the lipid corona would increase, as shown in Fig. 5c&d.
Conclusion
This study demonstrates lipid corona could be formed on NPs during incubation with serum and heavy cream. The conventional organic solvent extraction protocol was found suitable for lipid recovery from NPs, and the physical properties of the NPs were not changed before and after extraction, eliminating the potential impacts on lipid study from NP dissolution or aggregation. It was found in this study that, repeated extraction with 100% of CHCl3 could lead to the best lipid recovery from the NPs. LC separation condition was also optimized to cover the wide range of lipids found in these samples. The developed method was applied to study the lipid corona composition on three types of NPs incubated in serum and heavy cream. More lipids were recovered from heavy cream than from serum, which is probably due to the higher lipid content in heavy cream. The more hydrophobic CNF and PS formed similar lipid corona in both matrices, while the more hydrophilic TiO2 tended to adsorb less TAG. In addition, our result hints that, lipid adsorption on TiO2 is more selective than CNF and PS, because its lipid profile would be affected by the lipid composition of the matrix: if the matrix contains the competitive lipids, like LysoPL, adsorption of TAG, which is more abundant than others, would be affected. Specific interaction may occur between TiO2 and LysoPL which may have higher affinity to TiO2 than other lipid categories.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number U01ES027293, as well as by the Engineered Nanomaterials Resource and Coordination Core established at Harvard T. H. Chan School of Public Health (NIH grant # U24ES026946), both as parts of the Nanotechnology Health Implications Research Consortium. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with ethical standards.
Conflict of interest. The authors declare that they have no conflict of interest.
References
- 1.Bondarenko O, Juganson K, Ivask A, Kasemets K, Mortimer M, Kahru A. Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review. Arch Toxicol. 2013;87:1181–1200. doi: 10.1007/s00204-013-1079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Love SA, Maurer-Jones MA, Thompson JW, Tu-Shen Lin, Haynes CL. Assessing Nanoparticles Toxicity. Annu Rev Anal Chem. 2012;5:181–205. doi: 10.1146/annurev-anchem-062011-143134. [DOI] [PubMed] [Google Scholar]
- 3.Monopoli MP, Åberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotech. 2012;7:779–86. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 4.Lynch I, Dawson KA. Protein nanoparticle interactions. Nano Today. 2008;3:40–7. [Google Scholar]
- 5.Deng ZJ, Mortimer G, Schiller T, Musumeci A, Martin D, Minchin RF. Differential plasma protein binding to metal oxide nanoparticles. Nanotechnology. 2009;20:455101. doi: 10.1088/0957-4484/20/45/455101. [DOI] [PubMed] [Google Scholar]
- 6.Ashby J, Pan S, Zhong W. Size and surface functionalization of iron oxide nanoparticles influence the composition and dynamic nature of their protein corona. ACS Appl Mater Interfaces. 2014;6:15412–19. doi: 10.1021/am503909q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashby J, Schachermeyer S, Pan S, Zhong W. Dissociation-based screening of nanoparticle protein interaction via flow field-flow fractionation. Anal Chem. 2013;85(15):7494–501. doi: 10.1021/ac401485j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA. Physical-chemical aspects of protein corona: relevance to in vitro and in vivobiological impacts of nanoparticles. J Am Chem Soc. 2011;133:2525–34. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 9.Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW, Hall JB, McNeil SE. Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomed Nanotech Biol Med. 2009;5:106–17. doi: 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nanotech. 2013;8:772–81. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 11.Choi HS, Ashitate Y, Lee JH, Kim SH, Matsui A, Insin N, Bawendi MG, Semmler-Behnke M, Frangioni JV, Tsuda A. Rapid translocation of nanoparticles from the lung airspaces to the body. Nature Biotechnol. 2010;28:1300–3. doi: 10.1038/nbt.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milani S, Baldelli BF, Pitek AS, Dawson KA, Rädler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano. 2012;6:2532–41. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 13.Martel J, Young D, Young A, Wu CY, Chen CD, Yu JS, Young JD. Comprehensive proteomic analysis of mineral nanoparticles derived from human body fluids and analyzed by liquid chromatography-tandem mass spectrometry. Anal Biochem. 2011;418:111–25. doi: 10.1016/j.ab.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Tenzer s, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5:7155–67. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 15.Muller RH, Mader K, Gohla S. Solid lipid nanoparticles (SLN) for controlled drug delivery a review of the state of the art. Eur J Pharm & Biopharm. 2000;50:161–177. doi: 10.1016/s0939-6411(00)00087-4. [DOI] [PubMed] [Google Scholar]
- 16.Divecha N, Irvine RF. Phospholipid signaling. Cell. 1995;80:269–78. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 17.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 18.Ishii I, Fukushima N, Ye X, Chun J. Lysophospholipid receptors: signaling and biology. Annu Rev Biochem. 2004;73:321–35. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 19.Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4(7):e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C, Kong H, Qu F, Li Y, Yu Z, Gao P, Peng S, Xu G. Application of plasma lipidomics in studying the response of patients with essential hypertension to antihypertensive drug therapy. Mol BioSyst. 2011;7(12):3271–9. doi: 10.1039/c1mb05342f. [DOI] [PubMed] [Google Scholar]
- 21.Scherer M, Montoliu I, Qanadli SD, Collino S, Rezzi S, Kussmann M, Giusti V, Martin FP. Blood plasma lipidomic signature of epicardial fat in healthy obese women. Obesity. 2015;23:130–7. doi: 10.1002/oby.20925. [DOI] [PubMed] [Google Scholar]
- 22.Kien CL, Bunn JY, Poynter ME, Stevens R, Bain J, Ikayeva O, Fukagawa NK, Champagne CM, Crain KI, Koves TR, Muoio DM. A lipidomics analysis of the relationship between dietary fatty acid composition and insulin sensitivity in young adults. Diabetes. 2013;62:1054–63. doi: 10.2337/db12-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tonks KT, Coster AC, Christopher MJ, Chaudhuri R, Xu A, Gagnon-Bartsch J, Chisholm DJ, James DE, Meikle PJ, Greenfield JR, Samocha-Bonet D. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity. 2016;24:908–16. doi: 10.1002/oby.21448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvayre R, Auge N, Benoist H, Negre-Salvayre A. Oxidized low-density lipoprotein-induced apoptosis. Biochim Biophys Acta. 2002;30:213–21. doi: 10.1016/s1388-1981(02)00343-8. [DOI] [PubMed] [Google Scholar]
- 25.Leonarduzzi GC, Arkan M, Başağa H, Chiarpotto E, Sevanian A, Poli G. Lipid oxidation products in cell signaling. Free Rad Biol & Med. 2000;28:1370–1378. doi: 10.1016/s0891-5849(00)00216-1. [DOI] [PubMed] [Google Scholar]
- 26.Heller DN, Murphy CM, Cotter RJ, Fenselau C, Uy OM. Constant Neutral Loss Scanning for the Characterization of Bacterial Phospholipids Desorbed by Fast Atom Bombardment. Anal Chem. 1988;60:2787–2791. doi: 10.1021/ac00175a029. [DOI] [PubMed] [Google Scholar]
- 27.Völlmin JA. Separation and identification of urinary steroids on glass capillary columns in a combination of gas chromatograph and mass spectrometer. Chromatographia. 1970;3:238–241. [Google Scholar]
- 28.Bang DY, Kang D, Moon MH. Nanoflow Liquid Chromatography/Tandem Mass Spectrometry for the characterization of Intact Phosphatidylcholine Lipids: Soybean, Bovine Brain, and Liver. J Chromatography A. 2006;1104:222–229. doi: 10.1016/j.chroma.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Bang DY, Ahn E, Moon MH. Shotgun Analysis of Phospholipids from Mouse Liver and Brain by Nanoflow Liquid Chromatography/Tandem Mass Spectrometry. J Chromatography B. 2007;852:268–277. doi: 10.1016/j.jchromb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Bang DY, Lim S, Moon MH. Effect of ionization modifiers on the simultaneous analysis of all classes of phospholipids by nanoflow liquid chromatography/tandem mass spectrometry in negative ion mode. J Chromatogr A. 2012;1240:69–76. doi: 10.1016/j.chroma.2012.03.073. [DOI] [PubMed] [Google Scholar]
- 31.Byeon SK, Lee JC, Chung BC, Seo HS, Moon MH. High-Throughput and Rapid Quantification of Lipids by Nanoflow UPLC-ESI-MS/MS: Application to the Hepatic Lipids of Rabbits with Non-Alcoholic Liver Disease. Anal Bioanal Chem. 2016;408:4975–4985. doi: 10.1007/s00216-016-9592-y. [DOI] [PubMed] [Google Scholar]
- 32.Lee JY, Min H, Moon MH. Simultaneous Profiling of Lysophospholipids and Phospholipids from Human Plasma by Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal Bioanal Chem. 2011;400:2953–2961. doi: 10.1007/s00216-011-4958-7. [DOI] [PubMed] [Google Scholar]
- 33.Byeon SK, Kim J, Lee JY, Chung BC, Seo HS, Moon MH. Top-down and Bottom-up Lipidomic Analysis of Rabbit Lipoproteins under Different Metabolic Conditions using Flow Field-Flow Fractionation, Nanoflow Liquid Chromatography and Mass Spectrometry. J Chromatogr A. 2015;1405:140–148. doi: 10.1016/j.chroma.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 34.Beltran-Huarac J, Zhang Z, Pyrgiotakis G, Deloid G, Vaze N, Demokritou P. Development of reference metal and metal oxide engineered nanomaterials for nanotoxicology research using high throughput and precision flame spray synthesis approaches. NanoImpact. 2018;10:26–37. doi: 10.1016/j.impact.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyrgiotakis G, Luu W, Zhang Z, Vaze N, DeLoid G, Rubio L, Graham WAC, Bell DC, Bousfield D, Demokritou P. Development of high throughput, high precision synthesis platforms and characterization methodologies for toxicological studies of nanocellulose. Cellulose. 2018;25:2303–2319. [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen JM, Beltran-Huarac J, Pyrgiotakis G, Demokritou P. Effective delivery of sonication energy to fast settling and agglomerating nanomaterial suspensions for cellular studies: Implications for stability, particle kinetics, dosimetry and toxicity. NanoImpact. 2018;10:81–86. doi: 10.1016/j.impact.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JY, Lim S, Park S, Moon MH. Characterization of oxidized phospholipids in oxidatively-modified low density lipoproteins by nanoflow liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2013;1288:54–62. doi: 10.1016/j.chroma.2013.02.086. [DOI] [PubMed] [Google Scholar]
- 38.Folch J, Lees M, Solane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Bio Chem. 1957;226:497–509. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.