Abstract
Mesenchymal stromal cells are involved in the pathogenesis of myelodysplastic syndromes and acute myeloid leukemia, but the underlying mechanisms are incompletely understood. To further characterize the pathological phenotype we performed RNA sequencing of mesenchymal stromal cells from patients with myelodysplastic syndromes and acute myeloid leukemia and found a specific molecular signature of genes commonly deregulated in these disorders. Pathway analysis showed a strong enrichment of genes related to osteogenesis, senescence, inflammation and inhibitory cytokines, thereby reflecting the structural and functional deficits of mesenchymal stromal cells in myelodysplastic syndromes and acute myeloid leukemia on a molecular level. Further analysis identified transforming growth factor β1 as the most probable extrinsic trigger factor for this altered gene expression. Following exposure to transforming growth factor β1, healthy mesenchymal stromal cells developed functional deficits and adopted a phenotype reminiscent of that observed in patient-derived stromal cells. These suppressive effects of transforming growth factor β1 on stromal cell functionality were abrogated by SD-208, an established inhibitor of transforming growth factor β receptor signaling. Blockade of transforming growth factor β signaling by SD-208 also restored the osteogenic differentiation capacity of patient-derived stromal cells, thus confirming the role of transforming growth factor β1 in the bone marrow microenvironment of patients with myelodysplastic syndromes and acute myeloid leukemia. Our findings establish transforming growth factor β1 as a relevant trigger causing functional inhibition of mesenchymal stromal cells in myelodysplastic syndromes and acute myeloid leukemia and identify SD-208 as a candidate to revert these effects.
Introduction
Myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) are blood stem cell disorders that are characterized by hematopoietic insufficiency causing relevant morbidity and mortality. For a long time both entities have been considered to be hematopoietic-cell autonomous diseases originating from the accumulation of genetic and epigenetic alterations within the hematopoietic stem and progenitor cell (HSPC) population.1,2 Recently, it has become apparent that these myeloid disorders do not exclusively arise from HSPC but also involve the entire bone marrow microenvironment and, in particular, mesenchymal stromal cells (MSC).3,4 In this context, it was demonstrated in several mouse models that genetic perturbation of mesenchymal cells can induce MDS and promote the emergence of AML.5,6 Vice versa, malignant myeloid cells can also act on niche elements such as MSC leading to a self-reinforcing mechanism supporting leukemic cells at the expense of normal hematopoiesis.7–9 Investigating the pathogenic role of MSC in humans, we and others have recently shown that primary MSC from the bone marrow of MDS and AML patients are structurally, genetically and functionally altered.4,10–15 For instance, we found that these MSC have impaired growth capacity and a decreased ability to undergo osteogenic differentiation, accompanied by a specific methylation signature. Along with impaired stromal support of healthy HSPC this suggests that MSC alterations contribute substantially to the inadequate hematopoiesis in MDS and AML.12,13 Furthermore, these findings imply significantly deregulated bi-directional crosstalk between malignant myeloid cells and MSC. In the current analysis, we used discovery-based strategies, such as RNA sequencing, followed by candidate-testing approaches. We identified transforming growth factor β (TGFβ) signaling as a relevant mechanism responsible for the functional inhibition of MSC in MDS and AML and showed that this was pharmacologically reversible upon TGFβ blockage.
Methods
Patients, healthy controls and cell preparation
Bone marrow samples were obtained from a total of 28 patients with newly-diagnosed MDS or AML (median age 59 years; range, 25-89 years) and 16 age- and sex-matched healthy controls (median age 68 years; range, 39-86 years; P=0.14). Details of the patients’ characteristics are given in Online Supplementary Table S1. The study was approved by our local institutional review board (approval number: 4777) and all individuals gave written informed consent to participation in it.
MSC were derived from the mononuclear cell fraction of the specimens and cultured as described previously.12,13 All experiments were carried out using MSC derived from passages 3–4. Furthermore, CD34+ HSPC were obtained from the bone marrow of healthy controls by density gradient separation and subsequent immunomagnetic selection (Miltenyi Biotec, Bergisch Gladbach, Germany) as described elsewhere.16
RNA sequencing
Transcriptome sequencing libraries were prepared from isolated total RNA using the TruSeq RNA sample preparation kit (Illumina, San Diego, CA, USA), and single-read 50 bp sequencing was performed in a HiSeq-2000 device (Illumina). Reads were then trimmed by removing stretches of bases with a quality score <30 at their ends, and subsequently mapped using Tophat2.0.6 against the hg19 assembly of the human genome.17 Finally, differential expression was quantified using DESeq2 and Cuffdiff 2.0, and subjected to diverse testing corrections.18,19 Genes with a q-value <0.05 were considered differentially expressed. Principal component analysis plots and heat maps were created in R using the FactoMineR and pheatmap packages, respectively. For gene set enrichment analysis (GSEA, Broad Institute, Boston, MA, USA),20 the fragments per kilobase of transcript per million mapped reads (FPKM) values obtained from Cuffdiff 2.0 for the different samples were compared either with the gene sets contained in the Molecular Signature Database or with self-made gene lists. The Signal2Noise metric and 1000 gene set-based permutations were applied to all analyses. Ingenuity pathway analysis software (Qiagen, Hilden, Germany) was used to predict the potential extrinsic factors capable of generating the different RNA sequencing data sets.
Cell culture conditions and reagents
Healthy MSC were cultured in Dulbecco modified Eagle medium low glucose supplemented with 30% fetal bovine serum and 1% penicillin/streptomycin/L-glutamine (all from Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) in the presence of TGFβ1 (10 ng/mL, PeproTech, Rocky Hill, NJ, USA) and/or SD-208 (0.5 μM, Tocris/R&D Systems, MN, USA), an established small molecule pyridopyrimidine inhibitor of the TGFβ receptor I kinase.21–23 The inhibitor was diluted in dimethyl sulfoxide (10 mM). The concentration of TGFβ1 used in our experiments is within the range of TGFβ1 levels previously detected in patients with MDS24,25 and has demonstrated inhibitory effects on healthy and MDS-derived hematopoietic cells.26–28 Furthermore, it was previously shown that TGFβ1 at this concentration mediates pathophysiologically relevant effects in other myeloid malignancies such as AML, chronic myeloid leukemia and myeloproliferative syndromes.9,29 Depending on the experimental setting, incubation time ranged from 3 to 28 days corresponding to the duration of the culture of MSC investigated by RNA sequencing. Detailed information is given in the legends of the respective figures.
Subsequently, to investigate the effects of TGFβ1 on the functionality of cells, pre-incubated MSC underwent the phenotypic and functional analyses described below.
Phenotypic and functional characterization of mesenchymal stromal cells
The morphology and growth properties of primary and pre-incubated MSC were characterized by light microscopy. For quantification of growth potential, absolute cell numbers and cumulative population doubling were determined as described elsewhere.12,13 Furthermore, surface expression of established stromal cell markers CD73, CD90 and CD105,30 hematopoietic antigens CD34 and CD45 as well as Jagged1 were measured by flow cytometry using a FACSCalibur (BD Biosciences, Heidelberg, Germany) and FCS Express V3 software (De Novo Software, Los Angeles, CA, USA) for data analysis. Antibody specifications are provided in Online Supplementary Table S2. Osteogenic differentiation capacity was investigated and visualized by Alizarin red staining as reported elsewhere.12,13 In addition, mRNA expression of markers of osteogenesis, osterix and osteocalcin, and other candidate genes was measured by quantitative polymerase chain reaction (PCR) on a StepOne Plus Realtime PCR Cycler (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) as described before.12,13
Long-term culture-initiating cell assay
After pre-incubation with TGFβ1 or its antagonist, SD-208, 0.8×106–1.2×106 MSC were cultured on 96-well plates (Costar, Corning, USA) and irradiated with 30 Gray using Gulmay RS225 X-ray equipment. Subsequently, 6×103 healthy CD34+ cells were plated on these MSC feeder layers and then further processed using the same conditions and reagents as in our previous work.12,13
Statistical analysis
Statistical analyses were performed using Prism 5.01 (GraphPad Software Inc., La Jolla, USA). For inter-individual comparisons the two-sided unpaired Student t-test was employed, while for intra-individual analyses the Wilcoxon signed rank test was used. For all experiments means and the standard error of mean (SEM) are given. Statistical significance was established at P≤0.05.
Data access
RNA sequencing expression data have been stored in the Gene Expression Omnibus database (GSE107490).
Results
RNA sequencing analysis of myelodysplastic syndrome- and acute myeloid leukemia-derived mesenchymal stromal cells reveals a specific gene expression profile reflecting their phenotypic and functional deficits
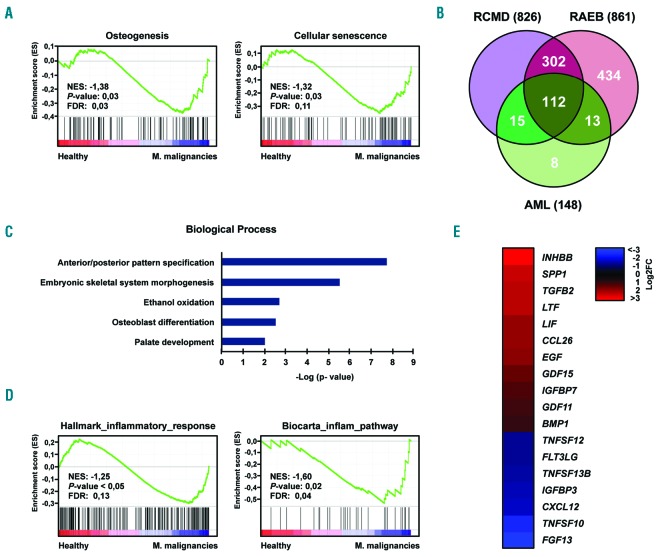
We have previously shown that MSC from MDS and AML patients display common functional deficits and DNA methylation changes.12,13 In order to determine whether these similarities extend to the gene expression level and contribute to the altered phenotype, we performed RNA sequencing. Sequencing data were obtained for MSC from three healthy donors (all male, median age 68 years) and nine patients with myeloid malignancies [all male, median age 60 years, P=1.0; 3 MDS patients (n. 4-6) with refractory cytopenia with multilineage dysplasia (RCMD), 3 MDS patients (n. 9-11) with refractory anemia with excess blasts (RAEB), and 3 AML patients (n. 9-11)]. Data analysis revealed 1673 genes that were significantly (q<0.05) differentially expressed, relative to controls, in the MSC from the three subentities (Online Supplementary Figure S1, Online Supplementary Table S3). GSEA confirmed a significant overlap of the deregulated genes with those involved in osteogenesis or cellular senescence, two processes that we previously found to be deregulated in MDS- and AML-related MSC (Figure 1A and Online Supplementary Tables S4 and S5).12,13 Pairwise comparisons between healthy samples and each of the different disease conditions defined a set of 112 deregulated genes (q<0.05) in common, mostly related to general developmental processes and osteogenesis (Figure 1B,C, Online Supplementary Figure S2, and Online Supplementary Tables S3 and S6). Further analysis revealed a remarkable abundance of gene signatures associated with inflammatory responses (Figure 1D) as well as altered expression of a variety of cytokines (Figure 1E and Online Supplementary Figure S3). Overall, the expression of 18 cytokines was significantly altered (11 upregulated, 7 downregulated) with several of them known to be involved in the regulation of hematopoiesis. Collectively, these results are consistent with the malignant phenotypes previously reported in MDS- and AML-related MSC, and further suggest a related pathomechanism for the MSC from the three entities.
Figure 1.
RNA sequencing analysis of mesenchymal stromal cells from patients with myeloid malignancies and from healthy controls. (A) GSEA plots showing the specific deregulation of gene sets associated with osteogenesis and cellular senescence in MSC from patients with MDS and AML. For both plots, the normalized enriched score (NES), false discovery rate (FDR) and P-values are given. (B) Venn diagram representing the number of genes differentially expressed (q<0.05) in MSC from patients with RCMD, RAEB or AML with respect to healthy MSC, as well as those genes deregulated in common in the three different myeloid entities. (C) Gene ontology analysis of the 112 genes deregulated in common in RCMD-, RAEB- and AML-derived MSC. (D) Examples of GSEA plots demonstrating an inflammatory reaction in MSC from patients with MDS or AML. For both plots, NES, FDR and P-values are given. (E) Heat map illustrating the expression changes of the set of cytokines found to be statistically significantly (q<0.05) deregulated in the MSC from MDS and AML patients. A total of 173 cytokines involved in the regulation of hematopoiesis, some of which previously described in MDS,10 were analyzed. FC: fold-change in expression.
RNA sequencing analysis suggests that transforming growth factor β1 signaling is a common cause of abnormal gene expression patterns in myelodysplastic syndrome- and acute myeloid leukemia-derived mesenchymal stromal cells
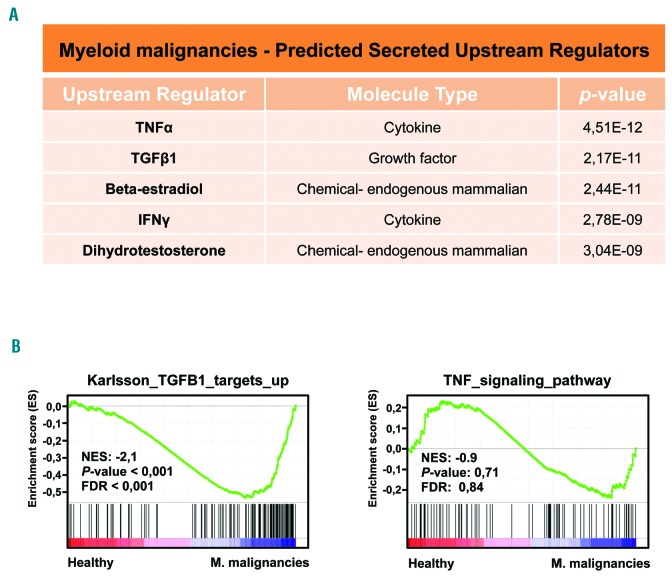
Our previous work suggested that the phenotypic abnormalities observed in AML-derived MSC may be triggered by an extrinsic factor.13 We hypothesized that the conserved aberrant gene expression patterns are caused by an extrinsic factor. To identify this molecule, we used ingenuity pathway analysis,29 which enables the prediction of upstream regulators for a given RNA sequencing data set. This computational approach identified the inhibitory cytokines tumor necrosis factor a (TNFa) and TGFβ1 as the two most probable (P values of 2.17×10−11 or lower) secreted factors for the induction of the aberrant gene expression patterns in the MSC derived from RCMD, RAEB and AML patients (Figure 2A). These two molecules also appeared among the most probable secreted factors in analyses of RNA sequencing data sets from pairwise comparisons (Online Supplementary Figure S4). To analyze whether only one of these two molecules could be sufficient to induce the gene expression deregulation observed in the MSC from the patients, we used GSEA. The results showed that our gene set was significantly enriched in the TGFβ1 signature but not in the TNFa signature (Figure 2B). Our RNA sequencing experiments therefore suggest that increased TGFβ1 signaling in the bone marrow may lead to the aberrant gene expression patterns observed in the MSC from these myeloid malignancies and, ultimately, to their functional inhibition.
Figure 2.
RNA sequencing identified transforming growth factor β1 as the most probable secreted upstream regulator inducing the aberrant gene expression patterns. (A) Ingenuity pathway analysis29 predicted that TNFα and TGFβ1 are the most probable secreted upstream regulators of the gene expression aberrations observed in the MSC from MDS and AML patients. (B) GSEA confirmed that overactivation of TGFβ1 signaling, but not TNFa, in the bone marrow caused the aberrant gene expression patterns. For both plots, the normalized enriched score (NES), false discovery rate (FDR) and P-values are given.
Transforming growth factor β1 induces functional deficits in healthy mesenchymal stromal cells recapitulating the phenotype of these cells in myelodysplastic syndrome and acute myeloid leukemia
Having identified TGFβ1 as a candidate factor for the induction of the observed phenotypic alterations and functional deficits of MSC in MDS and AML, we experimentally addressed this hypothesis using an in vitro culture system. For this purpose, we cultured healthy MSC in the presence or absence of TGFβ1 to investigate whether healthy MSC adopt a phenotype that is similar to the phenotype of patient-derived MSC. Furthermore, in this experimental system we also used SD-208, which specifically abrogates the signaling downstream of the TGFβ receptor (Online Supplementary Figure S5).
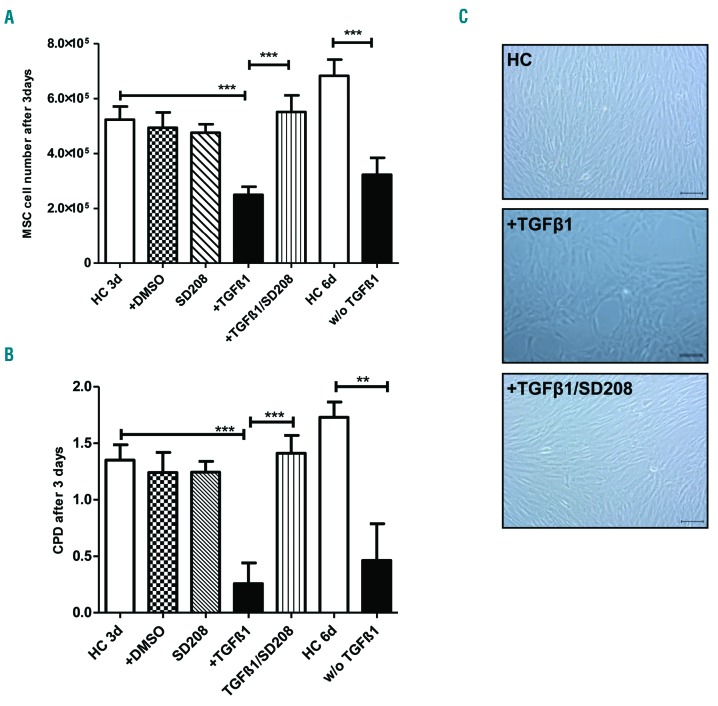
As a first read-out, we quantified the proliferative potential and the growth capacities of MSC. TGFβ1 significantly inhibited the proliferation and growth properties of healthy MSC, as indicated by the absolute cell number and cumulative population doubling of the cells treated in this way being similar to those observed in primary patient-derived MSC (Figure 3A,B and Online Supplementary Figure S6). This suppressive effect also persisted when healthy MSC were primed with TGFβ1 for 3 days and subsequently cultured in control media for another 3 days (Figure 3A,B). In line with these findings, microscopic analysis of healthy MSC exposed to TGFβ1 revealed a disorganized cellular architecture, which differed sharply from the fibroblastoid feeder layer of the control MSC (Figure 3C). The inhibitory effects of TGFβ1 on growth and proliferation of MSC were abrogated by the TGFβ receptor I inhibitor SD-208 (Figure 3A-C).
Figure 3.
Transforming growth factor β1 impairs growth properties of healthy mesenchymal stromal cells. (A) Bar charts showing the absolute MSC numbers after 3 days pre-incubation of 2×105 MSC with the respective factors (10 ng/mL). HC: healthy control; DMSO: dimethylsulfoxide; w/o TGFβ1: healthy MSC were treated with TGFβ1 for 3 days, then the medium was changed and the MSC were cultured for 3 additional days without TGFβ1. (B) Bar charts depicting cumulative population doubling (CPD). (C) Representative micrographs showing the morphology of healthy MSC after incubation in the presence of the respective compounds. Scale bars represent 100 μm. For all experiments results are expressed as mean ± SEM. Asterisks indicate P-values **P<0.01, ***P<0.001.
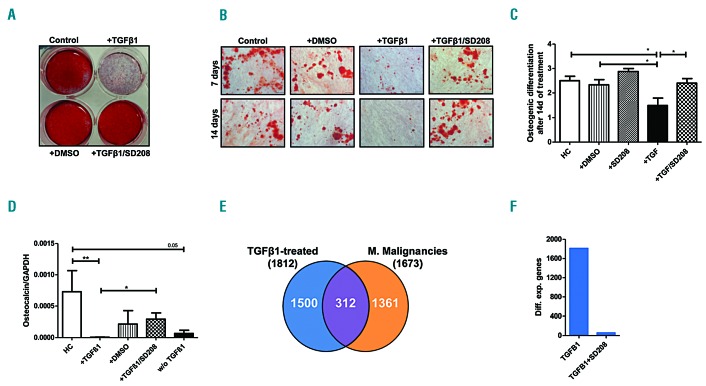
In subsequent experiments we investigated the influence of TGFβ1 on osteogenic differentiation, which has been previously shown to be significantly impaired in MDS- and AML-derived MSC.12,13 Using established culture conditions and staining methods12,13 we found that TGFβ1 significantly impaired osteogenic differentiation capacity (Figure 4A-B, Online Supplementary Figure S7). In agreement with this result, mRNA expression of the bone formation marker osteocalcin was significantly reduced after exposure to TGFβ1 (Figure 4D). Similar to its effect on MSC growth, SD-208 also abolished the TGFβ1-mediated suppression of osteogenic differentiation (Figure 4A-C). RNA sequencing of healthy MSC after incubation with TGFβ1 revealed a specific gene expression pattern substantially overlapping with the expression profile of primary patient-derived MSC (Figure 4E, Online Supplementary Figure S8 and Online Supplementary Table S7). In this regard, we decided to check whether the expression of PITX2, HOXB6 and TBX15, three genes physiologically involved in cell differentiation and skeletal morphogenesis, and which we had previously linked to the impaired osteogenesis in MDS and AML,12,13 could also be affected in the TGFβ1-treated healthy MSC. Although only PITX2 showed a statistically significant change in the RNA sequencing experiment (fold change=3.57, q<0.001), the dysregulation of the three genes could be further verified by quantitative real-time PCR (Online Supplementary Figure S9). SD-208 again reverted the TGFβ1-induced gene expression changes, including those for the three marker genes. (Figure 4F and Online Supplementary Figure S9).
Figure 4.
Transforming growth factor β1 suppresses the osteogenic differentiation capacity of healthy mesenchymal stromal cells and induces a specific gene expression profile. Healthy MSC (n=6) were pre-incubated with TGFβ1 and/or SD-208 for up to 28 days. Medium was changed every 3 days and supplemented with TGFβ1 at a concentration ranging from 5 ng/mL to 10 ng/mL and/or SD-208 (0.25 μM to 0.5 μM). Subsequently, osteogenic differentiation was induced for 14 days and visualized by Alizarin red staining as described previously.12,13 (A) Overview of a representative experiment. (B) Representative micrographs of healthy MSC after exposure to the respective factors with scale bars indicating 100 μm. (C) For the purpose of quantification, osteogenic differentiation capacity was graded according to microscopic analysis of staining intensity as follows: 0 = absent; 1 = weak; 2 = moderate; 3 = intensive as previously described.12 (D) Messenger RNA expression of osteocalcin was measured by quantitative real-time PCR analysis of healthy MSC (n=5) after 3 days of incubation. HC: healthy control; DMSO: dimethylsulfoxide; w/o TGFβ1: healthy MSC were treated with TGFβ1 for 3 days, then the medium was changed and the MSC were cultured for 3 additional days without TGFβ1. For all experiments results are expressed as mean ± SEM. Asterisks indicate P-values *P<0.05, **P<0.01. (E) After a 28-day incubation period healthy MSC (n=2) were subjected to RNA sequencing analysis. The Venn diagram illustrates a substantial overlap of 312 genes deregulated in both the TGFβ1-treated MSC and the patient-derived MSC. (F) Bar charts demonstrate that most of the differential gene expression provoked by TGFβ1 in healthy MSC (1812 genes, q<0.05) is abrogated by SD-208 (only 58 genes remained differentially expressed, q<0.05).
Transforming growth factor β1 impairs stromal hematopoietic support function
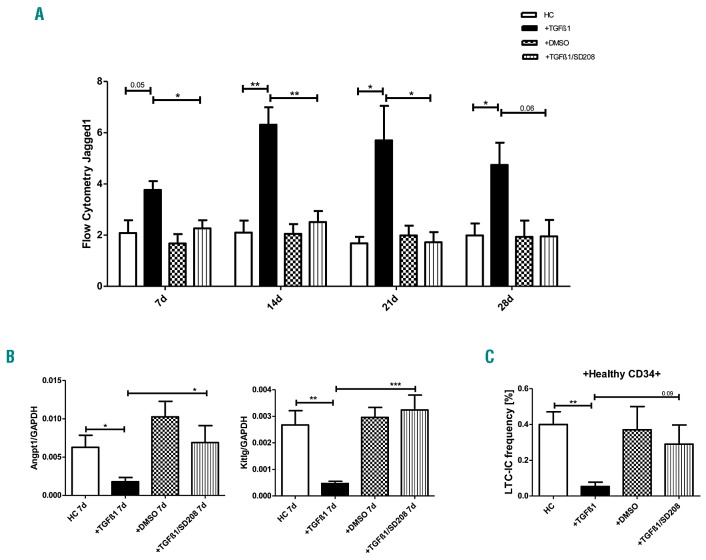
We have previously described the deregulation of Jagged1, Angiopoietin-1 and Kit-ligand, three signaling molecules physiologically involved in the regulation of HSPC, in patient-derived MSC.12,13 We now examined whether TGFβ1 can also induce alterations of these factors and of three candidate cytokines (CCL26, SPP1 and LIF) that were shown to be deregulated in primary patient-derived MSC. Indeed, quantitative PCR and flow cytometry detected an expression pattern (Figure 5A,B, Online Supplementary Figures S10 and S11) that is congruent with the expression previously detected in primary MDS- and AML-derived MSC.12,13 Again, these effects were reversible by adding SD-208 (Figure 5A,B).
Figure 5.
Impaired hematopoietic support capacities of healthy mesenchymal stromal cells exposed to transforming growth factor β1. Healthy MSC (n=5) were pre-incubated with TGFβ1 and/or SD-208 for up to 28 days. Medium was changed every 3 days and supplemented with TGFβ1 and/or SD-208 at a concentration ranging from 5 ng/mL to 10 ng/mL (SD-208: from 0.25 μM to 0.5 μM). Cells were harvested on days 7, 14, 21, and 28. (A) Bar charts illustrate protein expression of Jagged1 as measured by flow cytometry at the given time points. The mean fluorescence intensity43 is shown. HC: healthy control; DMSO: dimethylsulfoxide. (B) mRNA expression of Angiopoietin-1 (Angpt1) and Kit-ligand (Kitlg) was measured by quantitative real-time PCR after 7 days of incubation. (C) Bar charts showing LTCIC frequencies of healthy CD34+ HSPC cultured on healthy MSC which had been previously incubated with TGFβ1 and/or SD-208 for 28 days. For all experiments results are expressed as mean ± SEM. Asterisks indicate P-values *P<0.05, **P<0.01, ***P<0.001.
In light of the profound molecular alterations that were induced by TGFβ1 in healthy MSC, we were further interested in whether TGFβ1 also suppressed stromal hematopoietic support. We therefore used the established long-term culture-initiating cell (LTC-IC) assay and cultured healthy CD34+ HSPC on human MSC feeder layers which had previously been incubated with TGFβ1 or with TGFβ1 together with SD-208 for 28 days. As indicated in Figure 5C, TGFβ1 significantly inhibited the ability of healthy MSC to support CD34+ HSPC in the LTCIC assay, an effect that could be reversed by inhibiting TGFβ1 signaling through SD-208. In summary, TGFβ1 induces a phenotype and functional deficits recapitulating those observed in primary MSC obtained from patients with MDS or AML. Our data thus strongly support the hypothesis derived from RNA sequencing data analysis, namely that TGFβ1 contributes significantly to the functional inhibition of MSC in these two myeloid malignancies.
Inhibition of transforming growth factor β1 signaling restores osteogenic differentiation and hematopoietic support capacity of myelodysplastic syndrome-and acute myeloid leukemia-derived mesenchymal stromal cells
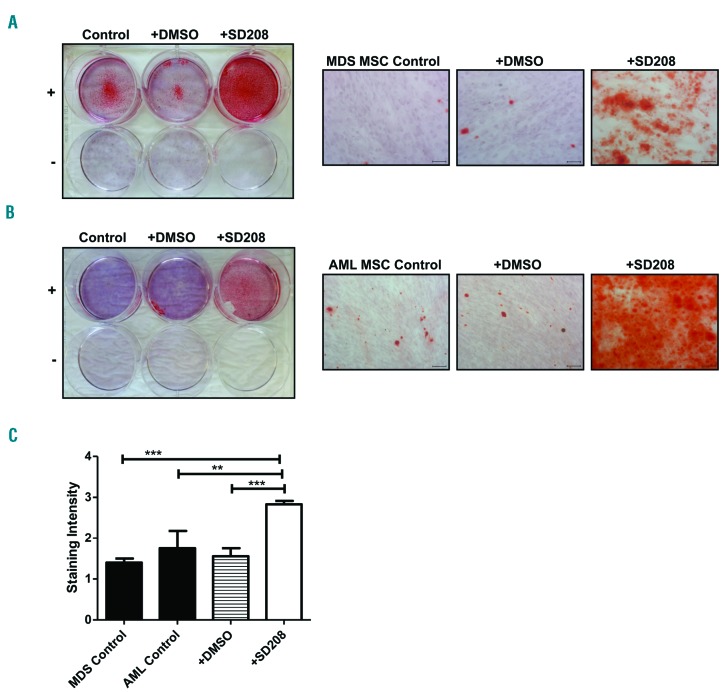
In a final set of experiments we directly addressed the impact of TGFβ1 on MSC functionality in human MDS and AML. To abrogate the potential influence of TGFβ1 on MSC functions we again used SD-208. This inhibitor had already been demonstrated to abolish TGFβ signaling in the above-mentioned experiments, as well as previously in MDS-derived CD34+ cells.12,13 Given the substantial inhibition of osteogenic differentiation in primary patient-derived MSC, we focused on the potential effects of TGFβ1 on osteogenesis. For this purpose we cultured MSC, freshly isolated from the bone marrow of patients with MDS and AML, in the presence of SD-208 and documented that osteogenic differentiation capacity could be significantly rescued compared to that occurring in the control condition of patient-derived MSC cultured in the absence of SD-208 (Figure 6A-C). In addition, treatment of MDS- and AML-derived MSC with SD-208 substantially increased these cells’ support of healthy CD34+ cells, as reflected by their 2.1-fold higher LTC-IC frequency in comparison to that of untreated MDS- and AML-derived MSC (Online Supplementary Figure S12, P=0.078). These findings clearly indicate that TGFβ1 inhibits osteogenic differentiation and hematopoietic support capacity of MSC in human MDS and AML and that this effect can be pharmacologically reverted by SD-208.
Figure 6.
Blockage of transforming growth factor β1 signaling by SD-208 restores the osteogenic differentiation capacity of myelodysplastic syndrome- and acute myeloid leukemia-derived mesenchymal stromal cells. Osteogenic differentiation of MDS- and AML-derived MSC was induced and visualized by Alizarin red staining. SD-208 (0.5 μM) was added to the osteogenic medium during this procedure. (A) Overview of a representative experiment investigating MDS-derived MSC (n=5, MDS patients n. 1, 2, 7, 12, 13). Representative micrographs of MDS-derived MSC after exposure to SD-208 with scale bars indicating 100 μm. (B) Overview of a representative experiment investigating AML-derived MSC (n=4, AML patients n. 1, 5, 12, 13). Representative micrographs of AML-derived MSC after exposure to SD-208 with scale bars indicating 100 μm. (C) Differences in osteogenic differentiation between patient-derived MSC (black bars, MDS n=5; AML n=4) either native or treated with SD-208 (white bar) were quantified by a previously described score.12 In detail, differentiation was graded according to microscopic analysis of staining intensity as follows: 0 = absent; 1 = weak; 2 = moderate; 3 = intensive. For all experiments results are expressed as mean ± SEM. Asterisks indicate P-values **P<0.01, ***P<0.001.
Discussion
In our previous work we demonstrated that MSC from patients with MDS and AML exhibit impaired growth and osteogenic differentiation capacity as well as altered expression of hematopoietic signaling molecules such as Angiopoietin-1, Kit-ligand and Jagged1. Comparable with recent results from Bhagat et al.,31 these structural and functional deficits were associated with a specific methylation pattern and resulted in impaired stromal support for HSPC.12,13 These findings were compatible with confirmatory results published by other groups in the last years.4,11,14,15,32,33 and directly linked alterations of MSC to the pathophysiology of MDS and AML.
To elucidate the underlying molecular mechanisms, in this study we performed RNA sequencing of MSC from patients with early MDS, advanced MDS and AML. Our data analysis defined a specific common molecular profile which clearly separated MSC of these myeloid diseases from those of healthy controls. Pathway analyses further identified an enrichment of genes involved in general developmental processes, cellular senescence and in particular genes essential for skeletal morphogenesis, including the three candidate genes PITX2, HOXB6 and TBX15 that are commonly deregulated in MDS- and AML-derived MSC.12,13 On the molecular level these results provide an excellent explanation for the phenotypic and functional alterations of MDS- and AML-derived MSC that were previously reported by our group and by others.11–14
Additional findings suggested an ongoing stromal response to an inflammatory environment. This is in line with the results from two other groups who also reported an inflammatory stress response in MSC derived from patients with low-risk MDS.10,14 Our data now expand this finding to MSC from patients with advanced MDS and AML, and suggest that the pro-inflammatory signaling within the bone marrow microenvironment influences the stromal compartment in addition to HSPC, thereby contributing to insufficient hematopoiesis.
Ingenuity pathway analysis and GSEA predicted that the inhibitory cytokine TGFβ1 (but not TGFβ2, data not shown) is an upstream regulator of the observed functional and molecular alterations in MDS- and AML-derived MSC. Again, this confirms and expands the results from Chen et al., who recently identified signatures related to TGFβ signaling in primary MSC from patients with low-risk MDS.10 This molecular similarity between our culture-expanded MSC and the non-expanded, FACS-sorted MSC reported by Chen et al. also indicates that the most relevant pathophysiological mechanisms seem to be well preserved even after culture. Apart from its pathophysiological relevance this also has important methodological implications. The use of non-expanded FACS-sorted MSC does not allow subsequent direct functional testing of candidate genes because of the low number of cells, but also requires culture or cell lines.10 In contrast, we were able to test the effects of TGFβ1 on stromal cell functionality in the same culture system previously used to isolate and characterize MDS- and AML-derived MSC. This methodological issue should ideally be further investigated by a direct comparison of matched FACS-sorted, unexpanded and culture-propagated MSC from the same individual patients.
We did not detect any relevant effects on MSC functions after exposure to TNFa (data not shown). Considering the limited clinical efficacy of TNFa blockade by etanercept and infliximab in MDS patients,34–36 we believe that TNFa plays, at most, a minor role. In contrast, exposure to TGFβ1 suppressed growth and osteogenic differentiation of healthy MSC and induced changes in the expression of candidate genes (PITX2, HOXB6, TBX15, Kit-ligand, Angiopoietin-1, and Jagged1). Together with impaired stromal hematopoietic support, the phenotype of healthy MSC therefore resembles that of primary MDS- and AML-derived MSC after exposure to TGFβ1.12,13 The inhibitory effect of TGFβ1 on normal HSPC is well documented.37 In addition, overactivation of TGFβ signaling in CD34+ HSPC has been shown to be an important mechanism mediating hematopoietic insufficiency in MDS.23,38,39 Growth differentiation factor-11, another ligand of the TGFβ superfamily, inhibits maturation of erythroid precursors and elevated levels of this factor have been found in patients with MDS, implicating it in the development of anemia.40 In addition to suppression of HSPC and erythroid progenitors, our data from RNA sequencing and functional experiments show for the first time that activated TGFβ signaling contributes significantly to ineffective hematopoiesis in MDS and AML via functional inhibition of MSC.
Envisaging targeted stromal therapies, it is of particular interest that we could pharmacologically revert the suppressive effects of TGFβ1 on healthy MSC in vitro using SD-208. This small molecule pyridopyrimidine inhibitor of TGFβ receptor I abrogates the signaling downstream of TGFβ receptor I, independently of the specific ligand. SD-208 has been shown to stimulate hematopoiesis in a mouse model of bone marrow failure as well as when MDS HSPC were cultured in vitro.23 In agreement with these findings, SD-208 was also able to restore the osteogenic differentiation capacity and, with a clear trend, the hematopoietic support capacity of MDS- and AML-derived MSC in our analysis. Targeting TGFβ signaling to improve hematopoiesis is the aim of two ligand-trapping approaches with luspatercept and sotatercept, currently under clinical investigation in MDS.40,41 Of particular interest, the effects of the latter are not mediated via CD34+ HSPC but rather via the stromal compartment,41 thus further supporting the role of TGFβ-mediated stromal inhibition in MDS and AML. At this point it remains unclear whether TGFβ1-mediated functional inhibition of MSC in MDS and AML is a cell-intrinsic mechanism or is extrinsically mediated by other TGFβ1-producing cells within the bone marrow microenvironment. While Chen et al. recently detected overexpression of TGFβ1 in FACS-sorted unexpanded MSC of patients exclusively with low-risk MDS, we did not find overexpression of TGFβ1 in culture-expanded MSC derived from patients with early or advanced MDS or AML (data not shown). On the other hand, we did find mRNA overexpression of TGFβ1 in CD34+ HSPC of patients with MDS or AML by performing quantitative PCR as well as by analyzing data from 173 AML samples in comparison to 967 samples from 28 different healthy tissues included in The Cancer Genome Atlas database (Online Supplementary Figures S13 and S14). This suggests that TGFβ1 released from CD34+ HSPC may contribute, at least in part, to the inhibition of MSC in MDS and AML. In further support of an extrinsic mechanism, it was recently shown that myeloid-derived suppressor cells also overproduce TGFβ1 and thereby suppress hematopoiesis in MDS.42 Overall, these data imply that different cells may be involved as sources of TGFβ1 and TGFβ1-related inhibition of hematopoiesis in MDS and AML. Indeed, since there may even be variations between early and advanced MDS as well as between MDS and AML this issue needs to be addressed in future studies.
In conclusion, our RNA sequencing analysis revealed a specific molecular signature of MDS- and AML-derived MSC which reflected and explained these cells’ structural and functional deficits. By subsequent functional testing we confirmed that TGFβ1 is a relevant trigger for the molecular alterations and functional inhibition of MSC in MDS and AML. Our results also further support targeting of this signal pathway as a promising treatment approach.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SCHR 1470/1-1), the Research Committee of the Medical Faculty, Heinrich-Heine-University, Duesseldorf, Germany and the Leukämie Lymphom Liga e. V., Duesseldorf, Germany to TS.
We would like to thank Dennis Sohn from the Laboratory of Molecular Radio-oncology, Clinic and Polyclinic for Radiation Therapy and Radio-oncology for technical assistance regarding LTC-IC assays.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/9/1462
References
- 1.Shastri A, Will B, Steidl U, Verma A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129(12): 1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129(12):1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medyouf H. The microenvironment in human myeloid malignancies: emerging concepts and therapeutic implications. Blood. 2017;129(12):1617–1626. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder T, Geyh S, Germing U, Haas R. Mesenchymal stromal cells in myeloid malignancies. Blood Res. 2016;51(4):225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kode A, Manavalan JS, Mosialou I, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506(7487):240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464(7290):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frisch BJ, Ashton JM, Xing L, et al. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood. 2012;119(2):540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanoun M, Zhang D, Mizoguchi T, et al. Acute myelogenous leukemia-induced sympathetic neuropathy promotes malignancy in an altered hematopoietic stem cell niche. Cell Stem Cell. 2014;15(3):365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schepers K, Pietras EM, Reynaud D, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13(3):285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Zambetti NA, Bindels EM, et al. Massive parallel RNA sequencing of highly purified mesenchymal elements in low-risk MDS reveals tissue-context-dependent activation of inflammatory programs. Leukemia. 2016;30(9):1938–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer RA, Wobus M, List C, et al. Mesenchymal stromal cells from patients with myelodyplastic syndrome display distinct functional alterations that are modulated by lenalidomide. Haematologica. 2013;98(11):1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geyh S, Oz S, Cadeddu RP, et al. Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia. 2013;27(9):1841–1851. [DOI] [PubMed] [Google Scholar]
- 13.Geyh S, Rodriguez-Paredes M, Jager P, et al. Functional inhibition of mesenchymal stromal cells in acute myeloid leukemia. Leukemia. 2016;30(3):683–691. [DOI] [PubMed] [Google Scholar]
- 14.Medyouf H, Mossner M, Jann JC, et al. Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell. 2014;14(6):824–837. [DOI] [PubMed] [Google Scholar]
- 15.von der Heide EK, Neumann M, Vosberg S, et al. Molecular alterations in bone marrow mesenchymal stromal cells derived from acute myeloid leukemia patients. Leukemia. 2017;31(5):1069–1078. [DOI] [PubMed] [Google Scholar]
- 16.Schroeder T, Czibere A, Zohren F, et al. Meningioma 1 gene is differentially expressed in CD34 positive cells from bone marrow of patients with myelodysplastic syndromes with the highest expression in refractory anemia with excess of blasts and secondary acute myeloid leukemia. Leuk Lymphoma. 2009;50(6):1043–1046. [DOI] [PubMed] [Google Scholar]
- 17.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruns I, Cadeddu RP, Brueckmann I, et al. Multiple myeloma-related deregulation of bone marrow-derived CD34(+) hematopoietic stem and progenitor cells. Blood. 2012;120(13):2620–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi T, Hideshima T, Nguyen AN, et al. Transforming growth factor beta receptor I kinase inhibitor down-regulates cytokine secretion and multiple myeloma cell growth in the bone marrow microenvironment. Clin Cancer Res. 2004;10(22):7540–7546. [DOI] [PubMed] [Google Scholar]
- 23.Zhou L, Nguyen AN, Sohal D, et al. Inhibition of the TGF-beta receptor I kinase promotes hematopoiesis in MDS. Blood. 2008;112(8):3434–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiyama T, Matsunaga T, Terui T, et al. Involvement of transforming growth factor-beta and thrombopoietin in the pathogenesis of myelodysplastic syndrome with myelofibrosis. Leukemia. 2005;19(9):1558–1566. [DOI] [PubMed] [Google Scholar]
- 25.Zorat F, Shetty V, Dutt D, et al. The clinical and biological effects of thalidomide in patients with myelodysplastic syndromes. Br J Haematol. 2001;115(4):881–894. [DOI] [PubMed] [Google Scholar]
- 26.Blank U, Karlsson S. TGF-beta signaling in the control of hematopoietic stem cells. Blood. 2015;125(23):3542–3550. [DOI] [PubMed] [Google Scholar]
- 27.Garbe A, Spyridonidis A, Mobest D, et al. Transforming growth factor-beta 1 delays formation of granulocyte-macrophage colony-forming cells, but spares more primitive progenitors during ex vivo expansion of CD34+ haemopoietic progenitor cells. Br J Haematol. 1997;99(4):951–958. [DOI] [PubMed] [Google Scholar]
- 28.Sitnicka E, Ruscetti FW, Priestley GV, Wolf NS, Bartelmez SH. Transforming growth factor beta 1 directly and reversibly inhibits the initial cell divisions of long-term repopulating hematopoietic stem cells. Blood. 1996;88(1):82–88. [PubMed] [Google Scholar]
- 29.Krause DS, Fulzele K, Catic A, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19(11):1513–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. [DOI] [PubMed] [Google Scholar]
- 31.Bhagat TD, Chen S, Bartenstein M, et al. Epigenetically aberrant stroma in MDS propagates disease via Wnt/beta-catenin activation. Cancer Res. 2017;77(18):4846–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandran P, Le Y, Li Y, et al. Mesenchymal stromal cells from patients with acute myeloid leukemia have altered capacity to expand differentiated hematopoietic progenitors. Leuk Res. 2015;39(4):486–493. [DOI] [PubMed] [Google Scholar]
- 33.Kim JA, Shim JS, Lee GY, et al. Microenvironmental remodeling as a parameter and prognostic factor of heterogeneous leukemogenesis in acute myelogenous leukemia. Cancer Res. 2015;75(11): 2222–2231. [DOI] [PubMed] [Google Scholar]
- 34.Bachegowda L, Gligich O, Mantzaris I, et al. Signal transduction inhibitors in treatment of myelodysplastic syndromes. J Hematol Oncol. 2013;6:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baron F, Suciu S, Amadori S, et al. Value of infliximab (Remicade®) in patients with low-risk myelodysplastic syndrome: final results of a randomized phase II trial (EORTC trial 06023) of the EORTC Leukemia Group. Haematologica. 2012;97(4):529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deeg HJ, Gotlib J, Beckham C, et al. Soluble TNF receptor fusion protein (etanercept) for the treatment of myelodysplastic syndrome: a pilot study. Leukemia. 2002;16(2):162–164. [DOI] [PubMed] [Google Scholar]
- 37.Isufi I, Seetharam M, Zhou L, et al. Transforming growth factor-beta signaling in normal and malignant hematopoiesis. J Interferon Cytokine Res. 2007;27(7):543–552. [DOI] [PubMed] [Google Scholar]
- 38.Bhagat TD, Zhou L, Sokol L, et al. miR-21 mediates hematopoietic suppression in MDS by activating TGF-beta signaling. Blood. 2013;121(15):2875–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, McMahon C, Bhagat T, et al. Reduced SMAD7 leads to overactivation of TGF-beta signaling in MDS that can be reversed by a specific inhibitor of TGF-beta receptor I kinase. Cancer Res. 2011;71(3): 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suragani RN, Cadena SM, Cawley SM, et al. Transforming growth factor-beta superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat Med. 2014;20(4):408–414. [DOI] [PubMed] [Google Scholar]
- 41.Iancu-Rubin C, Mosoyan G, Wang J, et al. Stromal cell-mediated inhibition of erythropoiesis can be attenuated by Sotatercept (ACE-011), an activin receptor type II ligand trap. Exp Hematol. 2013;41(2):155–166.e17. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Eksioglu EA, Zhou J, et al. Induction of myelodysplasia by myeloid-derived suppressor cells. J Clin Invest. 2013;123(11):4595–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.