Abstract
Metabolic syndrome and obesity are increasing epidemics that significantly impact the peripheral nervous system and lead to negative changes in sensation and peripheral nerve function. Research to understand the consequences of diet, obesity and fuel usage in sensory neurons has commonly focused on glucose metabolism. Here, we tested whether mouse sensory neurons and nerves have the capacity to metabolize fat-based fuels (palmitoyl-CoA) and if these effects were altered by feeding of ketogenic (90% kcal fat) diet compared to control (14% kcal fat). Male C57Bl/6 mice were placed on diets for 10 weeks, and upon sacrifice, dorsal root ganglion (DRG) and sciatic nerves (SN) were placed in an OROBOROS O2K oxygraph to examine diet-induced alterations in metabolism (respiration) of palmitoyl-CoA and H2O2 emission (fluorescence). In addition, RNAseq was performed on the DRG of mice fed a control or ketogenic diet for 12 weeks and genes associated with mitochondrial respiratory function were analyzed. Our results suggest that the sciatic nerves from mice fed a ketogenic diet display reduced O2 respiration and H2O2 emission when metabolizing palmitoyl-CoA compared to mice fed a control diet. Assessments of mRNA gene expression changes reveal alterations in genes encoding the NADH dehydrogenase complex and Complex IV, which could alter reactive oxygen species (ROS) production. These novel findings highlight the ability of sensory neurons and axons to oxidize fat based fuel sources, and that these mechanisms are adaptable to dietary changes.
Introduction
The metabolism of fatty acids and mitochondrial function has been studied in numerous tissues and disease models to reveal the important role metabolism and mitochondria have in chronic disease states (Lee et al., 2003; Reddy & Rao, 2006; Osborn & Olefsky, 2012). Studies examining the nervous system have primarily focused on brain metabolism of glucose or ketones and have largely neglected to examine the metabolism of fatty acids. Similarly, peripheral nervous system metabolism has been extremely understudied, and to our knowledge, has only been examined for alterations in glucose metabolism (Chowdhury et al., 2010; Akude et al., 2011). Glucose metabolism may have only been studied in the nervous system due to the understanding that neurons in the central nervous system (CNS) are lacking the isoform of carnitine palmitoyltransferase channels which allow long chain fatty acid transport into the mitochondria for proper fatty acidy oxidation (Price et al., 2002; Wolfgang et al., 2006). However, the peripheral nervous system and specifically, dorsal root ganglion (DRG), are known to have the proper carnitine carrier and carnitine transferase enzymes required for mitochondrial fatty acid metabolism (Tonazzi et al., 2013). Emerging research has begun to recognize the key metabolic impact that peripheral nerves undergo in high-fat diet-induced obesity (Groover et al., 2013; Guilford & Wright, 2013; Cooper et al., 2016).
Ketone-based metabolism produces less damaging reactive oxygen species (ROS) compared to glucose metabolism at a mitochondrial and cellular level (Veech, 2004). Fluorescent emission of H2O2 has been utilized to examine the presence of ROS, as H2O2 is the end product of the free radical O2 molecule after processing by superoxide dismutase (Gomes A, 2005). In neurons, reductions in ROS due to caloric restriction/ketone signaling may occur due to alterations in Complex I of the respiratory chain. Additionally, ketones inhibit ROS production through an increase in NAD/NADH ratios while metabolizing beta-hydroxybutyrate as compared to glutamate (Maalouf et al., 2007). Further, this alteration in NAD/NADH ratio is protective as it engages Sirt1 to inhibit apoptotic and inflammatory signaling while putatively promoting increased mitochondrial biogenesis and reduced acetylation of mitochondrial proteins (Maalouf et al., 2007). Improved mitochondrial respiration and/or reduced H2O2 emission may offset detrimental mitochondrial changes associated with neuronal injury to improve neuronal health and healing.
One feature of metabolic dysfunction is an increase in mitochondrial dysfunction and increased ROS (Figueroa-Romero et al., 2008), which has been proposed to contribute to sensory dysfunction and neuropathy. Here, we show that mitochondria in sciatic nerves (SN) from mice fed a high fat, low carbohydrate (ketogenic) diet adapt to fat-based fuel sources with reduced oxidative stress compared to mice fed a traditional diet. This reduction in ROS production, as shown by reduced H2O2 emission, suggests that peripheral neurons can use a fat-based fuel source and can adapt to this fuel source when fed a ketogenic diet.
Experimental Procedure
Ethical Approval
All animal use was in accordance with the NIH guide for the care and use of laboratory animals 8th Ed. and conformed to the principles specified in a protocol approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee (#2016–2324). Seven-week-old male C57BL/6 CR #027 mice were purchased from Charles River (Wilmington, Mass) and maintained on a 12:12h light/dark cycle at the University of Kansas Medical Center. All mice were given ad libitum access to food and water and were fed either a standard control diet (8604; Envigo, Madison Wisconsin; 14% kcals from fat, 32% protein, and 54% carbohydrate) or a ketogenic diet (96355; Envigo; 90.5% kcals from vegetable shortening (hydrogenated) and corn oil fat, 9.2% protein, and 0.3% carbohydrate). Animals were kept on all diets for 10 weeks at which point all body weight, composition and metabolic measurements were performed. A separate cohort of animals was kept on the above diets for 12 weeks while being utilized for additional experiments including glucose, insulin and RNAseq analysis. The RNAseq cohort was utilized for numerous experiments and though it was kept on the above diets for 12 weeks, we have no expectation of significant alterations in gene expression between 10 and 12 weeks of dietary intervention. All animals were anaesthetized with isoflourene until achievement of loss of consciousness and were then sacrificed by decapitation while under anesthesia in accordance with the approved methods of the Institutional Animal Care and Use committee prior to all tissue and end point blood collection.
Body Composition
Body composition to assess fat and lean mass was measured using the EchoMRI-100 (EchoMRI, Houston, TX). The body composition was determined immediately before sacrifice.
Glucose Measurements
Mice underwent assessment for blood glucose (glucose diagnostic reagents; Sigma, St. Louis, MO) by tail vein nick following a 3-hour fast. Additionally, at the time of sacrifice following a 3 hour fast, blood was drawn from the chest cavity and allowed to clot for 30 min on ice, spun at 3,000g for 30 min at 4 °C and serum drawn off and frozen at −80°C until insulin was analyzed by ELISA (Alpco; Salem, NH).
Respiration and H2O2 emission
Respiration and H2O2 emission was measured using high-resolution respirometry and fluorescence (Oroboros Oxygraph-2k; Oroboros Instruments; Innsbruck, Austria). Protocols were based on O2k manual titrations for the substrate-uncoupler-inhibitor titration (SUIT) protocols for mitochondrial preparations. Isolated lumbar DRG or both SN were removed as previously described (Malin et al., 2007) and wet weight of both DRG and SN were determined prior to placement in respiration chambers. Both DRG and SN were permeabilized with 10μl digitonin (50mg/ml), briefly vortexed and placed on ice for 10 min. Nerves were then placed in respiration chambers with media (modified MiR05; sucrose, 110 mM; KMES, 60 mM; EGTA, 0.5 mM; MgCl2, 3 mM; KH2PO4, 10 mM; HEPES, 20 mM; Glucose, 20mM; adjusted to pH 7.1 with KOH at 37°C; and 1 g/L fatty acid free BSA) at a temperature of 37°C and a stir rate of 750 RPM for assessment of respiration. Basal oxygen flux was measured by addition of palmitoyl-CoA (2mM) and malate (0.8M) coenzyme A (12.5mM) and carnitine (250mM) to the chambers and measured after steady-state was achieved. Palmitoyl-CoA and carnitine were added to the chamber separately to allow observation of differences in fatty acid transport into the mitochondria, as opposed to palmitoyl-carnitine which some use for fatty acid oxidation analysis which would bypass alterations in transport which may arise between groups. State 3 respiration through complex I and electron-transferring flavoprotein (ETF) was then quantified by addition of ADP (0.5M). State 3 respiration of both complexes I and II was assessed by the addition of succinate (1M). Non-mitochondrial respiration was examined by the addition of cytochrome c (). Maximum uncoupled respiration was observed with the addition of FCCP (1μM repeated injections until stable maximal respiration achieved).
H2O2 flux methodology was based on methods utilized for simultaneous H2O2 flux and O2 respirometry in cells and isolated mitochondria (Makrecka-Kuka et al., 2015). Due to the utilization of permeabilized whole tissue and the inability to inject mitochondria or lysate, H2O2 calibration occurred prior to the addition of tissue. 10 μM Amplex UltraRed, 1U/ml horseradish peroxidase, and 10U/ml superoxide dismutase (superoxide dismutase from bovine liver, Sigma-Aldrich, catalog number S8160) were added to the chamber. After steady state was achieved H2O2 was added (0.1 μM H2O2) and the subsequent fluorescent response was calibrated for standardization. Permeabilized tissue was then added with basal respiratory components for O2 respiration (Malate, CoA, Carnitine, and Palmitoyl-CoA) and allowed to reach steady state. After steady state was achieved all subsequent analysis was completed as described above for State 3 and maximal (FCCP uncoupled) H2O2 emission. For all analysis only stable respiratory states were analyzed, while alterations induced by the addition of chemicals or tissues were excluded from analysis. Both O2 and H2O2 flux values were corrected for wet neuronal weight prior to digitonin treatment.
RNAseq
Total RNA was extracted from lumbar DRG 4,5,6 from the second cohort of mice with TruSeq Stranded mRNA Library Prep kit (Illumina, CA). Six biological replicates were used for each dietary intervention group, for a total of 18 samples. Pair-end sequencing was performed using the Illumina platform with read length of 100 bp and an insert size of 160 bp. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used to access sequence quality, with manual inspection of the results before proceeding to subsequent steps. The raw gene count data was obtained using RSEM (Li & Dewey, 2011) using the default parameters. Within RSEM, we used bowtie2 (Langmead & Salzberg, 2012) as mapping tool and the mouse mm10 genome assembly as the reference genome.
The Bioconductor package “edgeR” (Robinson et al., 2010) was used for preprocessing the raw gene count data. Gene count data were normalized by library size and genes with non/low transcription were removed. With regard to the latter, genes with expression level >1 cpm in at least 2 out of the 24 samples were retained, resulting in a total of ~16,000 genes that were considered for subsequent statistical analyses. The resulting genes were then used to perform KEGG pathway analysis to examine all genes associated with known pathways using the Bioconductor package “gage” (PMID:19473525). Mitochondrial oxidative phosphorylation pathways were examined for differences between ketogenic and control-fed mice and visualized by the Bioconductor package “pathview” (PMID: 23740750). These alterations shown before are relative gene expression differences and have not been analyzed for statistically significant alterations, but are instead utilized to show full complete pathway trends.
Statistical Analyses
All data is presented as mean ± SEM. Data was tested for normality utilizing D’Agostino & Pearson and then analyzed using simple t-test. Statistical significance was defined as p <0.05 and all statistics were run using either GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA) or R version 3.4.4 (https//cran.r-project.org).
Results
Measures of Obesity and Prediabetes
Ketogenic-diet fed mice weighed significantly more than control-diet fed mice following 10 weeks of diet (p<0.001) (Table 1). Ketogenic-diet fed mice also had increased fat mass as compared to control-fed mice (p<0.0001) (Table 1). No changes in lean mass were observed in any group (Table 1). In addition, no significant alterations in blood glucose levels were observed (Table 1).
Table 1.
Mice fed a ketogenic diet for 10 weeks have increased body weight and increased fat mass compared to control-diet fed mice (n=8 for all groups). No alterations in lean mass, fasting blood glucose, or insulin were observed between groups (n=8 for all groups).
| 10wk Weight (g) | 10wk Fat Mass (g) | 10wk Lean Mass (g) | 10wk Glucose (mg/dl) | 12wk Insulin (ng/ml) | |
|---|---|---|---|---|---|
| Control-Fed | 28.88*** | 4.51=**** | 22.81 | 120.90 | 1.11 |
| Ketogenic-Fed | 39.28 | 15.23 | 21.48 | 131.60 | 0.99 |
p<0.05
p<0.01
p<0.001
p<0.0001)
Mitochondrial Respiration of Palmitoyl-CoA in DRG
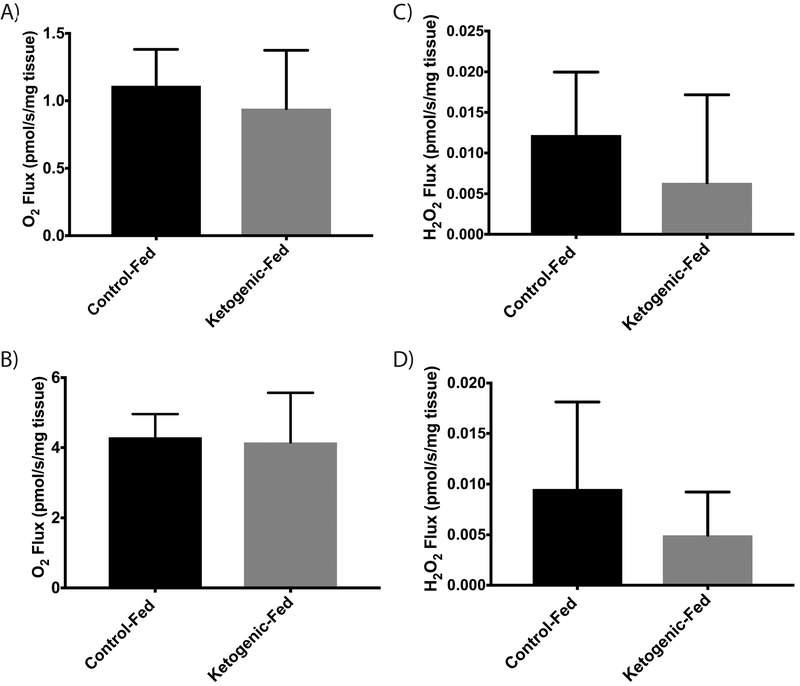
DRG harvested from mice either consuming a ketogenic or control diet for 10 weeks displayed no differences in ADP-stimulated (State 3) or maximal uncoupled respiration of palmitoyl-CoA (Fig. 1a & 1b). DRG also show no differences in H2O2 flux, a measure of ROS production, during metabolism of palmitoyl-CoA regardless of diet among groups (Fig. 1c & 1d). No alteration in respiration was observed with the addition of cytochrome c (not reported).
Figure 1. Oxidative Respiration and ROS Production in DRG.
(a) ADP dependent (State III) respiratory rate of DRG following 10 weeks of diet (b) Maximal respiratory rate of DRG (FCCP uncoupled) (c) State III rate of ROS production in DRG (d) ROS production during maximal respiratory rate in DRG (FCCP uncoupled) (n=8 for all groups). All data presented as mean ± SD
Mitochondrial Respiration of Palmitoyl-Coa in Sciatic Nerve
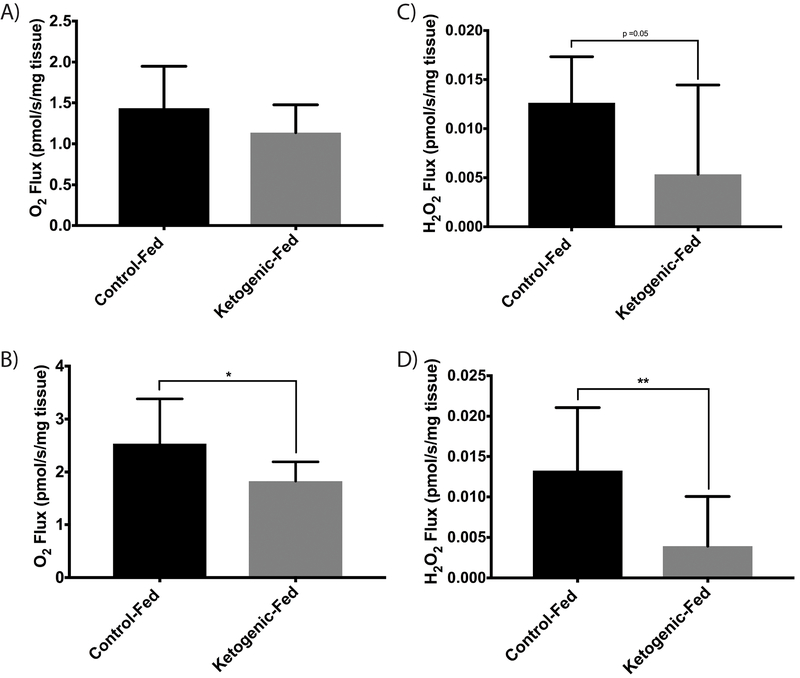
Sciatic nerves from control-diet fed mice had elevated maximal uncoupled respiration compared to ketogenic-diet fed mice (p=0.02), though they failed to reach a significant alteration in ADP-stimulated (State 3) respiration levels (p=0.13) (Fig. 2a & 2b). Sciatic nerves displayed no significant change in ADP-stimulated (State 3) H2O2 flux, though ketogenic-diet fed animals did approach statistical significance relative to control-diet fed mice (p=0.05) (Fig. 2c). Sciatic nerves from ketogenic-diet fed mice (p=0.002) had decreased H2O2 flux during maximal uncoupled respiration, a sign of reduced ROS production when metabolizing palmitoyl-CoA compared to control-diet fed mice; (Fig. 2d). No alteration in respiration was observed with the addition of cytochrome c (not reported).
Figure 2. Oxidative Respiration and ROS Production in Sciatic Nerve.
(a) ADP dependent (State III) respiratory rate of SN following 10 weeks of diet challenge (b) Maximal respiratory rate of SN (FCCP uncoupled) (c) State III rate of ROS production in SN (d) ROS production during maximal respiratory rate in SN (FCCP uncoupled) (n=8 for all groups). All data presented as mean ± SD * p<0.05; **p<0.01
Gene Expression in Oxidative Phosphorylation Complexes
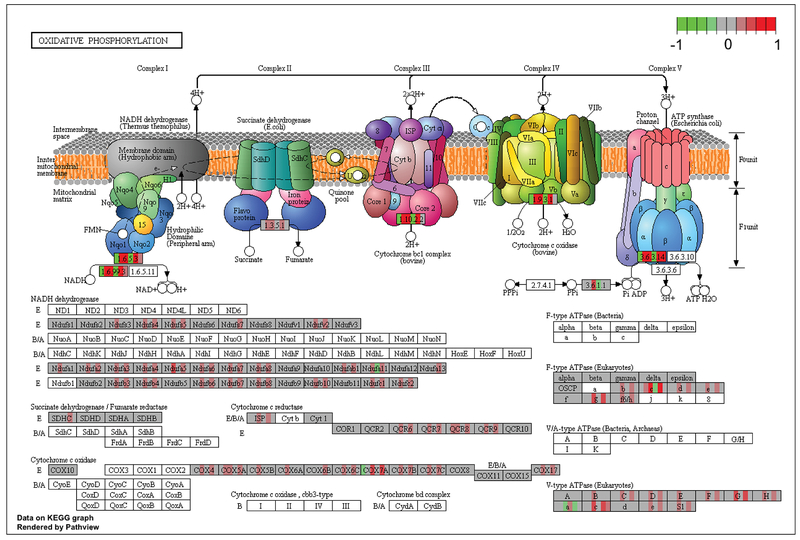
Figure 3 illustrates the relative changes in gene expression of oxidative phosphorylation complex genes in DRG of mice fed a ketogenic diet compared to control diet-fed mice. The oxidative phosphorylation pathway was statistically significantly enriched in ketogenic-diet fed mice relative to control-diet fed animals (q value =5.08e-05). Qualitative mRNA analysis of genes in the electron transport/oxidative phosphorylation pathway suggests that a ketogenic diet may increase gene expression from the NADH dehydrogenase (Complex I), cytochrome c reductase (Complex III) cytochrome c oxidase (Complex IV) compared to control-diet fed mice (Fig. 3). Per differential expression analyses, a number of mRNAs were observed to be altered, including Complex I associated genes: Ndufs, Ndufa, Ndufb, Ndufc; Complex III associated genes: ISP, QCR6, 7, 8, 9; Complex IV associated genes: COX4, 5a, 5b, 6c, 7a, 7b, 7b (Fig. 3).
Figure 3. Gene Expression of Oxidative Phosphorylation Complexes.
Relative gene expression of oxidative phosphorylation complex genes in DRG of mice fed a ketogenic diet compared to a control diet. For each mRNA, individual (n=6) mice fed a ketogenic-diet are represented by individual colored bars placed over each gene name for their expression relative to mice fed a control diet. Green indicates a decrease in relative expression, red represents an increase in relative expression, and gray represents no relative change compared to control-diet fed mice (n=6 for all groups). The color schema −1(green) ~1(red) represents a log2 fold change.
Discussion
Mitochondrial function has become an important target in complications or diseases that affect the central and peripheral nervous system. It is now becoming evident that the growing epidemic of obesity and metabolic syndrome is a likely contributor to neural dysfunction, and mitochondrial function is a key mediator in this process (Figueroa-Romero et al., 2008). Even though in contains a high level of fat, a ketogenic diet has become popular as a disease intervention. In our own studies, we have shown that feeding mice a ketogenic diet positively impacts peripheral nerve function and pain behaviors in mice, independent of traditional metabolic benefits and physiological improvements (Cooper et al., 2018a). As a clinical example, diabetes-associated peripheral neuropathy is thought to develop in part due to mitochondrial deficits (Chung et al., 2014). It is not known how a ketogenic diet will impact diabetic neuropathy in human patients. Here, we show that a ketogenic diet appears to lead to adaptations in sensory neuron and axonal mitochondria to metabolize fat based fuel sources with reduced oxidative stress compared to mice fed a control diet. As we have shown previously, mice on a ketogenic diet once again did not lose weight and instead gained weight relative to controls, this is contrary to many groups findings and may be due to the C57/Bl6 background substrain we utilized as we have recently seen key variations by substrain selection in the physiological effects of a high-fat diet in C57BL/6 mice (Cooper et al., 2018a; Cooper et al., 2018b). Additionally, a ketogenic diet appears to alter gene expression of key components of the electron transport chain in the cell bodies of peripheral neurons. Together these results reveal modifications in peripheral nerve metabolism related to a ketogenic diet that should prove useful in context of using a ketogenic diet as a dietary intervention.
There are numerous mechanisms that contribute to the development of peripheral neuropathy that can arise from poor metabolic health. One contributor to abnormal peripheral nerve function may be and increase oxidative stress associated with mitochondrial dysfunction (Figueroa-Romero et al., 2008). This idea suggests that during mitochondrial metabolism, an increase in ROS leads to increased lipid peroxidation and impaired mitochondrial function in the axon and axonal degeneration (Fernyhough et al., 2010). Here, we have shown that peripheral neurons, axons, and support cellscan metabolize fats and that control-diet fed mice show higher ROS production than ketogenic-diet fed mice. This suggests that peripheral neurons can adapt and metabolize fuels without the increase in ROS when challenged with dietary changes.
Based on the literature, an additional detrimental change in mitochondrial metabolism includes reduced enzymatic activity of Complex’s I and IV (Fernyhough et al., 2010). Consistent with this view, our RNAseq analysis of oxidative phosphorylation pathways in DRG displayed increases in gene expression of NADH Dehydrogenase (Complex I) genes, as well as a potential increase in cytochrome c oxidase (Complex IV) genes in ketogenic-diet fed mice relative to control-diet fed mice. Though RNAseq displayed alterations in key oxidative phosphorylation pathway genes, mitochondrial respiration and ROS production was unaltered in DRG. This disparity between gene and functional measures should be further examined in future work. Future research should re-examine the associated genes in our RNAseq analysis and subsequent protein alterations of Complex I and IV driven by a ketogenic diet in peripheral neurons to better understand the adaptation of peripheral neurons to a ketogenic diet.We interpret these changes to suggest that, contrary to control-diet fed mice, ketogenic-diet fed mice increase genes associated with complex I and IV in order to increase mitochondrial activity which is reduced in hyperglycemic conditions. This is important since it has been reported that mitochondrial function and gene expression is negatively altered in the sensory neurons of animal models of type 1 diabetes (Chowdhury et al., 2010).
Ketones can inhibit ROS production through an increase in NAD/NADH ratios and this could be a potential mechanism occurring in vivo allowing for improved metabolizing of fats (Maalouf et al., 2007; Fisher-Wellman & Neufer, 2012). An additional potential mechanism may be an alteration in ETF levels as ETF can metabolize the FADH2 produced in PCoA metabolism, future studies should examine the potential role of ETF in fat metabolism changes in peripheral nerves. In our study, the decrease in mitochondrial H2O2 flux in sciatic nerves during steady state and maximal respiration suggest that ketogenic-diet fed mice have a decrease in ROS production when metabolizing fats. This metabolic improvement may identify the key adaptation which neurons adapt to metabolizing fats when fed a ketogenic diet. The diet-induced differences in mitochondria were only observed in the sciatic nerve, suggesting that mitochondria in distal components of peripheral nerve react to dietary changes differently than proximal mitochondria in the neuronal soma. This also agrees with the view that distal segments of peripheral nerves have a different susceptibility to metabolic dysfunction (Hinder et al., 2013).
Conclusion
The present work highlights the impact of dietary changes on peripheral nerve mitochondrial. We provide evidence that peripheral neurons and axon associated cells can use a fat-based fuel source and can adapt to this fuel source when fed a ketogenic diet leading to reduced ROS production. A ketogenic diet may be benefitical to neurons by lowering fat oxidation and the associated ROS production which could improveperipheral nerve disease associated with poor mitochondrial function. These approaches may benefit peripheral axons and combat axonal degeneration, motor and sensory dysfunction, and pain that is experienced by millions of patients.
New Findings.
-
What is the central question of this study?
Do peripheral sensory neurons metabolize fat-based fuel sources, and does a ketogenic diet modify these processes?
-
What is the main finding and its importance
We show that peripheral axons from mice fed a ketogenic diet respond to fat-based fuel sources with reduced respiration and H2O2 emission compared to mice fed a control diet. These results add to our understanding of the responses of sensory neurons to neuropathy associated with poor diet, obesity and metabolic syndrome. These findings should be incorporated into current ideas of axonal protection and may identify how dietary interventions may change mitochondrial function is settings of sensory dysfunction.
Acknowledgments
Funding
This work was supported by National Institutes of Health (NIH) grants RO1NS43314 (DEW), P20 GM103418 from the Idea Network of Biomedical Research Excellence (INBRE) Program (DEW, CM, JPT), the Kansas IDDRC, U54 HD 090216 (DEW), Veterans Affairs Merit Review 1I01Bx002567–01 (JPT).
Footnotes
Competing Interests
None of the authors of this work have competing or conflicting interests in the associated work.
References
- Akude E, Zherebitskaya E, Chowdhury SK, Smith DR, Dobrowsky RT & Fernyhough P (2011). Diminished superoxide generation is associated with respiratory chain dysfunction and changes in the mitochondrial proteome of sensory neurons from diabetic rats. Diabetes 60, 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury SK, Zherebitskaya E, Smith DR, Akude E, Chattopadhyay S, Jolivalt CG, Calcutt NA & Fernyhough P (2010). Mitochondrial respiratory chain dysfunction in dorsal root ganglia of streptozotocin-induced diabetic rats and its correction by insulin treatment. Diabetes 59, 1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Prasad K & Lloyd TE (2014). Peripheral neuropathy: clinical and electrophysiological considerations. Neuroimaging Clin N Am 24, 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Menta BW, Perez-Sanchez C, Jack MM, Khan ZW, Ryals JM, Winter M & Wright DE (2018a). A ketogenic diet reduces metabolic syndrome-induced allodynia and promotes peripheral nerve growth in mice. Exp Neurol 306, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, O’Meara B, Jack MM, Elliot D, Lamb B, Khan ZW, Menta BW, Ryals JM, Winter MK & Wright DE (2018b). Intrinsic Activity of C57BL/6 Substrains Associates with High-Fat Diet-Induced Mechanical Sensitivity in Mice. J Pain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Ryals JM, Wu PY, Wright KD, Walter KR & Wright DE (2016). Modulation of diet-induced mechanical allodynia by metabolic parameters and inflammation. J Peripher Nerv Syst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernyhough P, Roy Chowdhury SK & Schmidt RE (2010). Mitochondrial stress and the pathogenesis of diabetic neuropathy. Expert Rev Endocrinol Metab 5, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Romero C, Sadidi M & Feldman EL (2008). Mechanisms of disease: the oxidative stress theory of diabetic neuropathy. Rev Endocr Metab Disord 9, 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher-Wellman KH & Neufer PD (2012). Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab 23, 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A FE, Lima JLFC. (2005). Fluorescence probes used for detection of reactive oxygen species. J. Biochem. Biophys. Methods., 45–80. [DOI] [PubMed] [Google Scholar]
- Groover AL, Ryals JM, Guilford BL, Wilson NM, Christianson JA & Wright DE (2013). Exercise-mediated improvements in painful neuropathy associated with prediabetes in mice. Pain 154, 2658–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford BL & Wright DE (2013). Chewing the fat: genetic approaches to model dyslipidemia-induced diabetic neuropathy in mice. Exp Neurol 248, 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinder LM, Vivekanandan-Giri A, McLean LL, Pennathur S & Feldman EL (2013). Decreased glycolytic and tricarboxylic acid cycle intermediates coincide with peripheral nervous system oxidative stress in a murine model of type 2 diabetes. J Endocrinol 216, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Olson P & Evans RM (2003). Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 144, 2201–2207. [DOI] [PubMed] [Google Scholar]
- Li B & Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maalouf M, Sullivan PG, Davis L, Kim DY & Rho JM (2007). Ketones inhibit mitochondrial production of reactive oxygen species production following glutamate excitotoxicity by increasing NADH oxidation. Neuroscience 145, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrecka-Kuka M, Krumschnabel G & Gnaiger E (2015). High-Resolution Respirometry for Simultaneous Measurement of Oxygen and Hydrogen Peroxide Fluxes in Permeabilized Cells, Tissue Homogenate and Isolated Mitochondria. Biomolecules 5, 1319–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SA, Davis BM & Molliver DC (2007). Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2, 152–160. [DOI] [PubMed] [Google Scholar]
- Osborn O & Olefsky JM (2012). The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med 18, 363–374. [DOI] [PubMed] [Google Scholar]
- Price N, van der Leij F, Jackson V, Corstorphine C, Thomson R, Sorensen A & Zammit V (2002). A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics 80, 433–442. [DOI] [PubMed] [Google Scholar]
- Reddy JK & Rao MS (2006). Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol 290, G852–858. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ & Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonazzi A, Mantovani C, Colella M, Terenghi G & Indiveri C (2013). Localization of mitochondrial carnitine/acylcarnitine translocase in sensory neurons from rat dorsal root ganglia. Neurochem Res 38, 2535–2541. [DOI] [PubMed] [Google Scholar]
- Veech RL (2004). The therapeutic implications of ketone bodies: the effects of ketone bodies in pathological conditions: ketosis, ketogenic diet, redox states, insulin resistance, and mitochondrial metabolism. Prostaglandins Leukot Essent Fatty Acids 70, 309–319. [DOI] [PubMed] [Google Scholar]
- Wolfgang MJ, Kurama T, Dai Y, Suwa A, Asaumi M, Matsumoto S, Cha SH, Shimokawa T & Lane MD (2006). The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc Natl Acad Sci U S A 103, 7282–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]