Abstract
Epigenetic mechanisms have been proposed to mediate fear extinction in animal models. Here, MAOA methylation was analyzed via direct sequencing of sodium bisulfite-treated DNA extracted from blood cells before and after a 2-week exposure therapy in a sample of n = 28 female patients with acrophobia as well as in n = 28 matched healthy female controls. Clinical response was measured using the Acrophobia Questionnaire and the Attitude Towards Heights Questionnaire. The functional relevance of altered MAOA methylation was investigated by luciferase-based reporter gene assays. MAOA methylation was found to be significantly decreased in patients with acrophobia compared with healthy controls. Furthermore, MAOA methylation levels were shown to significantly increase after treatment and correlate with treatment response as reflected by decreasing Acrophobia Questionnaire/Attitude Towards Heights Questionnaire scores. Functional analyses revealed decreased reporter gene activity in presence of methylated compared with unmethylated pCpGfree_MAOA reporter gene vector constructs. The present proof-of-concept psychotherapy-epigenetic study for the first time suggests functional MAOA methylation changes as a potential epigenetic correlate of treatment response in acrophobia and fosters further investigation into the notion of epigenetic mechanisms underlying fear extinction.
Keywords: monoamine oxidase A, anxiety, extinction, epigenetics, DNA methylation
Introduction
An “epigenetic switch is the key to persistent extinction” (Stafford et al., 2011), a notion supported by experimental evidence suggesting that pharmacological modulation of epigenetic mechanisms such as DNA methylation or histone acetylation can produce lasting suppression of memories (for review, see Lattal et al., 2013). This seems to be particularly relevant for the psychotherapeutic treatment of anxiety disorders essentially entailing fear extinction (Whittle et al., 2014).
In female patients with panic disorder, we recently identified hypomethylation of a promoter/exon I/intron I region of the monoamine oxidase A (MAOA) gene on chromosome Xp11.4-p11.3 as a disease risk factor (Domschke et al., 2012), which was found to be reversible with a successful cognitive behavioral psychotherapeutic intervention (Ziegler et al., 2016). The MAOA gene is a prime candidate gene also for specific phobias based on evidence from animal studies (Dubrovina et al., 2006) and genetic association studies in patients with specific phobias (Samochowiec et al., 2004).
Thus, in the present proof-of-concept “psychotherapy-epigenetic” study, we for the first time investigated DNA methylation in the above-mentioned promoter/exon I/intron I region of the MAOA gene in female patients with acrophobia during a standardized 2-week cognitive behavioral psychotherapeutic intervention including exposure exercises using virtual reality technology, partly enhanced by repetitive transcranial magnetic stimulation (rTMS). In analogy to a previous study in panic disorder we predicted MAOA methylation to increase and thus to “normalize” along with treatment response. Additionally, the functional relevance of altered MAOA promoter/exon I/intron I methylation was investigated by means of luciferase-based reporter gene assays.
Methods
Sample
Twenty-eight female Caucasian patients (44.86 ± 13.67 years) with acrophobia (SCID-I) were recruited at the Department of Psychiatry, University of Würzburg, Germany. Exclusion criteria were any psychiatric comorbidities or previous treatment of acrophobia within the last 6 months, metal parts in the head, medical implants, increased intracranial pressure, pregnancy, current involvement in psycho- or pharmacotherapy, cardiovascular or neurological diseases, a history of tinnitus, or family history of epilepsy (for details see Herrmann et al., 2016). Additionally, 28 healthy female controls drawn from a larger, well-characterized pool of healthy probands (see Domschke et al., 2012) were matched to the sample of patients with acrophobia by age (39.61±6.68 years, t = -1.8, n.s.). This study was approved by the ethics committees of the Universities of Würzburg and Münster, Germany and was conducted according to the ethical principles of the Helsinki Declaration. Written informed consent was obtained from all participants.
Treatment
After receiving psycho-educative information in written form, patients performed 2 exposure sessions within 2 weeks in a 3- x 4-m, 5-sided Cave Automatic Virtual Environment operated by the Department of Psychology, University of Würzburg (PsyCave; see Mühlberger et al., 2015). Prior to the exposure session, 20 minutes of excitatory rTMS was applied over the medial prefrontal cortex in a lying position (10 Hz, verum or sham; CBT[-rTMS] and CBT[+rTMS]; see Herrmann et al., 2016).
Psychometric assessment of fear of heights was evaluated before (T0) and after (T1) the therapeutic intervention using the Acrophobia Questionnaire (AQ, Anxiety Subscale; Cohen, 1977) and the Attitude Towards Heights Questionnaire (ATHQ; Abelson and Curtis, 1989). Patients were instructed not to change any lifestyle patterns including smoking behavior from T0 to T1.
MAOA Methylation Analysis
In the patient sample, DNA was isolated from EDTA blood taken at T0 and T1 using the FlexiGene DNA Kit (Qiagen). After sodium bisulfite conversion (EpiTect 96 Bisulfite Kit, Qiagen), an amplicon comprising MAOA exon I as well as the adjacent promoter and intron I regions (chromosome X, GRCh38.p2 Primary Assembly, NCBI Reference Sequence: NC_000023.11, 43656260–43656613) was PCR-amplified following a published protocol (Domschke et al., 2012, 2015; Ziegler et al., 2016, 2017) and sequenced (ABI 3730XL, LGC Genomics). The obtained sequences were quantitatively analyzed using the Epigenetic Sequencing Methylation software (ESME; Lewin et al., 2004; cf. Domschke et al., 2012, 2013, 2014, 2015; Tadić et al., 2014; Ziegler et al., 2015, 2016, 2018). Electropherograms were robustly readable for 13 CpG sites (CpG1 = 43656 316; CpG2 = 43656327; CpG3 = 43656 362; CpG4 = 43656368; CpG5 = 43656370; CpG6 = 43656383; CpG7 = 43656386; CpG8 = 43656392; CpG9 = 43656398; CpG10 = 43656427; CpG11 = 43656432; CpG12 = 43656514; CpG13 = 43656553). Healthy controls had been assessed for MAOA methylation using the same methods as applied to the present patient sample (see Domschke et al., 2012). Quality control was performed as described previously (cf. Domschke et al., 2012; Ziegler et al., 2016). No participant had to be excluded from the reported analyses when applying quality control (data missing for CpG1 in 3 patients).
Additionally, all patients and controls were genotyped for the MAOA VNTR according to published protocols (Reif et al., 2012) and stratified into low (33/34/3a4/35) and high expression (44/45) genotype groups (cf. Reif et al., 2012; Domschke et al., 2015).
Functional Analysis
Functional analysis was accomplished using the pCpGfree-promoter-Lucia vector (InvivoGen) expressing a Lucia luciferase under a human elongation factor-1 (hEF1) promoter. The insert comprising CpGs 1–13 was PCR-amplified and ligated into the linearized vector using T4 DNA Ligase (Invitrogen). The vectors were methylated using the CpG methyltransferase M.SssI (NEB) and transfected to HEK293 cells (ECACC) using the TransFast reagent (Promega). Luciferase assays were performed using the Stop & Glo® Reagent (Promega) on a GloMax+ Luminometer (Promega), and Lucia luciferase activity was normalized using a co-transfected pGL4.74 Renilla luciferase control vector. All analyses were conducted in technical triplicates.
Statistical Analysis
Differences in MAOA methylation levels between patients and controls as well as between genotype groups were tested by means of independent samples t tests. The association between baseline methylation and treatment response as defined as the intra-individual relative change (in percent) in clinical symptom scores (AQ Anxiety) was assessed by means of linear regression analysis and controlled for genotype and TMS status. Changes in MAOA methylation levels following therapy were tested using repeated-measures ANOVA and controlled for grouped genotype and TMS randomization. Symptom changes were evaluated by means of paired sample t tests. A partial correlation was run to determine the relationship between individual changes in methylation (T1-T0) and symptom improvement (T1-T0) whilst controlling for baseline methylation (T0), genotype, and TMS status. Luciferase assay data were normalized to transfection efficiency by Renilla luciferase control and z-transformed to eliminate day and measurement specific fluctuations. All tests were carried out 2-sided, and an alpha level of <0.05 was considered significant. Given the present proof-of-concept approach and high correlation between the 13 individual CpG sites (data not shown), no correction for multiple testing was applied when analyzing single CpG sites.
Results
Mean scores on both questionnaires decreased significantly after the therapeutic intervention (AQ Anxiety T0: 57.05±18.11, T1: 43.47±18.91, t = 4.61, P < .001; ATHQ Total T0: 37.86±16.56, T1: 26.89±9.53, t = 4.59, P < .001; ATHQ Danger T0: 18.32±8.19, T1: 12.61±5.34, t = 5.19, P < .001). For this female patient subsample, no difference was discerned between the CBT(-rTMS) and CBT(+rTMS) groups regarding treatment response (data not shown), allowing for a joint epigenetic analysis of the 2 groups in the present context.
At T0, average MAOA methylation (0.432±0.021) did not correlate with age or disease severity according to AQ Anxiety or ATHQ Total/Danger scores (all P > .05). No influence of grouped genotype (low expression [33/34/3a4/35]: n = 17 patients and n = 19 controls; high expression [44/45]: n = 11 patients and n = 9 controls) was discerned on average MAOA methylation or questionnaire scores at T0 (all P > .05). Furthermore, patients and controls did not differ in genotype distribution (Χ2 = .311, P = .577).
Average MAOA methylation as well as methylation at CpGs 1–8 and 10–13 differed significantly between patients with acrophobia at baseline (T0) and healthy controls. Patients were characterized by significantly lower average methylation levels, specifically driven by single CpGs 1–8, 10, and 11 (for details see Table 1).
Table 1.
MAOA Methylation Levels in Patients with Acrophobia and Healthy Controls
| Patients (n = 28) Mean (SE) | Controls (n = 28) Mean (SE) | Statistics | ||
|---|---|---|---|---|
| t value | P value | |||
| Average methylation | 0.432 (0.004) | 0.4712 (0.008) | 4.488 | <.001 |
| CpG1 | 0.359 (0.009) | 0.4594 (0.011) | 7.040 | <.001 |
| CpG2 | 0.358 (0.010) | 0.3927 (0.011) | 2.366 | .022 |
| CpG3 | 0.370 (0.008) | 0.4097 (0.011) | 2.873 | .006 |
| CpG4 | 0.403 (0.008) | 0.5895 (0.020) | 8.756 | <.001 |
| CpG5 | 0.286 (0.009) | 0.3375 (0.016) | 2.800 | .008 |
| CpG6 | 0.361 (0.009) | 0.4246 (0.011) | 4.403 | <.001 |
| CpG7 | 0.441 (0.007) | 0.4964 (0.009) | 4.900 | <.001 |
| CpG8 | 0.318 (0.009) | 0.3751 (0.010) | 4.284 | <.001 |
| CpG9 | 0.467 (0.006) | 0.4836 (0.009) | 1.575 | .121 |
| CpG10 | 0.482 (0.005) | 0.5049 (0.007) | 2.758 | .008 |
| CpG11 | 0.282 (0.009) | 0.3138 (0.008) | 2.571 | .013 |
| CpG12 | 0.913 (0.007) | 0.8484 (0.020) | -3.082 | .003 |
| CpG13 | 0.569 (0.011) | 0.4896 (0.018) | -3.670 | .001 |
P-values from independent samples t test.
A positive change in AQ Anxiety scores indicating treatment response was seen in 20 patients (71.4%). While no significant association between baseline average MAOA methylation and treatment response could be discerned, differences were observed regarding CpG1 (β = .567, P = .003) and, approaching trend levels of significance, CpG5 (β = .340, P = .086) and CpG6 (β = .331, P = .091), with lower baseline methylation predicting impaired treatment response.
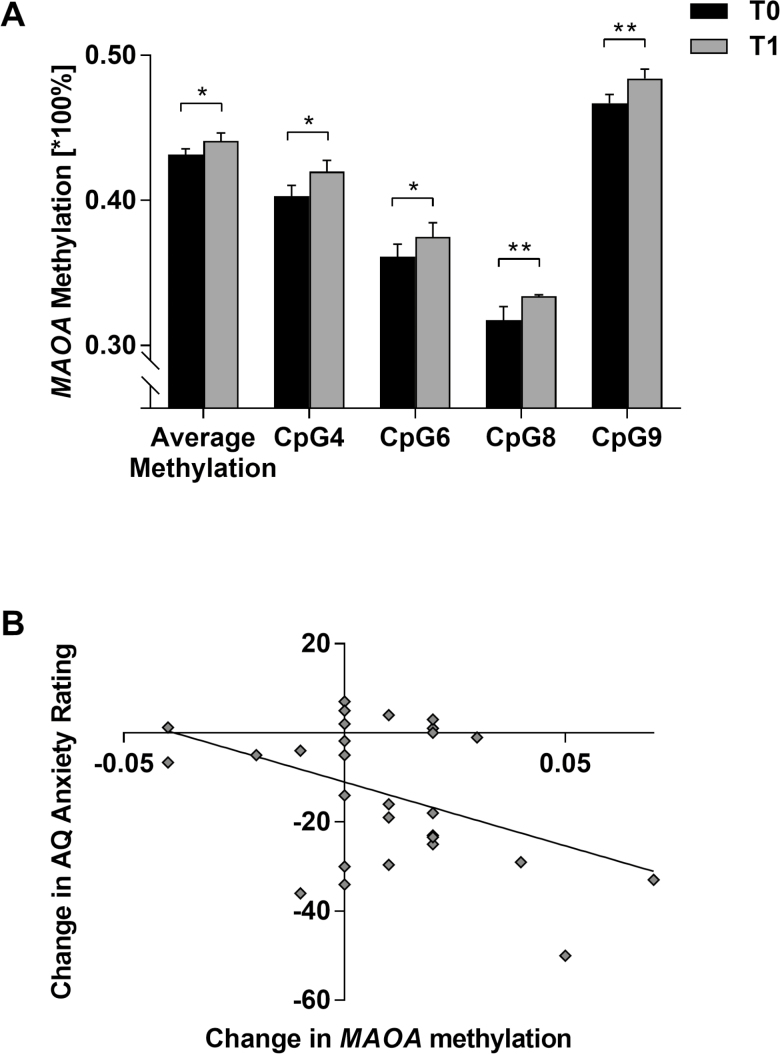
Following therapy, MAOA methylation increased significantly in the patient group for average methylation (T1: 0.441±0.029, F = 4.703, P = .040) and specifically at CpG4 (T1: 0.420±0.040, F = 4.708, P = .040), CpG6 (T1: 0.375±0.051, F = 4.954, P = .036), CpG8 (T1: 0.334±0.053, F = 10.204, P = .004) and CpG9 (T1: 0.484±0.035, F = 10.249, P = .004) (Figure 1A). Methylation at all other CpG sites did not change significantly from T0 to T1 (all P > .05).
Figure 1.
MAOA methylation and treatment response in acrophobia. (A) Significant changes in MAOA methylation from baseline (T0; black bars) to post therapy (T1; grey bars) in n = 28 female patients with acrophobia at single CpG sites 4, 6, 8, and 9 and for average methylation (CpGs 1–13); mean ± SE. *P < .05, **P < .01. (B) Correlation between reduction in anxiety severity (Acrophobia Questionnaire [AQ] Anxiety score from T0 to T1) and MAOA average methylation change from T0 to T1 (r = -0.449, P = .019).
In the main analyses regarding methylation change dependent on treatment response as measured by changes in AQ Anxiety or ATHQ Total/Danger scores, respectively, negative correlations indicated clinical symptom improvement to go along with an increase in average MAOA methylation (AQ Anxiety: r = -0.465, P = .019, Figure 1B; ATHQ Danger: r = -0.496, P = .012). This pattern held true for several individual CpG sites (AQ Anxiety: CpG2: r = -0.455, P = .022; CpG3: r = -0.398, P = .049; CpG7: r = -0.459, P = .021; CpG9: r = -0.399, P = .048; CpG10: r = -0.527, P = .007; ATHQ Total: CpG9: r = -0.402, P = .047; ATHQ Danger: CpG2: r = -.412, P = .041; CpG3: r = -0.401, P = .047; CpG7: r = -0.473, P = .017; CpG9: r = -0.541, P = .005; CpG11: r = -0.424, P = .035).
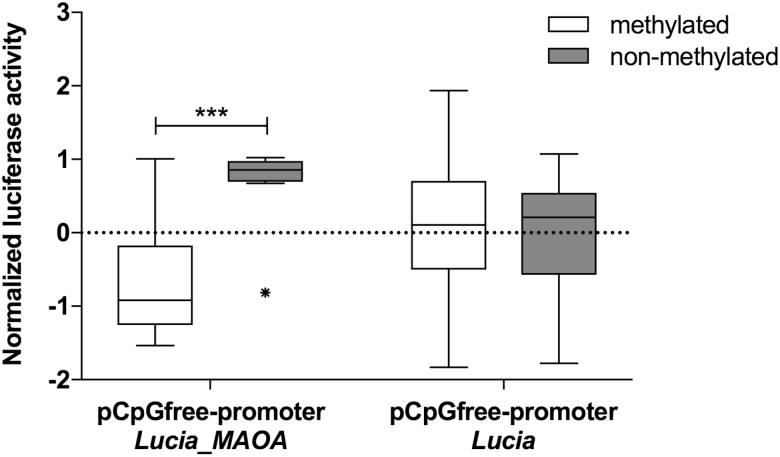
Applying in vitro luciferase assays, nonmethylated pCpGfree-promoter Lucia_MAOA vectors showed a significant increase in normalized Lucia luciferase activity compared with pCpGfree-promoter Lucia_MAOA vectors methylated with M.SssI prior to transfection (t = 4.157, p < .001; Figure 2). PCpGfree-promoter Lucia vectors without insert showed no significant difference between the methylated and the nonmethylated state (t = 0.394, p = .70; Figure 2).
Figure 2.
Functional analysis of MAOA promoter/exon I/intron I DNA methylation using luciferase-based reporter gene assays. Left: Normalized reporter gene activity was significantly decreased in the presence of pCpGfree-promoter Lucia_MAOA vectors containing the methylated insert spanning CpGs 1–13 compared with those carrying a nonmethylated insert; ***P < .001. Right: No significant difference in normalized reporter gene activity was discerned between methylated or non-methylated pCpGfree-promoter Lucia control vectors lacking the insert of the sequence spanning CpGs 1–13.
Discussion
The present finding of (I) decreased MAOA methylation in patients with acrophobia compared with healthy controls, (II) decreased methylation at a single MAOA CpG site to predict treatment response, and (III) increasing MAOA methylation along with treatment response further strengthens and extends the emerging body of evidence for a potential role of MAOA methylation in the pathogenesis of fear-related disorders. Additionally, in line with the hypothesis of epigenetically driven neuroplasticity underlying response to extinction-related psychotherapeutic interventions (Stafford et al., 2011), MAOA methylation might be worthwhile to be further investigated as a possible correlate of successful extinction treatment (cf. in panic disorder: Ziegler et al., 2016). Furthermore, using luciferase-based reporter gene assays, we demonstrated increased methylation of the therapy-modulated region to drive decreased gene expression, which is line with a previous functional in vitro assay (Checknita et al., 2015) and blood MAOA methylation correlating inversely with brain MAO-A levels in vivo using [(11)C]clorgyline positron emission tomography (Shumay et al., 2012). These functional results also support the notion that not only methylation of the promoter but also of exon I with its adjacent promoter and intron I regions seems to be linked to transcriptional repression (Brenet et al., 2011). In sum, we hypothesize that in acrophobia, decreased availability of monoamines conferred by decreased MAOA methylation might be reversible by remethylation during the course of a successful therapeutic intervention.
The present study thereby adds to the only recently burgeoning evidence for epigenetic mechanisms possibly constituting dynamic biological correlates of therapeutic interventions in mental disorders (for review, see Schiele and Domschke, 2018). In anxiety disorders, besides the above-mentioned study (Ziegler et al., 2016) only 2 other studies have investigated methylation dynamics with respect to treatment effects. Here, methylation changes in the serotonin transporter (5-HTT) gene as well as in the FK506-binding protein 5 (FKBP5) gene were found to be associated with response to psychotherapy in children with mixed primary anxiety disorder diagnoses and obsessive-compulsive disorder (Roberts et al., 2014, 2015).
A major limitation of the present study entailing the risk of false positive results is the relatively small sample size owed to the proof-of-concept design, strict inclusion/exclusion criteria, and inclusion of female patients only due to the X-chromosomal location of the MAOA gene and previous observations of female-specific effects of genetic as well as epigenetic MAOA variation in anxiety disorders (Domschke et al., 2012; Reif et al., 2012; Ziegler et al., 2016). Thus, despite comparability to previous treatment-epigenetic studies regarding size (e.g., Ziegler et al., 2016), replication in larger samples as well as in mixed samples including also male patients is warranted. Given that no control group was available paralleling the time course of treatment in the patient sample, the observed changes in MAOA methylation in patients cannot conclusively be attributed to the effects of the intervention but could rather be an inconsequential side effect arising from a significant environmental stimulus. Also, despite no statistically significant influence having been detected, psychotherapeutic and rTMS effects on DNA methylation changes along with clinical symptom improvement cannot be differentiated. In general, DNA methylation was measured in peripheral blood samples entailing cell composition effects driven by a vast amount of factors such as inflammation, diet, exercise, stress, or hormonal status as a possible confounder. Also, while patients were instructed not to change smoking patterns from T0 to T1, smoking patterns could still have confounded the present results. Furthermore, data from in vitro luciferase assays spanning 13 CpG sites do not allow for evaluating MAOA gene regulation in toto or at single CpG sites and thus are to be considered suggestive, but not conclusive, particularly since different CpG sites were found to predict or correlate with treatment response, respectively. Data on MAOA mRNA expression in apt tissue dependent on individual MAOA methylation status would aid in further defining the in vivo physiological consequences of MAOA promoter hypomethylation (cf. Hotamisligil and Breakefield, 1991). Along these lines, despite some evidence for their potential as viable markers of central processes (e.g., Shumay et al., 2012), peripheral methylation patterns do not necessarily allow for deductions regarding MAOA methylation status in brain tissue. Finally, localized changes such as the presently identified MAOA methylation patterns might reflect global changes in genomic DNA methylation either by CpG hyper- and hypomethylation necessitating investigation by means of epigenome-wide association studies in larger, sufficiently powered samples.
In sum, while warranting replication and considering all caveats mentioned above, the present psychotherapy-epigenetic study for the first time suggests functionally relevant MAOA methylation changes as a potential epigenetic correlate of response to cognitive behavioral psychotherapy in acrophobia and corroborates a previous independent finding in panic disorder (Ziegler et al., 2016). The emerging notion of epigenetic signatures as a core mechanism of action of fear extinction might aid in probing pharmacological treatment enhancers such as MAO-inhibitors or, in a more general sense, drugs inducing epigenetic changes as augmentation strategies for lasting extinction effects and is thereby hoped to contribute to a more effective treatment based on individual epigenetic information.
Funding
This work was supported by the German Research Foundation, SFB-TRR-58, projects C02 (to K.D.), C06 (to M.J.H.) and Z02 (to K.D., P.P., A.R., and J.D.), and the German Ministry of Research and Education (01EE1402F, PROTECT-AD, P5 to K.D. and J.D.).
Acknowledgments
We gratefully acknowledge the skillful technical support by Carola Gagel.
Statement of Interest
None.
References
- Abelson JL, Curtis GC(1989)Cardiac and neuroendocrine responses to exposure therapy in height phobics: Desynchrony within the “physiological response system.” Behav Res Ther 27:561–567. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM(2011)DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6:e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checknita D, Maussion G, Labonté B, Comai S, Tremblay RE, Vitaro F, Turecki N, Bertazzo A, Gobbi G, Côté G, Turecki G(2015)Monoamine oxidase A gene promoter methylation and transcriptional downregulation in an offender population with antisocial personality disorder. Br J Psychiatry 206:216–222. [DOI] [PubMed] [Google Scholar]
- Cohen DC.(1977)Comparison of self-report and overt-behavioral procedures for assessing acrophobia. Behav Therapy 8:17–23. [Google Scholar]
- Domschke K, Tidow N, Kuithan H, Schwarte K, Klauke B, Ambrée O, Reif A, Schmidt H, Arolt V, Kersting A, Zwanzger P, Deckert J(2012)Monoamine oxidase A gene DNA hypomethylation - a risk factor for panic disorder?Int J Neuropsychopharmacol 15:1217–1228. [DOI] [PubMed] [Google Scholar]
- Domschke K, Tidow N, Schrempf M, Schwarte K, Klauke B, Reif A, Kersting A, Arolt V, Zwanzger P, Deckert J(2013)Epigenetic signature of panic disorder: a role of glutamate decarboxylase 1 (GAD1) DNA hypomethylation?Prog Neuropsychopharmacol Biol Psychiatry 46:189–196. [DOI] [PubMed] [Google Scholar]
- Domschke K, Tidow N, Schwarte K, Deckert J, Lesch KP, Arolt V, Zwanzger P, Baune BT(2014)Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int J Neuropsychopharmacol 17:1167–1176. [DOI] [PubMed] [Google Scholar]
- Domschke K, Tidow N, Schwarte K, Ziegler C, Lesch KP, Deckert J, Arolt V, Zwanzger P, Baune BT(2015)Pharmacoepigenetics of depression: no major influence of MAO-A DNA methylation on treatment response. J Neural Transm 122:99–108. [DOI] [PubMed] [Google Scholar]
- Dubrovina NI, Popova NK, Gilinskii MA, Tomilenko RA, Seif I(2006)Acquisition and extinction of a conditioned passive avoidance reflex in mice with genetic knockout of monoamine oxidase A. Neurosci Behav Physiol 36:335–339. [DOI] [PubMed] [Google Scholar]
- Herrmann MJ, Katzorke A, Busch Y, Gromer D, Polak T, Pauli P, Deckert J(2016)Medial prefrontal cortex stimulation accelerates therapy response of exposure therapy in acrophobia. Brain Stimul 10:291–297. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Breakefield XO(1991)Human monoamine oxidase A gene determines levels of enzyme activity. Am J Hum Genet. 49:383–392. [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Wood MA(2013)Epigenetics and persistent memory: implications for reconsolidation and silent extinction beyond the zero. Nat Neurosci 16:124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin J, Schmitt AO, Adorján P, Hildmann T, Piepenbrock C(2004)Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics 20:3005–3012. [DOI] [PubMed] [Google Scholar]
- Mühlberger A, Kinateder M, Brütting J, Eder S, Müller M, Gromer D, Pauli P(2015)Influence of information and instructions on human behavior in tunnel accidents: a virtual reality study. JVRB 12:1–13. [Google Scholar]
- Reif A, Weber H, Domschke K, Klauke B, Baumann C, Jacob CP, Ströhle A, Gerlach AL, Alpers GW, Pauli P, Hamm A, Kircher T, Arolt V, Wittchen HU, Binder EB, Erhardt A, Deckert J(2012)Meta-analysis argues for a female-specific role of MAOA-uVNTR in panic disorder in four European populations. Am J Med Genet B Neuropsychiatr Genet 159B:786–793. [DOI] [PubMed] [Google Scholar]
- Roberts S, Lester KJ, Hudson JL, Rapee RM, Creswell C, Cooper PJ, Thirlwall KJ, Coleman JRI, Breen G, Wong CCY, Eley TC(2014)Serotonin transporter [corrected] methylation and response to cognitive behaviour therapy in children with anxiety disorders. Transl Psychiatry 4:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, et al. (2015)HPA axis related genes and response to psychological therapies: genetics and epigenetics. Depress Anxiety 32:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samochowiec J, Hajduk A, Samochowiec A, Horodnicki J, Stepień G, Grzywacz A, Kucharska-Mazur J(2004)Association studies of MAO-A, COMT, and 5-HTT genes polymorphisms in patients with anxiety disorders of the phobic spectrum. Psychiatry Res 128:21–26. [DOI] [PubMed] [Google Scholar]
- Schiele MA, Domschke K(2018)Epigenetics at the crossroads between genes, environment and resilience in anxiety disorders. Genes Brain Behav 17:e12423. [DOI] [PubMed] [Google Scholar]
- Shumay E, Logan J, Volkow ND, Fowler JS(2012)Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics 7:1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Lattal KM(2011)Is an epigenetic switch the key to persistent extinction?Neurobiol Learn Mem 96:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadić A, Müller-Engling L, Schlicht KF, Kotsiari A, Dreimüller N, Kleimann A, Bleich S, Lieb K, Frieling H(2014)Methylation of the promoter of brain-derived neurotrophic factor exon IV and antidepressant response in major depression. Mol Psychiatry 19:281–283. [DOI] [PubMed] [Google Scholar]
- Whittle N, Singewald N(2014)HDAC inhibitors as cognitive enhancers in fear, anxiety and trauma therapy: where do we stand?Biochem Soc T 42:569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, Dannlowski U, Bräuer D, Stevens S, Laeger I, Wittmann H, Kugel H, Dobel C, Hurlemann R, Reif A, Lesch K-P, Heindel W, Kirschbaum C, Arolt V, Gerlach AL, Hoyer J, Deckert J, Zwanzger P, Domschke K(2015)Oxytocin receptor gene methylation: converging multilevel evidence for a role in social anxiety. Neuropsychopharmacology 40:1528–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, Richter J, Mahr M, Gajewska A, Schiele MA, Gehrmann A, Schmidt B, Lesch K-P, Lang T, Helbig-Lang S, Pauli P Kircher T, Reif A, Rief W, Vossbeck-Elsebusch AN, Arolt V, Wittchen H-U, Hamm AO, Deckert J, Domschke K(2016)MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Transl Psychiatry 6:e773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, et al. (2018)Monoamine oxidase A gene methylation and its role in posttraumatic stress disorder - first evidence from the South Eastern Europe (SEE)-PTSD study. Int J Neuropsychopharmacol 21:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]