Abstract
Tumor mutational burden correlates with response to immune checkpoint blockade in multiple solid tumors, though in microsatellite stable tumors this association is weak and of limited clinical utility. Here, we uniformly analyzed whole exome sequencing (WES) of 249 tumors and matched normal tissue from patients with clinically annotated outcomes to immune checkpoint therapy, including radiographic response, across multiple cancer types to examine additional tumor genomic features that contribute to selective response. Our analyses identified genomic correlates of response beyond mutational burden, including somatic events in individual driver genes, certain global mutational signatures, and specific HLA-restricted neoantigens. However, these features were often inter-related, highlighting the complexity of identifying genetic driver events that generate an immunoresponsive tumor environment. This study lays a path forward in analyzing large clinical cohorts in an integrated and multifaceted manner to enhance our ability to discover clinically meaningful predictive features of response to immune checkpoint blockade.
Introduction
Immune checkpoint inhibitors, including monoclonal antibodies targeting the immune inhibitory proteins programmed cell death protein 1 (PD-1) and cytotoxic T lymphocyte associated antigen 4 (CTLA-4), significantly extend patient survival across many cancer types1. However, pre-treatment predictive biomarkers of response remain unclear. Immunohistochemical (IHC) assessment of tumor immune activity, including staining for the ligand to PD-1 (PD-L1) and the presence of CD8+ T cells, may help predict response to anti-PD-1 therapies1,2,3, but these histological snapshots may not fully represent a dynamic tumor-immune microenvironment4. Further, they are limited by variable sensitivity and specificity by tumor type5 and minimal predictive value for anti-CTLA-4 therapies.
Molecular analysis of patient tumors has revealed that a high burden of tumor-specific mutant peptides (neoantigens) generated from somatic nonsynonymous coding mutations may increase tumor immunogenicity and the likelihood of patient benefit from immune checkpoint therapy in both microsatellite stable and unstable tumor types6,7,8,9,10. However, not all studies of immunotherapy-treated cohorts have found a strong relationship between mutational burden and response11,12. Even in studies that identified a statistically significant association, the distributions of mutational burden between responders and non-responders overlapped substantially10, and the opportunities for therapeutic targeting of this feature to enhance response remain uncertain. Even the most optimal cutoff yields weak sensitivity (74%) and specificity (59.3%) to discriminate potential clinical benefit13, limiting the utility of tumor mutational burden as a clinical biomarker for individual patients.
Given the major limitations of mutational burden as a predictive biomarker, additional genomic studies have suggested that clonal mutations and neoantigens14, mutations and copy number alterations affecting particular genes and signaling pathways15,16,17,18,19, and overall tumor aneuploidy12,20 may have additional predictive value for response or resistance to immune checkpoint therapies. However, existing studies have focused on cancer types individually without identifying pan-cancer relationships; have been of limited sample size; and have had inconsistent computational methods. These limitations have restricted the power of these studies to identify meaningful associations and have likely contributed to difficulties in validating prior findings in independent prospective patient cohorts21.
We hypothesized that an expanded and uniformly analyzed cohort of clinically-annotated patient samples would provide greater power to detect significant associations between pre-treatment tumor characteristics and response to immune checkpoint therapies. Thus, we gathered raw tumor and germline pre-treatment whole exome sequencing data from tumors from immune checkpoint-treated patients from seven published studies (N=171)7,8,10,22,23,24,25 and combined it with data from 78 newly-sequenced pre-treatment tumors. By harmonizing clinical annotations and whole exome analyses across 249 samples and multiple cancer types, we aimed to 1) assess the generalizability of prior hypotheses regarding immune checkpoint blockade response to other histological or drug settings; 2) apply new computational techniques for inference of tumor biology and immunogenicity; and 3) determine whether our cohort was of sufficient size to reveal statistically robust associations about specific genetic mediators of selective response.
Results
Consolidation of a clinically annotated cohort of whole exome sequencing of tumors from patients treated with immune checkpoint blockade
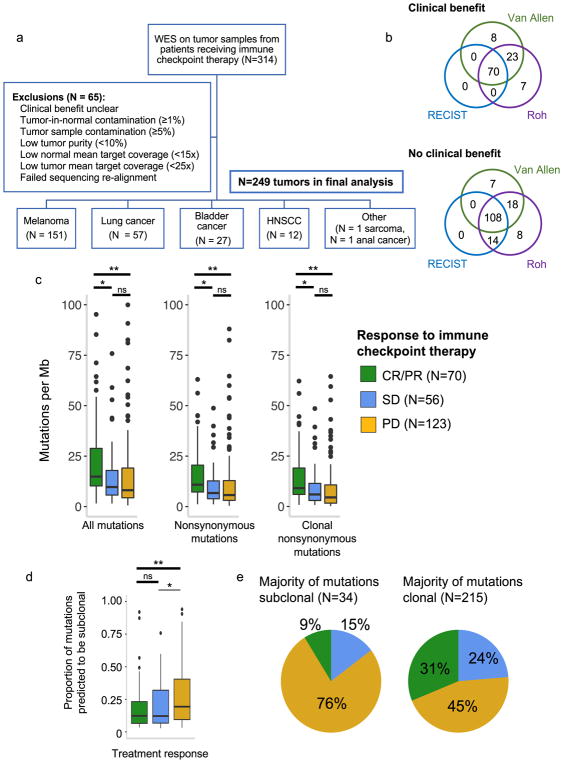
Whole exome sequencing (WES) from clinically annotated tumor samples with matched germline blood or adjacent normal tissue was available for 314 patients (Supplementary Table 1, Fig. 1a). Standard quality control measures were taken to ensure adequate power to detect tumor-specific mutations26 (Fig. 1a, Supplementary Fig. 1a–c). Our final analysis cohort included 249 patient tumors across six cancer types: melanoma (N=151), non-small cell lung cancer (NSCLC) (N=57), bladder cancer (N=27), head and neck squamous cell carcinoma (HNSCC) (N=12), anal cancer (N=1), and sarcoma (N=1) (Fig. 1a). These patients were treated with anti-PD-1 (N=74), anti-PD-L1 (N=20), anti-CTLA-4 (N=145), or a combination of anti-CTLA-4 and anti-PD-1/L1 therapies (N=10). A small minority of patients received anti-PD-1, anti-PD-L1, or anti-CTLA-4 therapy in combination with another immunotherapy, targeted therapy, or cytotoxic chemotherapy (N=7) (Supplementary Table 2). Across these 249 samples, average mean target sequencing coverage was 150-fold for tumor tissue and 119-fold for matched germline tissue. Mean estimated tumor purity was 58% (range 10%–97%) (Supplementary Table 1).
Fig. 1. Clinical cohort consolidation, response stratification, and mutational load investigation.
(a) Data quality control for 249 samples included in final analysis. (b) Comparison of three published response metrics for determining OR vs. NR. (c) Comparison of tumor mutational burden between CR/PR vs. PD (For ‘All mutations’, p=0.0005 for CR/PR vs PD, p=0.0054 for CR/PR vs SD, and p=0.434 for SD vs PD; for ‘Nonsynonymous mutations’, p=0.0003 for CR/PR vs PD, p=0.0063 for CR/PR vs SD, and p=0.3769 for SD vs PD; for ‘Clonal nonsynonymous mutations’, p=0.00005 for CR/PR vs PD, p=0.011 for CR/PR vs SD, and p=0.151 for SD vs PD). Outlying points from patients with mutations/Mb > 101 are not shown (2 CR/PR, 1 SD, 3 PD). (d) Intratumoral heterogeneity across response groups (n=249 biologically independent samples, p=0.001 for CR/PR vs. PD, p=0.5122 for CR/PR vs SD). (e) Clinical response to immune checkpoint therapy broken down by intratumoral heterogeneity. For (c, d), p-values calculated by two-sided Mann-Whitney U *p<0.05, **p<0.005, ns = not significant. Boxplots show the median, first and third quartiles, whiskers extend to 1.5 × the interquartile range, and outlying points are plotted individually.
In selecting a framework to assess clinical response in this study, we encountered multiple patient stratification methods from prior studies of response predictors to immune checkpoint therapy10,11,12,27. These varied mostly in their treatment of patients with stable disease (SD) by RECIST 1.127, who have minimal to no change in tumor burden following therapy. Applying three existing response definitions to our cohort, we observed substantial differences in patient classification into responder and nonresponder groups (Fig. 1b, Supplementary Table 3). Given the evolving viewpoints on classifying response to immune checkpoint blockade28, we adopted a conservative method of defining objective response (OR) as complete response (CR) or partial response (PR) by RECIST (major decrease in tumor burden following treatment) and no response (NR) as progressive disease (PD) by RECIST (major increase in tumor burden following treatment) for the main analyses. Patients with SD were considered separately, and analyses using two other response definitions10,12 that stratify SD patients into OR vs. NR by duration of overall survival (OS) or progression-free survival (PFS) are available in the Supplementary Materials.
Mutational burden and response to immune checkpoint therapy
In examining whole exome genetic features in this cohort, we began with tumor mutational burden, as this has been the most widely reproduced association with response to immune checkpoint therapy. We found that in this combined cohort, patients with CR/PR had significantly higher tumor mutational burdens compared to patients with PD (p<0.05 for all; Mann-Whitney U) (Fig. 1c). This finding persisted within cancer types (Supplementary Fig. 2), and was particularly prominent in patients treated with anti-PD-1/PD-L1 inhibitors (Supplementary Fig. 3a). Patients with SD tended to have mutational burdens intermediate between those with PD or CR/PR, with higher mutational loads in patients with SD with long OS compared to short OS (Supplementary Fig. 3b).
While these findings are consistent with the growing body of literature supporting the association between mutational burden and immune checkpoint therapy response, we noted that the ranges of mutational burdens between response groups overlapped considerably (Fig. 1c), and we found that tumor mutational burden had poor predictive power to differentiate CR/PR vs. PD as a single variable in this cohort (AUC = 0.66) (Supplementary Fig. 4). To build upon the utility of tumor mutational burden as a predictive variable for response to immune checkpoint therapy, past studies have determined that clonal (found in every cancer cell) mutations rather than subclonal (found in a subset of cancer cells) mutations are more strongly associated with response in lung adenocarcinomas and some melanomas, potentially due to stronger T cell responses to neoantigens generated from clonal vs. subclonal mutations14.
We queried our cohort for this association by using ABSOLUTE to infer mutational clonality26, and demonstrated that clonal nonsynonymous mutational burden strongly predicted CR/PR vs. PD across cancer types and response categorizations (Fig. 1c–d, Supplementary Fig. 2). Patients with a large proportion of subclonal mutations (>50%) – which we term high intratumoral heterogeneity – were substantially more likely to have PD than CR/PR across all tumors described herein (Fig. 1e) (p=0.0014; Fisher’s exact). Thus, while mutational burden begins to explain the variance in patient response to immune checkpoint therapy, intratumoral heterogeneity contributes additional biological insight.
Mutations in specific genes associated with response or resistance to immune checkpoint therapy
Given the complexity we observed in the association between mutational load and response to immune checkpoint therapy, we pursued additional analyses of exome-level features that could provide further nuance to this association. We next investigated whether somatic mutations in specific genes were associated with response to immune checkpoint therapy, hypothesizing that this analysis would identify genes driving biological processes generating large numbers of mutations or those creating an immunoresponsive phenotype independent of mutational burden.
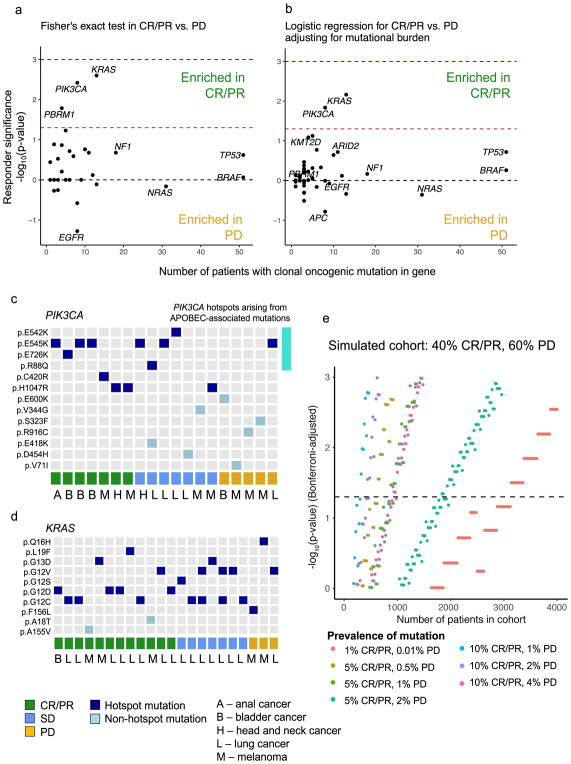
Given that response rates were similar across all cancer types (Supplementary Table 3), we first compared nonsynonymous mutations in CR/PR vs. PD across all genes; however, these associations were too weak to pass multiple hypothesis test correction (Supplementary Fig. 5). Thus, we limited our analysis to known hotspot mutations in cancer driver genes and to loss-of-function alterations in known tumor suppressors26 (Supplementary Table 4), as these events are more likely to have a significant impact on tumor biology. Clonal driver alterations in PIK3CA, KRAS, and PBRM1 were enriched in CR/PR, while clonal driver mutations in EGFR were enriched in PD (p<0.05 for all, Benjamini-Hochberg false discovery rate (FDR) q=0.18 for KRAS and PIK3CA) (Fig. 2a). After correcting for tumor mutational burden, KRAS and PIK3CA remained associated with CR/PR (p<0.05; logistic regression) (Fig. 2b), although these observations did not pass FDR correction and may be confounded by additional aspects of a tumor’s genetic profile.
Fig. 2. Mutations in specific genes associated with response to immune checkpoint therapy.
(a) Response-associated mutations in CR/PR vs. PD (Two-failed Fisher’s exact test, n = 70 biologically independent samples with CR/PR, n = 123 with PD). Dashed red line indicates Fisher’s exact p=0.05, and dashed dark red line indicates false discovery rate (FDR) q=0.05. (b) Response-associated mutations corrected for mutational burden. Sample size and dashed lines for p- and q-value cutoffs same as in (a). (c–d) Tile plot showing known hotspot and non-hotspot mutations in (c) PIK3CA and (d) KRAS by response group (bottom), with cancer type indicated by capital letters. Top four rows of (c) represent mutations arising in APOBEC-associated mutational contexts (See Supplementary Fig. 6). (e) Simulated statistical power calculation for detection of response-associated genes. Significance of association between response and presence of mutation in gene (Two-tailed Fisher’s exact test) is shown on the y-axis, for varying samples sizes (x-axis). Colors represent frequency of mutations and specificity of alteration to OR vs. NR. Dashed horizontal line represents Bonferroni-correct p=0.05 after correcting for multiple comparisons over the 116 cancer driver genes assessed in this study. Simulated cohort contains 40% CR/PR and 60% PD patients.
Driver mutations within a given gene may occur in different functional domains and have different phenotypic effects, often dependent on the cancer context. Thus, we next examined these cohort-wide gene associations for trends within cancer types. Of the patients with clonal hotspot mutations in PIK3CA, those with CR/PR had melanoma, HNSCC, anal cancer, or bladder cancer, while the majority of those with SD or PD had lung cancer (Fig. 2c, Supplementary Table 5, Supplementary Fig. 6). Hotspot mutations in KRAS occurred predominantly in CR/PR across multiple cancer types (Fig. 2d). Genetic events in PIK3CA and KRAS were too infrequent to fully clarify their relationships to response in this study, but these results demonstrate that single-gene associations with response to immune checkpoint therapy can provide additional information beyond mutational burden, in a manner that may be dependent on or independent of cancer type.
Despite combining data from multiple studies and cancer types, our analyses were still statistically underpowered to detect important relationships; thus, we sought to estimate the sample sizes needed for discovery of single-gene correlates of response with appropriate correction for multiple hypothesis testing26. We modeled statistical significance values for common or rare variants associated with CR/PR vs. PD at various sample sizes (Fig. 2e). In the best-case scenario, where a variant is both relatively common (~10% prevalence) and specific to responders, sample sizes of around 300 would be adequate to detect significant associations. Meanwhile, detection of rare response-associated variants (~1% frequency), even if highly specific, would necessitate sample sizes in the thousands. Thus, continuing to combine independent clinically-annotated cohorts will aid in increasing statistical power to detect common response-associated variants, but for detection of rarer events, applying insights from experimental studies for hypothesis-driven validation will be a crucial adjunct.
Integrated analysis of response- and resistance-associated mutations with mutational signatures
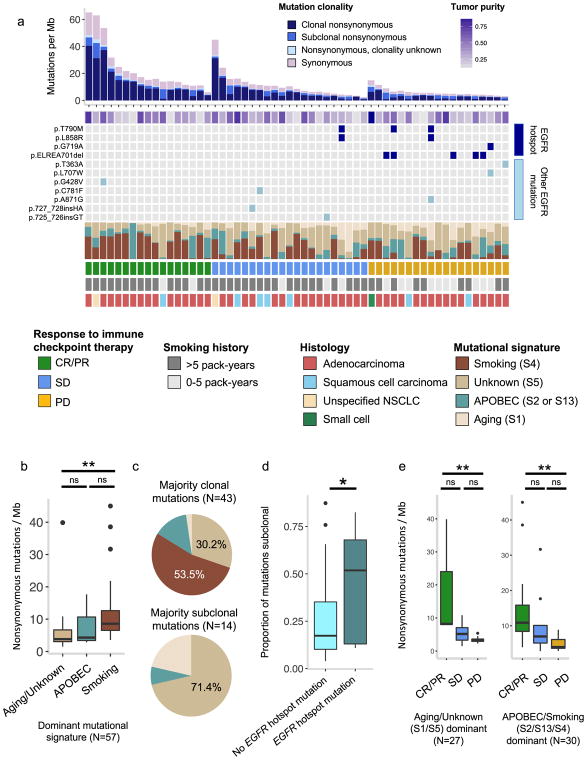
As a further step towards understanding the mutational processes that generate an immunoresponsive tumor environment, we next investigated whole exome signatures of mutagenic biological processes. We used a previously described non-negative matrix factorization technique to identify known mutagenic processes in lung cancer, melanoma, bladder cancer, and HNSCC26,29,30. Using this technique, the somatic mutations within a tumor are probabilistically assigned to underlying mutational signatures, which are patterns of somatic mutations thought to arise from carcinogenic processes (e.g. a predominance of C-to-A transversions in tobacco-smoking-associated cancers or C-to-T transitions in ultraviolet-light-associated tumors) (Supplementary Table 6, Supplementary Table 7). As has been seen previously in analyses of non-clinically-annotated NSCLC exomes31, pre-treatment NSCLC tumors in this cohort with a high proportion of smoking-associated mutations tended to have low intratumoral heterogeneity, high mutational burden, and frequent KRAS mutations (Fig. 3a–c). The proportion of smoking-associated mutations was also higher in CR/PR patients compared with PD (Supplementary Fig. 7a). Meanwhile, tumors with EGFR hotspot mutations trended towards enrichment in subclonal mutations (p=0.035), had low mutational burdens, were over-represented in never-smokers (Two-tailed Fisher’s exact test,p = 0.00017), and had generally poor responses to immune checkpoint therapy (Fig. 3a, d), which is also consistent with previously published results32,33. Thus, the association between KRAS and EGFR mutation status and response to immunotherapy (Fig. 2a–b) may be related not only to mutational burden but also to mutational signature and mutational clonality. After controlling for smoking history, mutational burden remained a significant predictor of response (Fig. 3e, Supplementary Fig. 7b). Concurrent consideration of mutational signatures, clonal architecture, and hotspot mutations in NSCLC enhances understanding of somatic mediators of immunotherapy response and resistance.
Fig. 3. Integrated analysis of EGFR mutational status, intratumoral heterogeneity, and mutational signatures in lung cancer.
(a) Stacked plot showing mutational burden (histogram, top), estimated tumor purity (tile plot, top), mutations in EGFR (tile plot, middle), mutational signatures (filled histogram, middle), and clinical response and clinical covariates (bottom). (b) Interaction between smoking-related mutational signatures and mutational burden. For aging/unknown vs smoking signatures p=0.0001; for aging/unknown vs APOBEC, p=0.427; for APOBEC vs smoking, p=0.158. (c) Proportion of patients with a given dominant mutational signature by clonal mutation composition. (d) Proportion of subclonal mutations in EGFR-mutant vs. EGFR-wildtype tumors (n = 57 biologically independent samples, p=0.035, unpaired two sample t-test). (e) Relationship between mutational burden and response by dominant mutational signature (for Aging/unknown dominant samples, CR/PR vs PD p=0.0044, CR/PR vs SD p=0.07, SD vs PD p=0.0597; for APOBEC/smoking dominant samples, CR/PR vs PD p=0.0023, CR/PR vs SD p=0.08, SD vs PD p=0.252). Two-sided Mann-Whitney U for (b, e), *p<0.05, **p<0.005, ns = not significant. Boxplots (b, d, e) show the median, first and third quartiles, whiskers extend to 1.5 × the interquartile range, and outlying points are plotted individually.
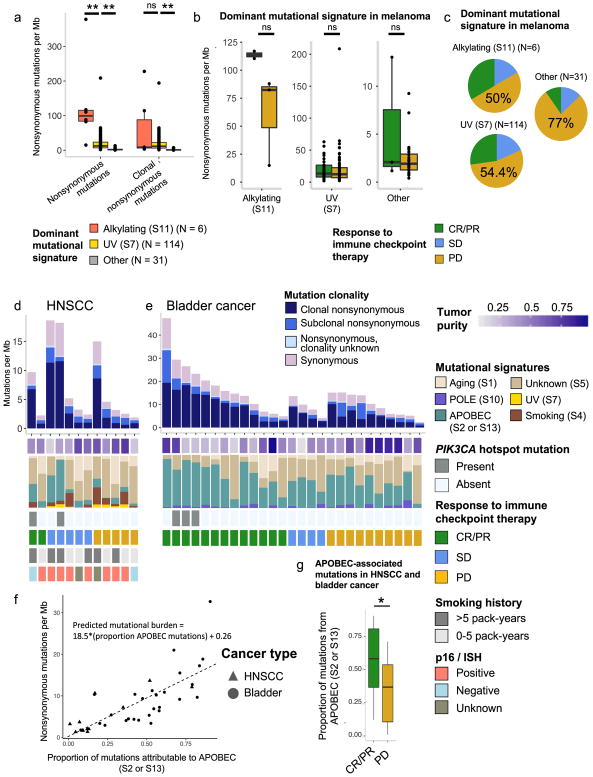
We also examined mutational signatures in melanoma tumors, which are dominated by exposure to ultraviolet (UV) light (S7), prior chemotherapeutic treatment with alkylating agents (S11), and other signatures not clearly associated with specific environmental exposures (S1, S5). Dominant mutational signature explained a large proportion of the variance in mutational burden and was highly correlated with intratumoral heterogeneity (Fig. 4a, Supplementary Fig. 8a). After stratifying by dominant mutational signature, no significant difference in mutational burden was observed between CR/PR vs. PD (p>0.05 for all) (Fig. 4b). The non-UV/non-alkylating group had a higher proportion of patients with PD compared to the other two groups (Fig. 4c), and was composed largely of mucosal, uveal, and acral lentiginous melanomas, though dominant mutational signature and histology did not overlap perfectly (Supplementary Fig. 8b). The observation that mutational load is not a significant predictor of response after correcting for dominant mutational signature in melanoma raises the possibility that in this cancer type, mutational burden itself may not directly mediate response but rather may serve as a proxy for an underlying biological process that both increases tumor immunogenicity and promotes accumulation of somatic mutations.
Fig. 4. Integrated analysis of mutational burden, intratumoral heterogeneity, and mutational signatures in melanoma, HNSCC, and bladder cancer.
(a) Nonsynonymous mutational burden (alkylating- vs. UV-dominant, p=0.0005; UV- vs. other-dominant, p=6.079e-14) and clonal mutational burden (alkylating- vs UV-dominant, p=0.938, UV- vs. other-dominant, p=6.404e-15) stratified by dominant mutational signature in melanoma patients only. b) Association between mutational burden and response within dominant mutational signature in melanoma (CR/PR vs PD in alkylating-dominant, p=0.2; CR/PR vs PD in UV-dominant, p=0.549; CR/PR vs PD in ‘Other’ dominant, p=0.689). (c) Likelihood of response by dominant mutational signature group in melanoma. (d–e) Stacked plots of mutational burden (histogram, top), tumor purity (tile plot, top), mutational signatures (filled histogram, middle), and histology and clinical response to immune checkpoint therapy (tile plots, bottom) for HNSCC (d) and bladder cancer (e). (f) Proportion of mutations attributable to APOBEC mutational signatures (S2/13) vs. nonsynonymous mutational burden (n = 39 biologically independent samples, p=2.66e-08 for slope; p=0.848 for intercept). Symbols indicate cancer type. (g) Proportion of mutations probabilistically attributable to the APOBEC mutational signature in CR/PR vs. PD in HNSCC and bladder cancer (p=0.019, n = 39). Two-sided Mann-Whitney U for all, *p<0.05, **p<0.005, ns = not significant. Boxplots (a, b, g) show the median, first and third quartiles, whiskers extend to 1.5 × the interquartile range, and outlying points are plotted individually.
In bladder cancer and HNSCC, similar analyses demonstrated association of APOBEC-associated signatures (S2 and S13) with higher mutational burdens (p=0.002 for bladder, p=0.03 for HNSCC, two-sided Mann Whitney U) and greater likelihood of CR/PR (p=0.019) (Fig. 4d–g). APOBEC signatures have been linked with tobacco exposure34, but viral infection and PIK3CA hotspot mutations are other potential etiologies35. These correlations may contribute to and/or confound the observed association between PIK3CA hotspot mutations and CR/PR described above (Fig. 2c, Supplementary Fig. 6). APOBEC mRNA expression has also been previously associated with increased PD-L1 IHC staining36 and high tumor mutational burden37 in urothelial carcinoma. Overall, genetic features – including mutational burden, intratumoral heterogeneity, tumor driver mutations, and mutational signatures – appear to have inter-related associations with response in this cohort. Determining which of these features ultimately drives response to immune checkpoint therapy in these patients will require further clinical and experimental study.
Copy number alterations associated with response or resistance to immune checkpoint therapy
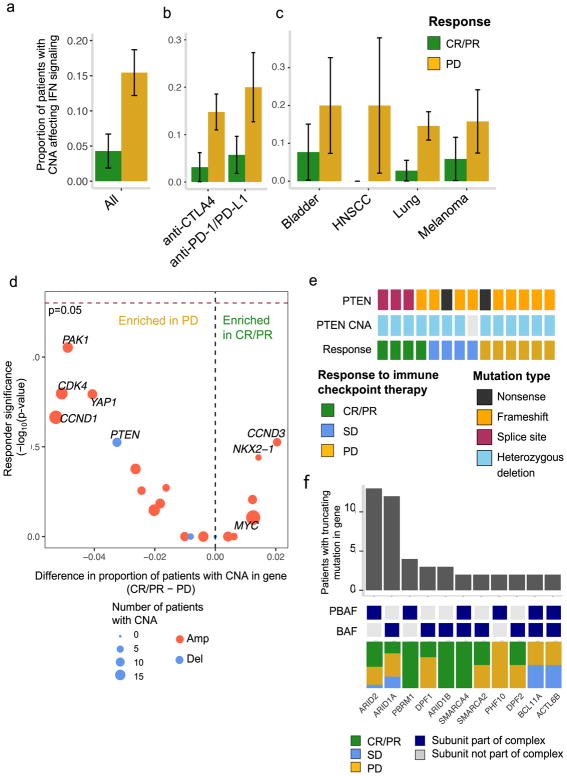
In addition to somatic mutations, copy number alterations (CNAs) may also contribute to selective response, and CNAs affecting the interferon-γ pathway have been implicated in intrinsic resistance to immune checkpoint therapies in melanoma17. In this cohort, we assessed CNAs expected to interfere with interferon-γ signaling after correcting for tumor purity26, which can strongly influence the number of called CNAs in cohorts with heterogeneous tumor purity and ploidy (Supplementary Fig. 9a–c). After correcting for tumor purity, interferon-γ-related CNA events were more infrequent than previously described, but consistent with prior studies, these events were enriched in PD vs. CR/PR (19/123 vs. 3/70; p=0.019, Fisher’s exact) (Fig. 5a). This relationship persisted within cancer types and therapy classes, though this study was insufficiently powered to detect a significant association in many of the subgroup analyses (Fig. 5b–c, Supplementary Fig. 10a–b, Supplementary Fig. 11a–b).
Fig. 5. Tumor copy number alterations associated with response to immune checkpoint therapy.
(a) Amplifications and deletions of genes in the interferon-γ signaling pathway in CR/PR vs. PD across all samples (Two-tailed Fisher’s exact p=0.019, 95% CI 0.05 to 0.885, n=193), by (b) drug class (n=120 anti-CTLA4-treated, n = 65 anti-PD1/PD-L1-treated) and (c) cancer type (n = 23 bladder, 7 HSNCC, and 125 melanoma). Error bars denote proportion of CR/PR or PD with CNA +/− standard error. *p<0.05. (d) Difference in proportion of CR/PR vs. PD harboring focal cancer driver CNAs. Negative log10(p-value) for a two-tailed Fisher’s exact test for enrichment of a gene-level CNA in CR/PR vs. PD is shown on the y-axis. Genes more commonly affected by CNAs in CR/PR are shown on the right, while those more commonly deleted or amplified in PD are shown on the left. Dashed red line indicates p=0.05. (n=193 biologically independent samples) (e) Truncating mutations in PTEN by response group. (f) Prevalence and response association of truncating alterations in genes encoding SWI/SNF subunits62. Dark blue tiles indicate membership in either PBAF or BAF, which are complexes within the SWI/SNF family. Only genes encoding SWI/SNF subunits harboring truncating mutations in at least two patients are shown.
Next, we analyzed focal CNAs affecting 63 commonly amplified and deleted tumor suppressors and oncogenes26 (Supplementary Tables 8–9) to nominate additional mediators of selective response or resistance. While these events were rare and no gene achieved statistical significance alone, amplifications of PAK1, YAP1, and CCND1 on chromosome 11q and amplifications of MDM2 and CDK4 on chromosome 12q were seen predominantly in PD patients (Fig. 5d), with the latter of note as CDK4/6 inhibition was recently associated with increased tumor immunogenicity38. Additionally, homozygous PTEN deletion occurred exclusively in patients with intrinsic resistance to immune checkpoint therapy (N=4 PD; p=0.30) (Fig. 5d), re-capitulating the observation of biallelic PTEN loss in resistance to immunotherapies in prior clinical and experimental studies 23,39. Clonal biallelic loss of PTEN via truncating mutation was not as clearly associated with PD, although many of these were splice site mutations of uncertain biological significance (Fig. 5e). Truncating PTEN mutations were also seen in resistant tumors from patients with tumor shrinkage following anti-CTLA-4 therapy at other sites, although these patients were excluded from the main analysis due to their mixed clinical response (Supplementary Fig. 12a–b, Supplementary Table 1). Thus, both mutations and CNAs can influence response to immune checkpoint therapy by activating or suppressing pathways that interact with tumor-immune signaling, though our analyses indicate that these features are elaborately interrelated, and require significantly increased cohort size and mechanistic validation for robust interpretation.
Validation of previously described response predictors for immune checkpoint therapies
Prior studies have identified additional pathways and genes associated with response or resistance to immune checkpoint therapy, which were not identified via the unbiased analyses presented thus far, potentially due to insufficient power after correcting for multiple hypothesis testing. We attempted a focused validation of these previous findings in this cohort, beginning with an analysis of loss-of-function alterations in PBRM1, a member of the PBAF form of the SWI/SNF complex. Prior work in clear-cell renal cell carcinoma (ccRCC), a cancer type not represented in this study, demonstrated that biallelic PBRM1 loss correlates with response to anti-PD-1/L1 therapy, while mutational load does not19. Additional functional data in melanoma have supported this association, and implicated other related genes in the SWI/SNF family of chromatin remodelers40. In this cohort, truncating mutations in PBRM1 and other related epigenetic regulators in the SWI/SNF complex were also over-represented in responders, with biallelic loss events occurring almost exclusively in the tumors of CR/PR patients (Fig. 2a–b, Fig. 5f, Supplementary Fig. 13). Other mutations that have previously been described in association with acquired resistance to immune checkpoint therapies include JAK/STAT pathway mutations and alterations in antigen-presentation machinery (e.g. beta-2-microglobulin, tapasin)41,42. Loss-of-function mutations and deletions in these pathways were rare in this cohort (Supplementary Fig. 14a–c), perhaps reflecting differences in genetic mechanisms of intrinsic vs. acquired resistance, and further work is required to determine the functional significance of these variants.
Neoantigens associated with response to immune checkpoint therapy
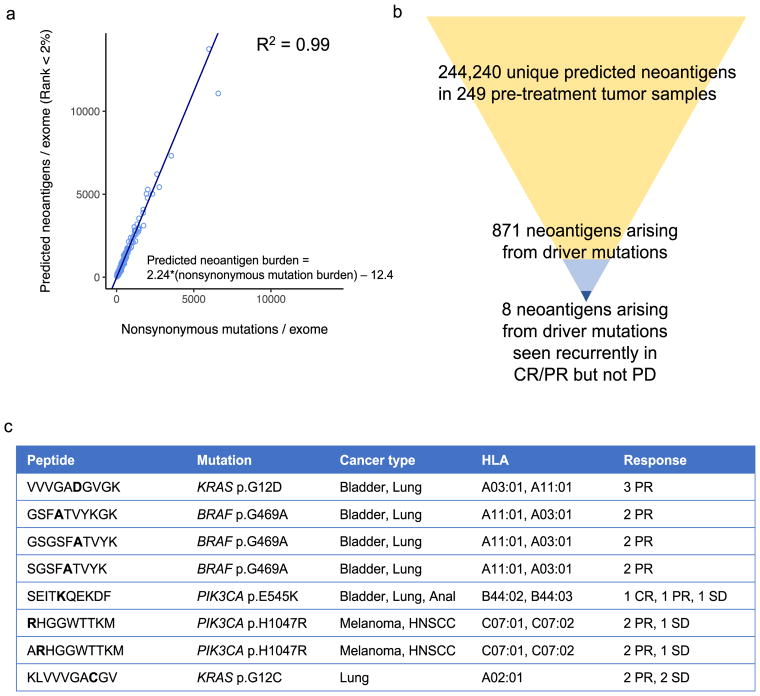
Given the complexities of computationally assessing the impact of oncogenic pathway alterations and tumor mutational burden on response to immune checkpoint therapy, we lastly examined mutations that could more directly influence tumor-immune interactions by generating tumor-specific neoantigens that induce a T cell-mediated anti-tumor response43. We inferred neoantigens in silico26 (Supplementary Table 10) and found that, on average, each nonsynonymous mutation generated 2.24 predicted neoantigens, with extremely high correlation (R2=0.99; p<0.00001) (Fig. 6a), making it difficult to disentangle the effects of mutational vs. neoantigen burden on response using in silico methods alone.
Fig. 6. Response-associated in silico predicted neoantigens.
(a) Relationship between predicted neoantigen burden (y-axis) and nonsynonymous mutational burden (x-axis). Linear regression excludes one outlier (Pat110) (n = 249 biologically independent samples). (b) Prioritization of clinically actionable predicted neoantigens by presence in cancer driver genes and exclusive presence in CR/PR. (c) Response-associated predicted neoantigens generated by cancer driver mutations.
Recent studies have demonstrated that personalized cancer vaccines targeting neoantigens specific to an individual’s tumor can lead to durable clinical benefit alone or in concert with immune checkpoint blockade therapies44,45. More than 99% of predicted neoantigens in this study arose from passenger mutations, which occur throughout the exome, are frequently found in subclonal tumor populations, and are largely unique to each patient’s tumor. However, 871 predicted neoantigens were generated by driver mutations, and eight of these “driver” neoantigens occurred recurrently in CR/PR but not in PD patients, in an HLA-dependent manner (Fig. 6b–c). Additionally, as expected given the known oncogenic effects of the mutations yielding these neoantigens, these eight neoantigens were clonal in all samples, suggesting that a T cell-mediated response, if present, would target all cancer cells14. Thus, driver alterations can generate tumor neoantigens and may contribute to provoking an effective immune response to checkpoint blockade therapy in HLA-matched patients, though further experimental study is required to clarify the biological significance of these putative neoantigens.
Discussion
Whole exome sequencing and analysis of 249 tumors from patients treated with anti-PD-1/L1 or anti-CTLA-4 therapies suggest that genomic features beyond mutational burden, including genetic driver events, tumor heterogeneity, and mutational signatures, may affect response to immune checkpoint blockade. In this work, we combined data from multiple institutions using a standardized computational pipeline and applied a uniform and well-accepted definition of radiographic response to cancer therapy to more robustly assess genetic predictors of response to immune checkpoint therapy.
In so doing, we validated past findings and expanded their generalizability to new cancer contexts, discovered new correlative biomarkers of response using a larger sample size with more statistical power, and investigated the relationships between predictive biomarkers in enhanced detail. For instance, biallelic PTEN loss was first clinically described in acquired resistance to anti-PD-1 therapy in metastatic uterine leiomyosarcoma, and these clinical results and prior pre-clinical findings suggest it may be relevant to intrinsic resistance and mixed response in metastatic melanoma as well23,39. Copy number alterations leading to loss of intact interferon-γ signaling were previously noted in metastatic melanoma treated with anti-CTLA-4 agents41, but this mechanism may also have relevance to anti-PD-1/L1-treated patients and in additional cancer types. PBRM1 loss was first implicated in increasing tumor intrinsic responsiveness to immune checkpoint therapy in ccRCC19, but shared biology of loss of PBRM1 or ARID2 – which both encode proteins within the PBAF form of the SWI/SNF complex – in melanoma, lung cancer, bladder cancer, and HNSCC may underlie the similar response association observed here. Notably, experimental descriptions of the mechanistic underpinnings of these genetic variants in influencing response or resistance to T cell-mediated killing have been instrumental in supporting computational results17,39,40, emphasizing the importance of cross-validation between clinically-relevant tumor variants and mechanistically-driven investigations.
Such findings show that comprehensive consideration of multiple genomic features may help place existing associations such as mutational burden in a broader biological context. In the melanoma tumors from this study, mutational burden was no longer a significant predictor of response after correcting for dominant mutational signature, a finding that warrants further experimental and translational inquiry. KRAS and EGFR mutations in lung cancer have previously been described as response- or resistance-associated, respectively, and this study demonstrates a relationship between these driver mutations and carcinogenic exposures, intratumoral heterogeneity, and mutational burden7,33. The global approach outlined herein will be essential in future investigations of predictors of immune checkpoint therapy response.
Power calculations suggest that combining hundreds or even thousands of clinically annotated patient samples will be necessary to reliably detect specific predictors of response to immune checkpoint therapies. While we preliminarily assessed response predictors specific to anti-CTLA-4 vs. anti-PD-1/L1 therapies, or within a cancer type, these findings are biased by available data from clinically-annotated cohorts; anti-CTLA-4 therapies were used predominantly in melanoma and anti-PD-1/L1 therapies dominated most other cancer types. Further studies more directly comparing therapy classes within the same tumor histology and vice versa will be necessary, as will consideration of response predictors for combinations of checkpoint inhibitors with or without targeted or cytotoxic chemotherapies. Exceptions to single-feature genomic associations between this study and prior works likely can be explained by complex context-dependent effects, and emphasize the need for caution to avoid over-interpretation of results.
While sample size and cohort heterogeneity remain major limitations of this work, this study describes a path forward for gathering insights from multiple clinically annotated patient cohorts. Our work advances hypotheses of biological mechanisms, suggests clinically relevant biomarkers, and highlights the importance of further, larger studies to reliably and robustly identify biomarkers of response and intrinsic resistance to immune checkpoint blockade.
Online Methods
Clinical cohort consolidation
Patients from the Dana-Farber Cancer Institute with metastatic bladder cancer, HNSCC, lung cancer, and melanoma treated with anti-PD-1, anti-PD-L1, anti-CTLA-4, or a combination of these therapies were identified, and pre-treatment tumor tissue and matched germline blood were obtained for genetic sequencing. These studies were conducted in accordance with the Declaration of Helsinki and approved by the Dana-Farber Cancer Institute Institutional Review Board (Protocols 11-104, 05-042, 02-180, 09-472, 02-021, 15-330). Electronic medical charts were reviewed to assess best response by RECIST (v1.1), duration of PFS, duration of OS, patient demographic characteristics, and other relevant clinical details (e.g. smoking history). “Current/former” smokers were those who reported >5 pack-year (packs per day * years smoking) of tobacco use. “Never” smokers were those who reported ≤5 pack-years46. Clinical information from studies conducted outside the Dana-Farber Cancer Institute were downloaded from online supplemental materials7,8. Where OS was not reported in these studies, OS was censored at PFS.
Response stratification
Patients were divided into objective responder (OR) and non-responder (NR) groups according to three published response metrics utilizing best response by RECIST, duration of PFS, and duration of OS10,11,12. For patients in the MSKCC melanoma cohort, all non-responder patients were presumed to have PD as their best response by RECIST. Tumor samples from progressing lesions from patients who had clinical benefit from immune checkpoint therapy were excluded from the main analysis. Tumors from patients with PFS or OS below 30 days were excluded from analysis, as these patients may have had disease that was too advanced to experience clinical benefit from immune checkpoint therapy.
DNA extraction and sequencing
For samples newly sequenced from the Dana-Farber Cancer institute, DNA extraction from formalin-fixed, paraffin-embedded (FFPE) tumor blocks was performed as previously described47. Exome sequencing and data processing to produce a BAM file was performed using established analytical pipelines at the Broad Institute23.
DNA sequencing quality control
These data were combined with raw sequencing data (BAM files) from previously published cohorts of tumor/normal sequencing from patients with metastatic melanoma8,10, lung cancer7,25, anal cancer24, and sarcoma23. All 314 samples with tumor and germline sequencing data and clinical annotations were processed through standard quality control pipelines. Samples with poor sequencing coverage (tumor mean target coverage < 25x, normal mean target coverage < 15x) or high sample contamination48 in tumor or normal tissue were excluded (Fig. 1a, Supplementary Table 1). Additionally, samples with germline sequencing from adjacent normal tissue were assessed for tumor-in-normal contamination using deTiN (https://github.com/broadinstitute/deTiN), and excluded if normal tissue contained ≥1% tumor nuclei (Fig. 1a, Supplementary Table 1). After mutation calling and somatic copy number alteration assessment, tumors with estimated purity below 10% were also excluded49 (Supplementary Fig. 1a–b). These quality control measures were taken because high sample contamination and low tumor purity can lead to systematic under-calling of somatic SNPs and CNAs and interference with accurate assessment of tumor mutational burden and identification of response-associated molecular features50.
Whole exome analysis
Somatic nucleotide polymorphisms (SNPs) were identified by MuTect50, with computational filtering of artifacts introduced by DNA oxidation during sequencing51 or FFPE-based DNA extraction using a filter-based method. Strelka52 was applied to detect small insertions and deletions (indels). Annotation of identified variants was done using Oncotator (http://www.broadinstitute.org/cancer/cga/oncotator). Mutational clonality was estimated by ABSOLUTE, which uses allelic fraction of called mutations and allelic copy number information to determine mutational clonality and overall tumor purity and ploidy49. Clonal mutations were defined as those with estimated cancer cell fraction (CCF) of 1 or those whose probability of being clonal exceeded the probability of being subclonal. Nonsynonymous mutational burden was normalized by megabases covered at adequate depth to detect variants with 80% power using MuTect given estimated tumor purity by ABSOLUTE (Supplementary Table 1). Number of bases covered at a given depth threshold in the tumor sample was determined using the GATK DepthOfCoverage module. Putative driver mutations were collected using cBioPortal standards53,54, including both 3D hotspots in tumor suppressors and oncogenes55 and any loss-of-function variant in a tumor suppressor.
For copy number analysis, copy ratios were calculated for each captured target by dividing the tumor coverage by the median coverage obtained in a set of reference normal samples. The resulting copy ratios were segmented using the circular binary segmentation algorithm56. Segments were considered amplified or deleted if the |log2(copy ratio)| exceeded 0.5 (Supplementary Fig. 9a). For samples with uniform sample purity and ploidy, this definition of amplifications and deletions is adequate to detect copy number alterations. However, in this sample of tumors, called copy number alterations were heavily influenced by sample purity (Supplementary Fig. 9a). The |log2(copy ratio)| > 0.5 definition was insufficiently sensitive in low-purity samples, which have artificially depressed copy ratios due to a high proportion of sequencing reads from normal tissue. Conversely, this definition may be excessively noisy in high-purity tumors.
Thus, to correct segment copy ratios for sample purity, segment copy ratios were re-scaled by sample purity and ploidy with values derived from ABSOLUTE. Segments were considered amplified or deleted if the |log2(purity-corrected copy ratio)| exceeded 0.5. Specifically, the purity-corrected copy ratio was derived by dividing the purity-corrected (or rescaled) total copy number (rCN) for a given segment by the sample ploidy. This procedure yielded improved false-negative rates in low-purity tumors and false-positive rates in high-purity tumors, such that the proportion of a tumor genome considered amplified or deleted was less closely associated with tumor purity (Supplementary Fig. 9b).
However, while using a |log2(purity-corrected copy ratio)| > 0.5 definition of deletions and amplifications was effective for detecting large CNAs with high sensitivity, it did not provide adequate specificity for detecting focal events that would be more likely to be genomic driver CNAs for a tumor. Thus, we applied a previously described concept called focality57 to identify CNAs that were large outliers in copy ratio, representing either homozygous deletions expected to completely eliminate tumor expression of a given gene or amplifications expected to greatly over-express a gene. In this process, the rCN from ABSOLUTE was used as input. For each segment in a tumor genome, the focality was calculated by considering the fraction of a sample’s genome with lower rCN than that segment (for amplified regions) or higher rCN (for deleted regions). Segments were considered deleted if their rCN was < 0.25 and their focality was > 0.995. Segments were considered amplified if focality exceeded 0.98 − 0.2 × log2 (rCN/5), and highly amplified if focality exceeded 0.98 − (1/7) × log2 (rCN/7). The results from this analysis are displayed in Supplementary Fig. 9c, where far fewer segments meet criteria for being called as amplifications or deletions under this focality-based definition. This focality-based definition of CNA was applied in Fig. 5 as well as Supplementary Figs. 10–12 to demonstrate putative driver CNAs in genes in the interferon-gamma signaling pathway, PTEN, and SWI/SNF family of chromatin regulators. Genes were considered amplified or deleted if all or part of the gene was in a segment with a called copy number alteration using this focality-based definition. For the interferon-gamma analysis, samples were defined as having a copy number alteration affecting interferon signaling if any of the regular interferon genes were deleted or any of the four interferon pathway inhibitors (SOCS1, SOCS3, PIAS1, and PIAS4) harbored high amplifications, as previously described17. All gene-level CNAs of interest were manually reviewed.
A similar focality procedure was applied to allelic copy number calls from ABSOLUTE to determine heterozygous deletions and amplifications to identify loss-of-heterozygosity events for Supplementary Figs. 12–13.
Mutational signature deconvolution was conducted using a non-negative matrix factorization technique as previously described30. Mutational signatures were chosen from those previously described in COSMIC (http://cancer.sanger.ac.uk/cosmic/signatures)(Supplementary Table 6). The vectors for the commonly observed mutational signatures for each cancer type were used as input for inference of their contribution to observed mutations. Thus, for example, the signatures selected for melanoma pertained to UV exposure, prior alkylating agents, and other exposures, while those used for mutational signature deconvolution in lung cancer included tobacco exposure.
For neoantigen prediction, the 4-digit HLA type for each sample was inferred using Polysolver58. Putative neoantigens were predicted for each patient by defining all novel amino acid 9-mers and 10-mers resulting from each somatic nonsynonymous point mutation and determining whether the predicted binding rank—a proxy for predicted binding affinity to the patient’s germline HLA alleles—was < 2%. Strong binders had rank < 0.5%, while weak binders had rank between 0.5% and 2% using NetMHCpan (v3.0)59,60,61.
Statistical analysis
Assessment of enrichment of binary molecular features (e.g. wildtype or mutant gene; CNA present or absent) with response (CR/PR vs. PD) was done with Fisher’s exact tests. Assessment of difference in means or medians for a continuous variable between two response groups was done with the non-parametric Mann-Whitney U test unless otherwise specified. Correction for multiple hypothesis testing was done controlling for false discovery rate (FDR) by the Benjamini-Hochberg method, unless otherwise noted. Receiver operating characteristic (ROC) analyses were done using the pROC and Epi packages in R.
For the power calculation analysis (Fig. 2e), correction for multiple hypothesis testing was modeled with a Bonferroni correction over the 116 genes with known cancer driver status assessed previously. Response rates were set at 40% CR/PR vs. 60% PD, which is a generous estimate for response rate in an unselected population. P-values were calculated using Fisher’s exact tests comparing the prevalence of mutations in a given gene in CR/PR vs. PD.
Alpha level for all comparisons was 0.05 unless indicated otherwise. All statistical analyses were done in R (v.3.3.2).
Supplementary Material
Acknowledgments
This work was supported by BroadIgnite (E.M.V), BroadNext10 (E.M.V., D.M.), and NIH K08CA188615 (E.M.V.). D.M. was supported by the Howard Hughes Medical Institute Medical Research Fellows Program. This research was also supported by the Center for Immuno-Oncology at the Dana Farber Cancer Institute, and Stand Up To Cancer – American Cancer Society Lung Cancer Dream Team Translational Research Grant (Grant number: SU2C-AACR-DT17-15). Stand Up To Cancer (SU2C) is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnotes
Accession Codes
Raw BAM files from previously-published cohorts are available at dbGap at the following accession numbers: phs000694.v3.p2, phs000980.v1.p1, phs001041.v1.p1, phs000452.v2.p1, phs001075.v1.p1. Newly sequenced samples are available at dbGap at accession number phs001565.v1.p1.
Author Contributions
D.M., C.A.M., N.I.V., A.T.W., D.L., D.A., and D.K. performed the analyses. B.S., M.M., M.M.A., N.C., G.H., and S.M. provided clinical annotations. L.S. and S.R. contributed to immunohistochemical profiling. K.K.W., J.E., M.M.A., D.B., R.H., D.S., F.S.H., T.K.C., J.B., P.A.J., P.H. and E.M.V. contributed to sample acquisition. E.M.V. supervised the study. D.M., C.A.M., N.I.V., D.L., and E.M.V. wrote the manuscript with contributions from all authors.
Competing Financial Interests Statement
A.T., D.L., D.M., M.M., N.I.V., C.A.M., D.A., D.K., S.M.W., L.M.S., A.T.W., P.P., K.K.W, S.J.R., J.B., P.A.J., N.G.C., R.H., and M.M.A. declare no conflicts of interest. T.K.C. has advisory roles with AstraZeneca, Bayer, Bristol-Myers Squibb, Cerulean, Elsa, Foundation Medicine, Genentech, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Prometheus Labs, Roche, and Elsai. T.K.C. receives research funding from AstraZeneca, Bristol-Myers Squibb, Exelixis, Genentech, GSK, Merck, Novartis, Peloton, Pfizer, Roche, Tracon, and Eisai. G.J.H. receives institutional support from Bristol-Myers Squibb and EMD Serono. B.S. is on the advisory board or has received honoraria from Novartis, Roche, Bristol-Myers Squibb and MSD Sharp & Dohme, research funding from Bristol-Myers Squibb and MSD Sharp & Dohme, and travel support from Novartis, Roche, Bristol-Myers Squibb and AMGEN. R.I.H. has advisory roles with Bristol-Myers Squibb, Pfizer, Merck, AstraZeneca, Genentech and Celgene. R.I.H. receives research funding from Bristol-Myers Squibb, Merck, Genentech, and Pfizer. S.S. is a consultant for AstraZeneca and Merck, and receives research funding from AstraZeneca, Bristol-Myers Squibb, Exelixis, and Roche. G.G. has an advisory role with MD Anderson, and receives research funding from IBM and Bayer AG. G.G. is listed as an inventor on patent applications regarding MuTect, ABSOLUTE and Polysolver. D.A.B. is a consultant for N of One. D.S. receives consulting fees from Amgen, GSK, BMS, Novartis, Roche, Amgen, Merck, AstraZeneca, Merck-Serono, and Pfizer. P.H. and J.E. are employees of Novartis. F.S.H is a consultant to Bristol-Myers Squibb, Merck, Novartis, EMD Serono, Sanofi, and Genentech, and receives institutional research support from Bristol-Myers Squibb. E.M.V. holds consulting roles with Tango Therapeutics, Invitae and Genome Medical and receives research support from Bristol-Myers Squibb and Novartis.
Life Sciences Reporting Summary:
Further information on experimental design is available in the Life Sciences Reporting Summary.
References
- 1.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahmer J, et al. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. New England Journal of Medicine. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P. Immune checkpoint therapy and the search for predictive biomarkers. Cancer J. 2016;22:68–72. doi: 10.1097/PPO.0000000000000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbognin L, et al. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): Sensitivity analysis of trials in melanoma, lung and genitourinary Cancers. PLOS ONE. 2015;10:e0130142. doi: 10.1371/journal.pone.0130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg JE, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. The Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roh W, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Science Translational Medicine. 2017;9 doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colli LM, et al. Burden of Nonsynonymous Mutations among TCGA Cancers and Candidate Immune Checkpoint Inhibitor Responses. Cancer Res. 2016;76:3767–3772. doi: 10.1158/0008-5472.CAN-16-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riaz N, et al. Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nature genetics. 2016;48:1327–1329. doi: 10.1038/ng.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson DB, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res. 2015;3:288–295. doi: 10.1158/2326-6066.CIR-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404.e399. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato S, et al. Hyper-progressors after immunotherapy: Analysis of genomic alterations associated with accelerated growth rate. Clinical Cancer Research. 2017 doi: 10.1158/1078-0432.ccr-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao D, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018 doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017:355. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sucker A, et al. Acquired IFNγ resistance impairs anti-tumor immunity and gives rise to T cell-resistant melanoma lesions. Nature Communications. 2017 doi: 10.1038/ncomms15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Allen EM, et al. Long-term benefit of PD-L1 blockade in lung cancer associated with JAK3 activation. Cancer Immunol Res. 2015;3:855–863. doi: 10.1158/2326-6066.CIR-15-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George S, et al. Loss of PTEN Is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouw KW, et al. Genomic evolution after chemoradiotherapy in anal squamous cell carcinoma. Clinical Cancer Research. 2016 doi: 10.1158/1078-0432.ccr-16-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garofalo A, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Medicine. 2016;8:1–10. doi: 10.1186/s13073-016-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Materials and methods are available as supplementary materials on Science Online.
- 27.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Wolchok JD, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
- 29.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature genetics. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamal-Hanjani M, et al. Tracking the evolution of non-small-cell lung cancer. New England Journal of Medicine. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 32.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizvi H, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand (PD-L)-Ligand 1 Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol. 2018 doi: 10.1200/JCO.2017.75.3384. JCO2017753384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell reports. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Mullane SA, et al. Correlation of APOBEC mRNA expression with overall survival and PD-L1 expression in urothelial carcinoma. Scientific Reports. 2016;6:27702. doi: 10.1038/srep27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goel S, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng W, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan D, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018 doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaretsky JM, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sade-Feldman M, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8:1136. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 46.Gettinger S, et al. Nivolumab Monotherapy for First-Line Treatment of Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2016;34:2980–2987. doi: 10.1200/JCO.2016.66.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Allen EM, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 53.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 2017;9:4. doi: 10.1186/s13073-016-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 57.Brastianos PK, et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shukla SA, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harbor perspectives in medicine. 2016:6. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.