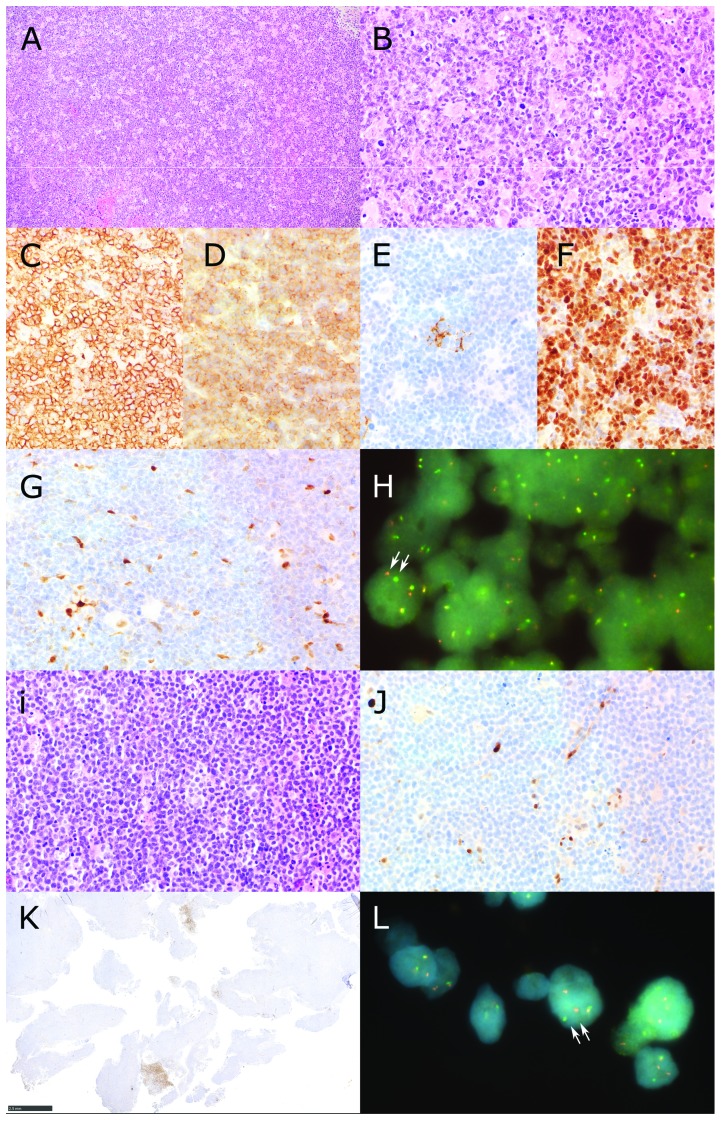
The t(11;14) translocation that juxtaposes the Cyclin D1 (CCND1) gene to the immunoglobulin heavy chain gene is considered a hallmark of mantel cell lymphoma (MCL).1 Immunohistochemical (IHC) analysis of CCND1 expression in formalin-fixed paraffin-embedded (FFPE) tissue sections of material from patients with suspected MCL is therefore the most obvious and effective diagnostic tool.2 We identified two cases of MCL based on morphology, expression of CD20, CD5 and SOX11 and absence of CD23 expression. Notably, both cases were negative for nuclear CCND1 staining by IHC (Figure 1G,J), despite being positive for the t(11;14)(q13;q32) translocation using fluorescence in situ hybridization (FISH, Figure 1H,L). Patient A was a 81-year-old female presenting with Ann Arbor stage IIIA. After six cycles of R-CHOP (rituximab, clorambucil, vincristine, prednisolone) and two additional cycles of rituximab, she achieved complete remission until last follow up 15 months after initial diagnosis. Patient B was an 87-year-old female with lymphadenopathy on both sides of the diaphragm, but staging remained incomplete. Due to comorbidities and patients’ refusal of chemotherapy, rituximab mono-therapy was initiated until last follow up after two months. Surgical excision biopsies of an inguinal (patient A) and cervical (patient B) lymph node were submitted for consultation. In order to investigate if the lack of CCND1 immunostaining was due to low CCND1 expression, we performed quantitative PCR analysis of CCND1 expression in total RNA from patient A and patient B compared to CCND1 expression observed in two classical CCND1-positive MCL cases and two normal lymph nodes. The expression of CCND2 was also investigate as it is known that CCND2 can compensate for CCND1 in some cases.3 As shown in Figure 2A, both patients have CCND1 expression levels similar or higher to that observed in classical MCL samples and much higher than the expression observed in normal lymph nodes. These data are in line with the presence of a t(11;14) translocation in biopsies of the two patients and suggest that the lack of CCND1 immunostaining in patient A and patient B is not due to a defect in mRNA expression.
Figure 1.
Identification of CCND1-negative MCL Patients. Patient A (panels A-H) showed a diffuse infiltrate of slightly pleomorphic centrocytes (A and B Hematoxilin and Eosin) with expression of CD20 (C), CD5 (D), absence of CD23 but scattered follicular dendritic cells (E) and expression of SOX11 (F). The lymphoma was high proliferative with Ki67 positivity up to 60%. Staining for Cyclin D1 (CCND1) revealed internal positive controls (macrophages and endothelial cells) but no staining of lymphoma cells (G). Fluorescence in situ hybridization using a break apart probe shows that the CCND1 gene of patient A has a chromosomal rearrangement (H). A cell nucleus where the segregation of the two fluorescent probes (red and green) is clearly visible is indicated by two arrows. Patient B (panels I-L) presented with a diffuse infiltrate with a classical cytology of mantle cell lymphoma (I, Hematoxilin and Eosin). There was expression of CD20, CD5 and SOX11 (data not shown). Also this patient presented a high proliferative lymphoma with Ki67 postivity of 70%. Expression of CCND1 was not detectable despite internal positive controls (macrophages and endothelial cells, J) except for the presence of small areas in the whole lymphomatous tissue containing CCND1 positive lymphatic cells with pleomorphic cytology (K). This could indicate intratumoral clonal heterogeneity in this lymphoma case affecting, for unknown reason, CCND1 reactivity. (L) Fluorescence in situ hybridization using a break apart probe shows that the CCND1 gene of patient B has a chromosomal rearrangement. A cell nucleus where the segregation of the two fluorescent probes (red and green) is clearly visible is indicated by two arrows (Original magnification A: 100×, B-G, i,J: 400×, H, L 1000×, K. 20×).
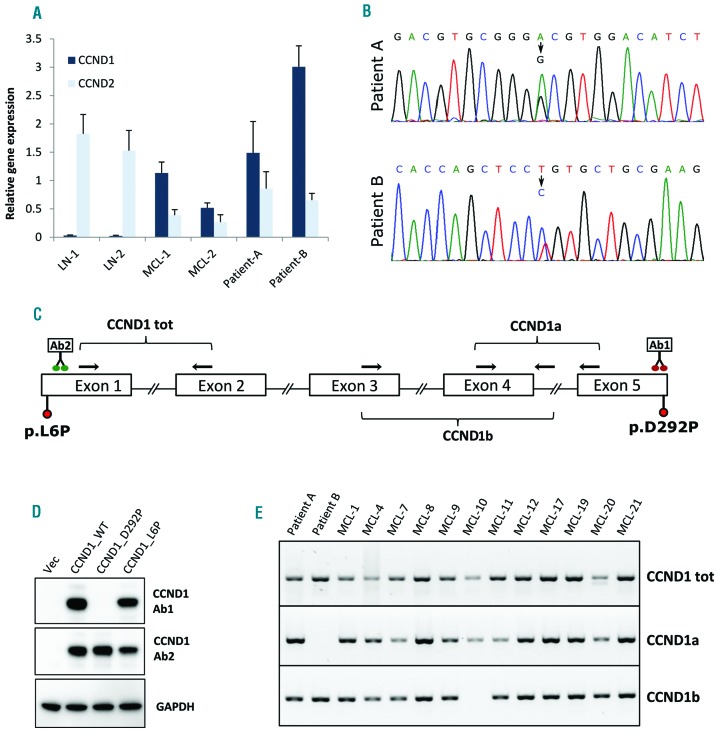
Figure 2.
Molecular analysis of CCND1 expression in MCL cases. (A) Analysis of CCND1 and CCND2 mRNA expression. RNA was extracted from FFPE material from two lymph nodes (LN1 & 2), two MCL cases positive for CCND1 expression in IHC (MCL-1 and 2) and from the two patients investigated in this study (patients A and B). After reverse transcription, cDNAs were analyzed by quantitative PCR using CCND1 and CCND2 specific primer assays (QIAGEN). GUSB was used as reference gene. (B) Sanger sequencing peaks showing the mutations identified in the two patients investigated in this study. (C) Schematic depiction of the CCND1 gene with the exon/intron structure, the position of the mutations identified in patients A and B and the position of the primers used to analyze the expression of the CCND1a and CCND1b isoformes. The location of the putative binding sites of the antibodies used in this study is also indicated (Ab1: Thermo Fisher Scientific, clone SP4; Ab2: Abcam, EP272Y). (D) Total lysates from HEK 293 cells transfected with plasmids expressing either wild type or mutated CCND1 were resolved on SDS PAGE and analyzed by western blot using two different antibodies against CCND1 (Ab1 and Ab2 as above) and against GAPDH as an internal reference protein. Cells transfected with an empty vector were also analyzed (Vec). (E) PCR analysis of CCND1 isoform expression in FFPE extracted RNA from patients A and B and from 12 CCND1-positive MCL samples. One microliter of cDNA was amplified in a 20 μl reaction containing 4 mM MgCl2, 1 X Q solution (Qiagen), 0.25 mM dNTPs, 0.25 μM each primer, 1.5 Units of AmpliTaq Gold™ DNA Polymerase (ThermoFisher Scientific) and 1 X reaction buffer II with 60 °C as annealing temperature and 45 cycles. For the amplification of total CCND1 we used primers D1EX1_fw and D1EX2_rev2, annealing respectively to exon 1 and exon 2 of the CCND1 gene. For the amplification of the CCND1a isoform we used primers D1EX4_fw2 and D1EX5_rev, annealing respectively to exon 4 and exon 5 of the CCND1 gene. For the amplification of the CCND1b isoform we used primers D1EX3_fw2 and D1EX4_rev, annealing respectively to exon 3 and to the beginning of intron 4 of the CCND1 gene. The sequence of the primers used is provided in Online Supplementary Table S1.
One possible explanation for the lack of CCND1 immunostaining in patient A and patient B, even in the presence of high CCND1 mRNA expression, is the presence of amino acid substitutions affecting CCND1 stability/folding or recognition from the antibody used in routine IHC. DNA form FFPE sections of both patients was extracted and each of the 5 exons of the CCND1 gene was PCR amplified and subject to Sanger sequencing. This analysis led to the identification of two different mutations in the CCND1 gene of the two patients (Figure 2B,C). In patient B, we found a T to C transition in position chr11:69,456,098 (GRCh37/hg19; NM_053056.2: c.17T>C) resulting in the change of a Leucine in position 6 to a Proline (NP_444284.1: p.L6P). This mutation has previously been described in a MCL case.4 According to the Combined Annotation Dependent Depletion (CADD) tool, the mutation is deleterious with a PHRED-scaled C-score of 26.5. In patient A, we identified an A to G transition in position chr11:69,466,037 (GRCh37/hg19; NM_053056.2: c.875A>G) resulting in the change of an Aspartic acid in position 292 to a Proline (NP_444284.1: p.D292P). This mutation affects a highly conserved amino acid in the C-terminal region of CCND1 and is also predicted to be deleterious with a PHRED-scaled C-score of 31. Mutations in D292 of CCND1 to date (February 2018) were not described in the COSMIC mutation database nor in the ICGC Data Portal, but a mutation in position 290 (p.V290G) was found in another MCL case before,5 and a mutation in position 291 (p.R291W) has been described in colorectal carcinoma.6 Mutations in residues T286 and P287 have been also documented in endometrial, melanoma, cervical and colorectal carcinomas, and were suggested to increase CCND1 stability and nuclear localization.7
In order to understand whether the mutations could explain the lack of immunostaining observed in patient A and patient B by IHC, we cloned the two CCND1 variants, together with WT CCND1 in a mammalian expression vector. All CCND1 variants were transfected in HEK 293 cells to achieve high expression level. Following transfection, cell extracts were analyzed by western blot in order to assess the expression of the different CCND1 protein variants. Interestingly, using the antibody currently used in routine diagnostic (Ab1), although we could readily detect a band of the molecular weight of CCND1 in extracts of cells transfected with WT and L6P mutated CCND1, no band was detected in extracts from cells transfected with the D292P mutant (Figure 2D). This was not due to a lack of CCND1 expression as a second antibody, directed against another region of CCND1 (Ab2), could recognize the CCND1 mutant in the same cell extracts. We conclude that mutation of Asp292, lying in the C-terminal end of CCND1, but not mutation of Leu6, lying in the N-terminal part of CCND1, impairs the binding of the CCND1 antibody currently used in routine diagnostic. This is in agreement with the fact that the antibody used in routine diagnostic (Thermo Fisher Scientific, Clone SP4) was produced using as immunogen a synthetic peptide derived from the C-terminus of CCND1. Unfortunately, the second antibody (Abcam EP272Y directed against the N-terminal part of CCND1) did not provide reliable IHC staining on FFPE tissue sections.
The data presented above clearly explain the lack of immunostaining observed in patient A by IHC, but do not explain why material from patient B is not reactive to the CCND1 antibody, given that the CCND1 variant p.L6P expressed by patient B is readily detected by the antibody in our western blot analysis (Figure 2, panel D). If the reason for the negative immunostaining observed in patient B was not lying in a structural defect of the CCND1 protein, we postulated that patient B could have aberrant CCND1 expression. The CCND1 gene is known to be subject to alternative splicing resulting in the expression of two different isoforms, CCND1a and CCND1b.8–10 Contrary to isoform CCND1a that is the result of the assembly of all five exons of the CCND1 gene (Figure 2C), isoform CCND1b derives from a splicing event skipping exon 5 and including part of intron 4. Given that the last 50 amino acids of the C-terminal region of the CCND1 protein are encoded in exon 5, the expression of isoform CCND1b should give rise to a protein that is missing the epitope recognized by the antibody used in routine CCND1 analysis by IHC. We therefore asked if patient B could have a defect in the expression of isoform CCND1a, a rare event previously described in MCL.11 If that were the case, we would also expect that other MCL cases, that are positive for CCND1 immunostaining in routine IHC, should all express the CCND1a isoform to some extent. To test our hypothesis we isolated RNA from FFPE material from 20 CCND1-positive MCL cases and performed RT-PCR analysis with isoform-specific primer sets (Figure 2C and Online Supplementary Table S1 for primers sequences). Given that RNA from FFPE material can result in poor performance in PCR analyses, we initially screened the samples for total CCND1 expression using primers designed in the first two exons of the CCND1 gene, as these exons are common to both isoforms. Of the 20 cases, only 12 showed a good signal for total CCND1 expression and were further analyzed for the expression of the two isoforms together with RNA from patients A and B. Strikingly, as shown in Figure 2E and in Online Supplementary Figure S1, the RNA from patient B was the only one among all samples analyzed that was completely missing the expression of the CCND1a isoform. No correlation with the genotype of the patient at the A870G polymorphism was observed, and no other mutation in splicing regulatory elements of the intron 4 were found in patient B. Nevertheless, based on this result, we speculate that the lack of immunostaining observed in patient B by IHC might be due to the fact that the CCND1 protein expressed by patient B, by missing the last 50 amino acids of the full length protein, is also missing the epitope recognized by the antibody used for the screening of CCND1 in routine diagnostic. Given that CCND1 variants, either carrying mutations in the C-terminal domain or lacking exon 5, have been shown to have increased oncogenic properties,12–14 we speculate that t(11;14)-positive MCL cases negative for CCND1 IHC staining could have a particularly aggressive behavior.
In summary, we describe for the first time MCL lacking CCND1 reactivity by IHC despite the presence of CCND1 translocations and provide possible molecular explanations for the observed phenomenon. The cases described present a potential pitfall in lymphoma diagnostics, since screening for MCL is usually done by IHC for CCND1. Our findings illustrate that testing for CCND1 translocations by FISH, together with SOX11 expression, might be useful in lymphomas morphologically and immunophenotypically resembling MCL but lacking CCND1 immunoreactivity by IHC in order not to miss these rare variants of MCL.
Supplementary Material
Acknowledgments
We thank Friederike Poche-de Vos, Gabriele Richartz and Reinhardt Golz for providing tissue specimen, Dana Germer and Reina Zühlke-Jenisch for excellent technical assistance. We also thank the Friedrich-Naumann-Stiftung für die Freiheit for supporting L.A.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th edition. IACR; 2017;285–290. [Google Scholar]
- 2.Klapper W. Histopathology of mantle cell lymphoma. Semin Hematol. 2011;48(3):148–154. [DOI] [PubMed] [Google Scholar]
- 3.Salaverria I, Royo C, Carvajal-Cuenca A, et al. CCND2 rearrangements are the most frequent genetic events in cyclin D1(-) mantle cell lymphoma. Blood. 2013;121(8):1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. [DOI] [PubMed] [Google Scholar]
- 5.Beà S, Valdés-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110(45):18250–18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannakis M, Mu XJ, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15(4):857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alt JR, Cleveland JL, Hannink M, et al. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14(24):3102–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betticher DC, Thatcher N, Altermatt HJ, et al. Alternate splicing pro duces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. [PubMed] [Google Scholar]
- 9.Knudsen KE, Diehl JA, Haiman CA, et al. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–1628. [DOI] [PubMed] [Google Scholar]
- 10.Marzec M, Kasprzycka M, Lai R, et al. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006; 108(5):1744–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrère N, Belaud-Rotureau M-A, Dubus P, et al. The relative levels of cyclin D1a and D1b alternative transcripts in mantle cell lymphoma may depend more on sample origin than on CCND1 polymorphism. Haematologica. 2005;90(6):854–856. [PubMed] [Google Scholar]
- 12.Solomon DA, Wang Y, Fox SR, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278(32):30339–30347. [DOI] [PubMed] [Google Scholar]
- 13.Gladden AB, Woolery R, Aggarwal P, et al. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2005;25(7):998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda Y, Oda K, Hiraike-Wada O, et al. Cyclin D1 harboring the T286I mutation promotes oncogenic activation in endometrial cancer. Oncol Rep. 2013;30(2):584–588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.