Abstract
Objective
In this systematic review, we present the molecular epidemiology and knowledge gaps of the carbapenem resistance in East Africa as well as the future probable research interventions that can be used to address the emergence of carbapenem resistance in the region.
Results
The 17 articles which presented concrete information about the prevalence of carbapenem resistance in East Africa were reviewed. Tanzania exhibited the highest level of carbapenem resistance at 35% while DRC had the lowest level at 0.96%. Uganda was the only country with studies documenting CR obtained amongst hospital environment isolates with incidence ranging from 21% in Pseudomonas aeruginosa to 55% in Acinetobacter baumannii. Carbapenem resistance was more exhibited in A. baumannii (23%), followed by P. aeruginosa (17%), Klebsiella pneumoniae (15%), Proteus mirabilis (14%) and Escherichia coli (12%) mainly isolated from respiratory tract, blood, urine and wound/pus. The regional genetic determinants of carbapenem resistance detected were blaIMP, blaVIM-1 blaSPM-l, blaNDM-1, blaOXA-23 blaOXA-24, blaOXA-58 and blaKPC.
Electronic supplementary material
The online version of this article (10.1186/s13104-018-3738-2) contains supplementary material, which is available to authorized users.
Keywords: East Africa, Molecular epidemiology, Carbapenem resistance
Introduction
In the recent past, carbapenems were potent against all multiple drug resistant (MDR) Gram negative bacteria and in combination with their negligible toxicity to the host, carbapenems became the preferred last resort antibiotics for the treatment of MDR Gram negative bacterial infections. Development of carbapenem resistance (CR) in Enterobacteriaceae is of great concern because there is no obvious next line of antibiotics to use against carbapenemase producing (CP) Enterobacteriaceae [1]. MDR has left less efficient antibiotics to take care of these expensive hard to treat life threatening infections [2–6].
Currently, the high prevalence of carbapenem resistant Enterobacteriaceae (CRE) isolates world over most importantly in Klebsiella pneumoniae and Escherichia coli isolates in hospitals, community-associated infections and animals is a huge burden to the health care system [3, 5–12], Additional file 2: Table S6. Genetic determinants of CR have been classified into: Ambler class A beta lactamases which include; KPC, GES/IBC, SME, NMC-A, IMI and SFC [12–15], Ambler class B beta lactamases which are termed as Metallo beta Lactamases consisting of NDM, VIM, IMP, SPM, GIM, SIM, KHM, AIM, DIM, SMB, TMB and FIM [7, 13]. IMP, VIM and NDM plasmid mediated Metallo beta lactamases are of worldwide occurrence possibly because the genes that code for them are located on mobile genetic elements [13] and carbapenem hydrolyzing class D beta lactamases (CHDLs) encompass various group of oxacillinases (OXA) with hydrolytic activity of amino and carboxy penicillins [16], Additional file 2: Tables S3–S5.
Studies have reported the existence of CP bacteria in East Africa but in general, there is no comprehensive data about the molecular epidemiology of CP organisms and its burden on the health care system [17, 18]. Furthermore, there is scanty information about CR prevalence in East African livestock yet MDR genes were observed in livestock commensal bacteria which are probably transmitted to humans through the food chain [19–21].
Comprehending the current status of CR throughout East Africa will influence decision making among stakeholders about the rational use of carbapenems. Therefore, this systematic review expounds the current molecular epidemiology of CP bacteria in East Africa, highlighting the carbapenemases genes, CR Knowledge gap and future research interventions to address CR in East Africa.
Main text
Methods
Literature review
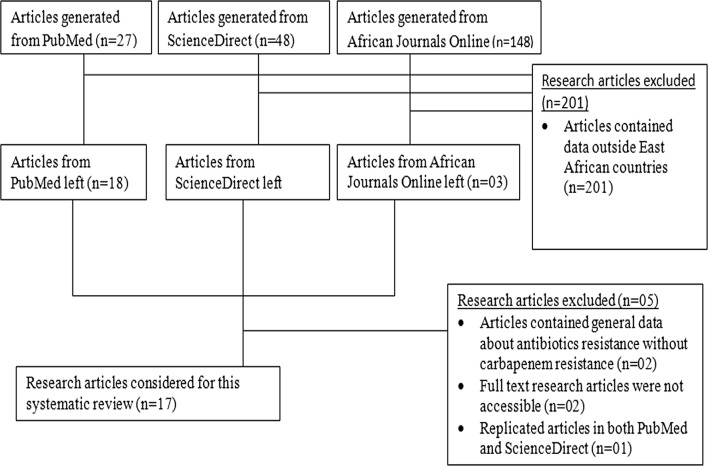
PubMed, ScienceDirect and African Journals Online databases were searched from March to December 2017. The search key words used were carbapenem resistance in East Africa to extract articles published only in English from 2008 onwards in an attempt to include up to date relevant CR data, Fig. 1.
Fig. 1.
Selection process of research articles for inclusion in this systematic review
Study selection criteria
Only full text research articles reporting the prevalence of CP bacteria isolated from patients and hospital environment in East African countries namely; Kenya, Uganda, Tanzania, Burundi, Rwanda, Ethiopia, Democratic Republic of Congo (DRC) and South Sudan were used. Only Studies elaborating bacteria study population, pathogens identified, phenotypic and genotypic methods used to detect CR were used. Patients’ populations of all ages were included while case reports and review articles were excluded from this systematic review as it has become conventional [22].
Data extraction
A database was created in which study location, publication year, sample collection period, bacterial species isolated, number of isolates tested for CR, CR prevalence, carbapenemase genes, methods used to identify resistant isolates and to type CR genetic determinants were included, Table 1.
Table 1.
Review of East Africa based carbapenem resistance studies
| Location | Number of isolates | CR isolates | CR prevalence (%) | Carbapenemase genes | Organism | Period | Methods used | Refs |
|---|---|---|---|---|---|---|---|---|
| Kenya | 416 | 57 | 13.7 | VIM-2 | P. aeruginosa | Jan 2006–Jun 2007 | PCR, PFGE and sequencing | [23] |
| Kenya | > 100 | 7 | – | NDM-1 | K. pneumoniae | 2007–2009 | PCR, PFGE and sequencing | [24] |
| Kenya | – | 16 | – | NDM-1 | A. baumannii | Jan 2009–Aug 2010 | PCR, PFGE, Sequencing and MBL Etest strips | [25] |
| Kenya | 190 | 44 | 23.2 | – | K. pneumoniae | 2002–2013 | Disk diffusion | [26] |
| Kenya | 195 | 25 | 12.8 | NDM-1 like | K. pneumoniae | 1994–2017 | WGS | [27] |
| Kenya | 17/219 | 1/219 or 1/17 | 0.5/5.9 | – | K. pneumoniae | 2010 | Disk diffusion | [28] |
| Kenya | 42 | 3 | 7 | – | E. coli | – | Disk diffusion | [29] |
| Uganda | 196 | 44 | 22.4 | OXA-48, IMP, KPC and NDM-1 | K. pneumoniae. E. coli, Enterobacter spp, Serratia marcescens, Proteus mirabilis, Citrobacter freundii, Klebsiella oxytoca and Pantoea agglomerans | Jan 2013–Mar 2014 | PCR. Disk diffusion and Modified Hodge test | [30] |
| Uganda | 658 | 68 | 10.3 | VIM and OXA-48 | E. coli, K. pneumoniae, Proteus mirabilis, Salmonella spp, Morganella morganii, Enterobacter sakazaki and Stenotrophomonas spp | Sept 2013–Jun 2014 | PCR and disk diffusion | [31] |
| Uganda | 869 (clinical) | 10 | 1.2 (10/869) 24 (10/42) |
IMP-like, VIM-like, SPM-like and NDM-1-like | P. aeruginosa (42/658 = 5%) | Feb 2007–Sep 2009 | PCR Phoenix Automated Microbiology System |
[32] |
| 9 | 1.1 (9/869) 31 (9/29) |
OXA-23,24, 58 like and VIM-like | A. baumannii (29/658 = 3%) | |||||
| 80 (environmental) | 15 | 18.8 (15/80) 33 (15/46) |
IMP-like, VIM-like, SPM-like and NDM-1-like | P. aeruginosa 57.5% (46/80) | ||||
| 6 | 7.5 (6/80) 55 (6/11) |
OXA-23,24, 58 like and VIM-like | A. baumannii 14% (11/80) | |||||
| Uganda | 736 (clinical) | 3 | 0.41 (3/736) 33 (3/9) |
– | P. aeruginosa (9/736 = 1.2%) | Sept 2012–Oct 2013 | Rep-PCR and disk diffusion | [33] |
| 1 | 0.14 (1/736) 14 (1/7) |
– | A. baumannii (7/736 = 0.95%) | |||||
| 100 (environmental) | 7 | 7 (7/100) 21 (7/33) |
– | P. aeruginosa (33/100 = 33%) | ||||
| 6 | 6 (6/100) 46 (6/13) |
– | A. baumannii (13/100 = 13%) | |||||
| Tanzania | 90 | 8 | 8.9 | VIM-2 | P. aeruginosa | May 2010–Jul 2011 | Sequencing, PGFE and Disc diffusing | [34] |
| Tanzania | 227 | 80 | 35 | VIM-, IMP-, NDM-KPC, OXA48 | K. pneumoniae, P. aeruginosa, E. coli, K. oxytoca A. baumannii, Citrobacter freundii, Serratia marcescens and Salmonella spp | 2007–2012 | PCR and disk diffusion | [35] |
| Rwanda | 55/154 | 5 in 154 or 5/55 | 2.9/8 | – | E. coli | Jul–Dec 2013 | Disk diffusion | [36] |
| Ethiopia | 33 | 4 | 12.1 | – | K. pneumoniae and Morganella morgani | Jan–Mar 2014 | Disc diffusion and Modified Hodge test |
[37] |
| Ethiopia | 267 | 5 | 1.87 | – |
K. pneumoniae
E. coli |
Dec 2012 | [38] | |
| DRC | 104/643 | 1 in 643 or 1/104 | 0.2/0.96 | – | Enterobacter spp. | Sept 2012–Aug 2013 | Disk diffusion | [39] |
Data analysis
Data analysis was performed using one-way ANOVA in XLSTAT version 2018.1 to establish the most prevalent carbapenem resistant bacteria type and their distribution variability within body systems. A P value of ≤ 0.05 indicated significant statistical difference.
Results
The search conducted between January and December 2017 generated 223 research articles; PubMed, Sciencedirect and African Journals Online liberated 27, 48 and 148 respectively. Using article abstracts and titles, 201 articles were excluded from this systematic review. Only 20 full text articles were accessible out of the 22 papers. Of the remaining 20 manuscripts, 17 presented concrete information about molecular epidemiology of CR in East Africa and consequently included in this review, Fig. 1. The search generated four manuscripts from Uganda, seven from Kenya, two from Ethiopia, and Tanzania, one from DRC and Rwanda, Table 1. Neither articles from Burundi nor from South Sudan met inclusion criteria for this systematic review. All studies were epidemiological hospital based cross sectional in nature and majority illustrated the prevalence and genetic determinants of CR as well as the methods employed to detect CP isolates, Table 1.
Resistance patterns
Clinical isolates
According to the molecular and antibiotics susceptibility assays employed in the articles incorporated into this systematic review, Tanzania exhibited the highest level of CR among enteric clinical isolates at 35% while DRC had the lowest level at 0.96% [35, 38], Table 1.
Hospital environment isolates
Uganda was the only regional country with two studies documenting CR obtained amongst hospital environment isolates [32, 33]. These studies reported hospital environment CR prevalence ranging from 21% in P. aeruginosa to 55% in A. baumannii, Table 1.
Body system harboring CP isolates
Only 12 articles analyzed in this review had detailed information about the samples from which CP bacteria were isolated, Table 2. The mean sample wise CP bacteria distribution was highest in respiratory tract samples (23%), followed by blood (22%), urine (19%), wound/pus (18%), stool and peritoneol fluid (10%), other samples (7%), ear swabs (6%) and cerebral fluid (3%), Additional file 1: Table S1. Seven studies reported CR in urine and blood isolates with prevalence ranging from 0.96% (DCR) to 39.2% (Tanzania) and 7% (Kenya) to 36.36% (Tanzania) respectively. Six articles documented respiratory tract CP bacteria with occurrence varying from 3.45% (Uganda) to 55.6% (Kenya) while five articles displayed CR in pus/wound isolates with a resistance incidence ranging from 7.14% (Uganda) to 33.04% (Tanzania), Table 2.
Table 2.
Sample wise distribution of CR bacteria isolates
| Country | Sample source | CR prevalence | Species | Refs |
|---|---|---|---|---|
| Kenya | Urine Blood Wounds Respiratory tract specimens various other specimens |
3/57 = 5% (urine) 4/57 = 7% (blood) 17/57 = 30% (wound/pus) 30/47 = 53% (respiratory) 3/57 = 5% (various other specimens) |
P. aeruginosa | [22] |
| Kenya | Blood (190) | 44/190 = 23.2% | K. pneumoniae | [24] |
| Kenya | Respiratory tract specimens Bone marrow aspirate cerebrospinal fluid Catheter tip Axillary swab Nasal swab Urine Blood Debrided tissue samples |
10/16 = 55.6% (respiratory) – – – – – – – – |
A. baumanii | [25] |
| Kenya | Blood (195) | 25/195 = 12.8% | K. pneumoniae | [27] |
| Kenya | Urine (121) | 1/17 = 5.9% | K. pneumoniae | [28] |
| Uganda | Urine Blood Stool Wound/pus Peritoneol fluid Others |
23% 27% 18% 14% 10% 8% |
E. coli, K. pneumoniae, Proteus mirabilis, Salmonella spp, Morganella morganii, Enterobacter sakazaki and Stenotrophomonas spp | [31] |
| Uganda | Blood (51), cerebral spinal fluid (49), Tracheal aspirates (163), Ear swabs (197), Sputum (204), Urine catheters (98) and Pus (107) | 3/42 = 7.14% (wound/Pus) 3/42 = 7.14% (Sputum) 3/42 = 7.14% (Tracheal) 1/42 = 2.4% (Ear swab) |
P. aeruginose | [32] |
| 4/29 = 13.8% (Tracheal) 3/29 = 10.35% (Ear swap) 1/29 = 3.45% (Sputum) 1/29 = 3.45% (Cerebral Spinal fluid) |
A. baumanii | |||
| Tanzania | Blood and pus | 5/90 = 5.6% Wound/Pus 3/90 = 3.3% Blood |
P. aeruginose | [34] |
| Tanzania | Pus (112), urine (56), blood (55), aspirate (3), and sputum (1). | 22/56 = 39.29% (Urine) 20/55 = 36.36% (Blood) 37/112 = 33.04% (wound/Pus) |
K. pneumoniae, P. aeruginosa, E. coli, K. oxytoca A. baumannii, Citrobacter
freundii, Serratia marcescens and Salmonella spp |
[35] |
| Ethiopia | Urine (24) Blood (9) |
4/24 = 17% (urine) 0/9 = 0% (blood) |
K. pneumoniae and Morganella morgani | [37] |
| Ethiopia | Feces (267) | 5/267 = 2% (feces) |
K. pneumoniae
E. coli |
[38] |
| DRC | Urine (104) | 1/104 = 0.96% (urine) | Enterobacter spp | [39] |
Distribution of CR among MDR enteric bacteria
One-way ANOVA displayed that distribution of CR among different bacteria species was not significantly different (P-value = 0.11 > 0.05). CR prevalence was highest in A. baumannii with an average of 23% followed by P. aeruginosa (17%), K. pneumonia (15%), P. mirabili (14%), E. coli (12%), C. freundii (8%), K. oxytoca (2%), M. morganii (2%), Salmonella spp E. sakazaki and Stenotrophomonas spp (1%). However, the most reported CP isolate across the region was K. pneumoniae (8 studies) followed by E. coli and P. aeruginosa (6 studies), A. baumannii (4 articles), M. morganii and Salmonella spp (2 articles), C. freundii, K. oxytoca and P. mirabilis (2 articles), E. sakazaki and Stenotrophomonas spp (1 article), Additional file 1: Table S2.
Prevalence of CR genetic determinants in East Africa
Uganda
CR genetic determinants in non-glucose fermenting bacteria reported at Mulago hospital were blaIMP-like (36%), blaVIM-like (32%), blaSPM-like (16%), blaNDM-1-like (4%) for P. aeruginosa and blaOXA-23-like (60%), blaOXA-24-like (7%), blaOXA-58-like (13%), and blaVIM-like (13%) for A. baumannii [32]. Carbapenemase genes in CRE at Mulago and Mbarara hospitals were also documented [30, 31]. At Mulago, the genes characterized included; blaVIM (10.7%), followed by blaOXA-48 (9.7%), blaIMP (6.1%), blaKPC (5.1%) and blaNDM-1 (2.6%). The highest number of genes appeared in Klebsiella pneumoniae (52.2%), followed by E. coli (28.4%), Enterobacter spp (7.5%), Serratia marcescens (4.5%), Proteus mirabilis (3.0%), Citrobacter freundii, Klebsiella oxytoca, and Pantoea agglomerans at 1.5% each while at Mbarara hospital, VIM and OXA-48 CR determinants were registered, Table 1.
Tanzania
Molecular analysis of CRE at a tertiary hospital in Mwanza established by multiplex PCR revealed that the principal CR genes were IMP (21.6%), followed by VIM (12.3%), OXA-48 (4.9%), then KPC (3.5%), and NDM (3.1%). CP E. coli had the highest prevalence (14%), followed by K. pneumoniae (10.57%), trailed by P. aeruginosa (10.13%), then Klebsiella oxytoca (1.76%), A. baumannii (1.3%), C. freundii (0.88%), Serratia marcescens (0.88%) and Salmonella spp. (0.44%) [35] while CP P. aeruginosa harbouring VIM CR gene were identified from Muhimbili National Hospital, using PCR [34], Table 1.
Kenya
CP K. pneumoniae, A. baumannii and Pseudomonas aeruginosa possessing NDM and VIM-2 genes respectively were isolated in Nairobi [21, 24, 25] while Whole Genome sequencing (WGS) was employed to identify NDM-1like CR genes in K. pneumoniae isolates at Kilifi County Hospital [27], Table 1.
Discussion
Geographical prevalence of CP bacteria
The most prevalent CRE across the region were K. pneumoniae and E. coli. CR in K. pneumoniae was reported by eight articles with mean prevalence of 15% in all East African countries except Rwanda and DRC while CP E. coli was accounted for in all countries apart from DCR by six studies with an average occurrence of 12%. This is in agreement with global data about CR. For example in USA, 11% of K. pneumoniae infections and 2% of E. coli infections were resistant to carbapenems [40] while in India, 13% of E. coli infections and 57% of K. pneumonia infections were caused by CP strains [41]. Additionally, high frequency of CR among the non-glucose fermenting P. aeruginosa (17%) and A. baumannii (23%) almost equal to that of CRE was registered in the region (Table 1 and Additional file 1: Table S2). This is in conformity with worldwide reports acknowledging that the magnitude of CP A. baumannii and P. aeruginosa is equal to that of CRE [40].
Prevalence of CR
The highest frequency of CR in the region was 35%. This prevalence correlates with other studies in India [43, 44] where the prevalence was 43% and 30% respectively. Contrary, this frequency is higher than CR levels reported by other studies in Nigeria (15.2%) and USA (4.5%) but lower than that of 68% reported by a broad study executed in 7 out of the 9 provinces of South Africa [45–47].
Carbapenem resistant bacteria in the hospital environment
The actual occurrence of environmental contamination by CP bacteria is not well researched yet hospital environments tainted with CP bacteria by infected patients are implicated as the main routes of transmission [48, 49]. Across East Africa, only two studies conducted in Uganda reported the existence of CP P. aeruginosa and A. baumannii isolated from hospital environment. The frequency varied from 21% in P. aeruginosa to 55% in A. baumannii, Table 1. Related studies which recovered CP bacteria from hospital environments in Israel and Brazil reported closely related results [50–52]. Horizontal transfer of mobile genetic elements from clinical pathogens to environmental bacteria can occur within the hospital environment hence promoting emergence of new resistant bacteria strains. Furthermore, resistant bacteria in hospital environment such as sewage may spill into the food chain, hence becoming one of the sources of community-acquired resistant pathogens [40].
Sample wise distribution of CP bacteria
This systematic review revealed that CP bacteria are highly distributed in the respiratory tract (23%), Blood (22%), urinary tract (19%) and wounds/pus (18%) in East Africa and this is in line with other investigations conducted in India [44, 53] and USA [46], where they reported high incidences of CR respiratory tract, urinary tract, blood and wound bacterial infections.
CR knowledge gap in East Africa
Various studies around the global have characterized the different variants of each genetic determinant of CR, Additional file 2: Tables S3–S5. Unfortunately, all these variants and their epidemiology are yet to be documented in East Africa, S-CRKGEA. Emergence of CR in K. pneumonia strains harbouring Extended Spectrum Beta-Lactamases-ESBLS (CTX-Ms or SHV-2) or plasmid borne AmpC enzymes (ACT-1, CMY-2, CMY-4 or DHA-1) in association to loss of outer membrane proteins (OMPs) as a result of truncated OMP gene [54, 55] is yet to be acknowledged in East Africa. Occurrence of CP bacteria in livestock and their environment was reported in Europe [9–12] while in East Africa no such research has ever been conducted.
Conclusion and recommendation
Identification of CP bacterial infections at their first appearance provides an opportunity to interfere before these CP organisms are spread more extensively [6]. Therefore, utilizing a robust molecular platform, the WGS, all genetic determinants of CR in humans, livestock and environment should be identified and documented hence bridging the knowledge gap about the molecular epidemiology of CP bacteria in East Africa. Antibiotics resistance stewardship team may profit from data generated by molecular testing of MDR organisms to enhance prevention of intra and inter health facility transmission and the possible cyclic transmission between livestock, humans and environment.
Limitations
Results of the 17 articles that illustrated significant CR across East Africa have been summarized by this review. However, in South Sudan and Burundi no studies have ever been conducted to investigate the epidemiology of CR. Furthermore, studies carried out in Rwanda, DRC, and Ethiopia were aimed at addressing general antimicrobial resistance hence providing very limited CR data while in Kenya, Uganda and Tanzania, more elaborate specific CR in enteric bacteria studies have be performed. Therefore this has led to a significant variation in knowledge about CR in the region.
Additional files
Additional file 1. One way ANOVA results. Table S1. Mean percentage of sample wise distribution of carbapenem resistant isolates generated by One-Way ANOVA. Table S2. Average percentage prevalence of the different carbapenem resistant bacteria computed by One-Way ANOVA.
Additional file 2. Class A, Class B and Class D genetic determinants of Carbapenem resistance. a. Table S3. Showing Ambler class A carbapenemase, their variants, organisms harbouring them and location. b. Table S4. MBLs, their variants, organisms harbouring them and location. c. Table S5. Carbapenem-hydrolyzing class D β-lactamases (CHDL), organisms harbouring them, their geographic distribution and location. d. Table S6. Shows the prevalence of carbapenem resistance in K. pneumonia in all WHO regions. e. Carbapenem resistance Knowledge gap in East Africa (S-CRKGEA).
Authors’ contributions
This work was carried out in collaboration between all authors. DKB and FB conceptualized this project and designed the format for this systematic review. KS and EW performed the literature search and data analysis. KS, DKB, EW and FB drafted the section of literature review. KS wrote the first draft of the manuscript and managed manuscript revisions. DKB, FB and EW participated in manuscript writing and revisions. All authors read and approved the final manuscript.
Acknowledgements
We are thankful to Dr. Charles Kato Drago for his tireless review of this systematic review paper.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Supplementary data is submitted with this manuscript in form of Tables.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The authors declare that there was no funding agency which extended financial assistance towards this Systematic review.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ACT
AmpC type
- AIM
Australian imipenemase
- AmpC
amino penicillin cephalosporinase
- ANOVA
analysis of variance
- bla
beta lactamase
- CHDL
carbapenem hydrolyzing class D beta lactamase
- CMY
cephamycins
- CP
carbapenemase producing
- CR
carbapenem resistance
- CRE
carbapenem resistant Enterobacteriaceae
- CTX
cephotaxime hydrolyzing capabilities
- DHA
Dhaharani Hospital
- DIM
Dutch imipenemase
- DRC
Democratic Republic of Congo
- ESBL
extended spectrum beta-lactamases
- FIM
florence imipenemase
- GES
Guiana extended Spectrum enzyme
- GIM
German imipenemase
- IBC
integron-borne cephalosporinase
- IMI
imipenem hydrolyzing lactamase
- IMP
imipenemase Metallo beta lactamase
- KHM
Kyorin University Hospital
- KPC
Klebsiella pneumoniae carbapenemase
- MBL
Metallo beta lactamase
- MDR
multi-drug resistant
- NDM
New Delhi Metallo beta lactamase
- NMC
not metalloenzyme carbapenemase
- OMP
outer membrane protein
- OXA
oxacillinases
- Ref
reference
- SFC
Serratia fonticola carbapenemase
- SHV
sulfhydryl variables
- SIM
Seoul imipenemase
- SMB
S. marcescens Metallo beta lactamase
- SME
Serratia marcescens enzyme
- SPM
Sao Paulo Metallo beta lactamase
- TMB
Tripoli Metallo beta lactamase
- VIM
Verona Integron encoded Metallo beta lactamase
Contributor Information
Kenneth Ssekatawa, Email: Kssekatee@gmail.com.
Dennis K. Byarugaba, Email: dkb@covab.mak.ac.ug
Edward Wampande, Email: ewampande@yahoo.co.uk.
Francis Ejobi, Email: ejobifrancis@gmail.com.
References
- 1.Livermore DM, Warner M, Mushtaq S, Doumith M, Zhang J, Woodford N. What remains against carbapenem resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents. 2011;37:415–424. doi: 10.1016/j.ijantimicag.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 2.CDC . Facility guidance for control of carbapenem-resistant Enterobacteriaceae (CRE) Atlanta: United States Department of Health and Human Services; 2015. [Google Scholar]
- 3.European Centre for Disease Prevention and Control-ECDC . Rapid risk assessment-carbapenem-resistant Enterobacteriaceae. Stockholm: ECDC; 2016. [Google Scholar]
- 4.European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC) EU summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J. 2015;13:4036. doi: 10.2903/j.efsa.2017.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu Rev Microbiol. 2011;65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 8.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 9.EFSA Scientific opinion on carbapenem resistance in food animal ecosystems. EFSA J. 2013;11(12):3501. [Google Scholar]
- 10.Hattie EW, Bugarel M, Bakker HC, Nightingale KK, Granier SA, Scott MH, Loneragan GH. Carbapenem-resistant bacteria recovered from faeces of dairy cattle in the high plains region of the USA. PLOS ONE. 2016 doi: 10.1371/journal.pone.0147363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mollenkopf DF, Stull JW, Mathys DA, Bowman AS, Feicht SM, Grooters SV, Daniels JB, Wittum TE. Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Am Soc Microbiol Antimicrob Agents Chemother. 2016;61:e01298-16. doi: 10.1128/AAC.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodford N, Wareham DW, Guerra B, Teale C. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother. 2013;69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 13.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013 doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambler RP, Coulson AFW, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A beta lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Queenan AM, Bush K. Carbapenemases: the versatile lactamases. Clin Microbiol Rev. 2007;20(23):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D -lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ampaire L, Muhindo A, Orikiriza P, Mwanga-Amumpaire J, Bebell L, Boum Y. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016;5(1):a432. doi: 10.4102/ajlm.v5i1.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa: a systematic review. J Antimicrob Chemother. 2015;70:23–40. doi: 10.1093/jac/dku356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byarugaba DK, Kisame R, Olet S. Multi-drug resistance in commensal bacteria of food of animal origin in Uganda. AJMR. 2011;5:1539–1548. [Google Scholar]
- 20.Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49–61. doi: 10.2147/IDR.S55778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warnes SL, Highmore CJ, Keevil CW. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: implications for public health. Am Soc Microbiol. 2012;3:6–12. doi: 10.1128/mBio.00489-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan DJ, Okeke IN, Laxminarayan R, et al. Non-prescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis. 2011;11(9):692–701. doi: 10.1016/S1473-3099(11)70054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitout JDD, Revathi G, Chow BL, Kabera B, Kariuki S, Nordmann P, Poirel L. Metallo beta-lactamase-producing Pseudomonas aeruginosa isolated from a large tertiary centre in Kenya. Clin Microbiol Infect. 2008;14:755–759. doi: 10.1111/j.1469-0691.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 24.Poirel L, Revathi G, Bernabeu S, Nordman P. Detection of NDM-1-producing Klebsiella pneumonia in Kenya. Am Soc Microbiol Antimicrob Agents Chemother. 2011;55:934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Revathi G, Siu LK, Po-Liang Lu, Huang L. First report of NDM-1 producing Acinetobacter baumannii in East Africa. Int J Infect Dis. 2013;17:1255–1258. doi: 10.1016/j.ijid.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Apondi OE, Oduor OC, Gye BK, Kipkoech MK. High prevalence of multi-drug resistant Klebsiella pneumoniae in a tertiary teaching hospital in western Kenya. Afr J Infect Dis. 2016;10:89–95. doi: 10.21010/ajid.v10i2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henson SP, Boinett CJ, Ellington MJ, Kagia N, Mwarumba S, Nyongesa S, Mturi N, Kariuki S, Scott JAG, Thomson NR, Morpeth SC. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017;307:422–429. doi: 10.1016/j.ijmm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayoyi OA, Kikuvi G, Bii C, Kariuki S. Prevalence, aetiology and antibiotic sensitivity profile of asymptomatic bacteriuria isolates from pregnant women in selected antenatal clinic from Nairobi, Kenya. Pan Afr Med J. 2017;26:41. doi: 10.11604/pamj.2017.26.41.10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndungu C, Muigai AWT, Kariuki S. Prevalence and antibiotic resistance patterns of Escherichia coli among hospitalised patients at thika district hospital. East Afr Med J. 2014;91:185–190. [Google Scholar]
- 30.Okoche D, Asiimwe BB, Katabazi FA, Kato L, Najjuka CF. Prevalence and characterization of carbapenem-resistant Enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLOS ONE. 2015 doi: 10.1371/journal.pone.0135745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ampaire ML, Katawera V, Nyehangane D, Yap BI, Bazira J. Epidemiology of carbapenem resistance among multi-drug resistant Enterobacteriaceae. Br Microbiol Res J. 2014;8:418–423. doi: 10.9734/BMRJ/2015/17055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kateete DP, Nakanjako R, Namugenyi J, Erume J, Joloba ML, Najjuka CF. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago Hospital in Kampala, Uganda (2007–2009) Springer Plus. 2016;5:1308. doi: 10.1186/s40064-016-2986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kateete DP, Nakanjako R, Okee M, Joloba ML, Najjuka CF. Genotypic diversity among multidrug resistant Pseudomonas aeruginosa and Acinetobacter species at Mulago Hospital in Kampala, Uganda. BMC Res Notes. 2017;10:284. doi: 10.1186/s13104-017-2612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyo S, Haldorsen B, Aboud S, Blomberg B, Maselle SY, Sundsfjord A, Langeland N, Samuelsen O. Identification of VIM-2-producing Pseudomonas aeruginosa from Tanzania is associated with sequence types 244 and 640 and the location of blaVIM-2 in an TniC integron. Agents Chemother Antimicrob. 2014 doi: 10.1128/AAC.01436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant gram negative clinical isolates from a tertiary hospital in Mwanza, Tanzania. Biomed Res Int. 2014 doi: 10.1155/2014/303104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am J Trop Med Hyg. 2015;92:865–870. doi: 10.4269/ajtmh.14-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae among ethiopian children. Infect Drug Resist. 2017;10:27–34. doi: 10.2147/IDR.S127177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desta K, Woldeamanuel Y, Azazh A, Mohammod H, Desalegn D, Shimelis D, et al. High gastrointestinal colonization rate with extended-spectrum β-lactamase-producing enterobacteriaceae in Hospitalized patients: emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS ONE. 2016;11(8):e0161685. doi: 10.1371/journal.pone.0161685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irenge LM, Kabego L, Vandenberg O, Chirimwami RB, Gala J-L. Antimicrobial resistance in urinary isolates from inpatients and outpatients at a tertiary care hospital in South-Kivu Province (Democratic Republic of Congo) BMC Res Notes. 2014;7:374. doi: 10.1186/1756-0500-7-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.CDC . Antibiotic resistance threats in the United States. Atlanta: Centers for Disease Control and Prevention; 2013. [Google Scholar]
- 41.Ne Gelband H, Miller-petrie M, Suraj P, Gandra S, Levinson J, Barter D, White A, Laxminarayan R. The state of the world’s antibiotics. Washington DC: CDDEP; The centre for disease dynamics, economics and Policy; 2015. [Google Scholar]
- 42.World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics; 2017. http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/. Accessed 16 June 2017.
- 43.Pitout JDD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43(7):3129–3135. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henkhoneng MPH, Sulochana DKH, Mamta DKSH, Damrolien S, Lilavati DN, Pratita DP. Prevalence of carbapenem resistance among gram negative bacteria in a Tertiary Care Hospital in North East India. J Dent Med Sci. 2014;13(12 Ver. III):56–60. [Google Scholar]
- 45.Oduyebo OO, Falayi OM, Oshun P, Ettu AO. Phenotypic determination of carbapenemase producing Enterobacteriaceae isolates from clinical specimens at a Tertiary Hospital in Lagos, Nigeria. Niger Postgrad Med J. 2015;22:223–227. doi: 10.4103/1117-1936.173973. [DOI] [PubMed] [Google Scholar]
- 46.Cai B, Echols R, Magee G, Ferreira JCA, Morgan G, Ariyasu M, Sawada T and Nagata TD (2017). Prevalence of carbapenem resistant gram-negative infections in the United States predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infectious Diseases-Infectious Diseases Society of America. [DOI] [PMC free article] [PubMed]
- 47.Singh-Moodley A, Perovic O. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis. 2016;16:1–12. doi: 10.1186/s12879-016-1858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Oliveira AC, Damasceno QS. Surfaces of the hospital environment as possible deposits of resistant bacteria: a review. Rev Esc Enferm USP. 2010;44(1118–1123):6. doi: 10.1590/s0080-62342010000400038. [DOI] [PubMed] [Google Scholar]
- 49.French GL, Otter JA, Shannon KP, Adams NM, Watling D, Parks MJ. Tackling contamination of the hospital environment by methicillin resistant Staphylococcus aureus (MRSA): a comparison between conventional terminal cleaning and hydrogen peroxide vapour decontamination. J Hosp Infect. 2004;57:31–37. doi: 10.1016/j.jhin.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Lerner A, Adler A, Abu-Hanna J, Meitus I, Navon-Venezia S, Carmeli Y. Environmental contamination by carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2012;51:177–181. doi: 10.1128/JCM.01992-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocha IV, Ferraz PDM, de Farias TGS, de Oliveira SR. Resistance of bacteria isolated from equipment in an intensive care unit. Acta Paul Enferm. 2015;28:433–439. doi: 10.1590/1982-0194201500073. [DOI] [Google Scholar]
- 52.Picão RC, Cardoso JP, Campana FH, Nicoletti AG, Petrolini FVB, Assis DM, Juliano L, Gales AC. The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn Microbiol Infect Dis. 2013;76:80–85. doi: 10.1016/j.diagmicrobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Nair PK, Vaz MS. Prevalence of carbapenem resistant Enterobacteriaceae from a tertiary care hospital in Mumbai, India. J Microbiol Infect Dis. 2013;3:207–210. doi: 10.5799/ahinjs.02.2013.04.0110. [DOI] [Google Scholar]
- 54.Crowley B, Bened VJ, Doménech-Sánchez A. Expression of SHV-2 b-lactamase and of reduced amounts of OmpK36 porin in Klebsiella pneumoniae results in increased resistance to cephalosporins and carbapenems. Antimicrob Agents Chemother. 2002;46:3679–3682. doi: 10.1128/AAC.46.11.3679-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang D, Guo Y, Zhang Z. Combined porin loss and extended spectrum beta lactamase production is associated with an increasing imipenem minimal inhibitory concentration in clinical Klebsiella pneumoniae strains. Curr Microbiol. 2009;58:366–370. doi: 10.1007/s00284-009-9364-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. One way ANOVA results. Table S1. Mean percentage of sample wise distribution of carbapenem resistant isolates generated by One-Way ANOVA. Table S2. Average percentage prevalence of the different carbapenem resistant bacteria computed by One-Way ANOVA.
Additional file 2. Class A, Class B and Class D genetic determinants of Carbapenem resistance. a. Table S3. Showing Ambler class A carbapenemase, their variants, organisms harbouring them and location. b. Table S4. MBLs, their variants, organisms harbouring them and location. c. Table S5. Carbapenem-hydrolyzing class D β-lactamases (CHDL), organisms harbouring them, their geographic distribution and location. d. Table S6. Shows the prevalence of carbapenem resistance in K. pneumonia in all WHO regions. e. Carbapenem resistance Knowledge gap in East Africa (S-CRKGEA).
Data Availability Statement
Supplementary data is submitted with this manuscript in form of Tables.