Abstract
Pressure-jump hardware permits direct observation of protein NMR spectra during a cyclically repeated protein folding process. For a two-state folding protein, the change in resonance frequency will occur nearly instantaneously when the protein clears the transition state barrier, resulting in a mono-exponential change of the ensemble-averaged chemical shift. However, protein folding pathways can be more complex and contain meta-stable intermediates. With a pseudo-3D NMR experiment that utilizes stroboscopic observation, we measure the ensemble-averaged chemical shifts, including those of exchange-broadened intermediates, during the folding process. Such measurements for a pressure-sensitized mutant of ubiquitin show an on-pathway kinetic intermediate whose 15N chemical shifts differ most from the natively folded protein for strands β5, its preceding turn, and the two strands that pair with β5 in the native structure.
Graphical Abstract
Direct observation of protein folding by NMR initially was limited to slower folding or unfolding systems,1–4 but more recent improvements in technology have significantly increased the accessible time resolution.5–6 In such experiments, sample conditions are changed abruptly from denaturing to those favoring the native state. This may be accomplished by a sudden jump in pH or temperature, dilution of denaturant, release of a co-factor, or sudden drop in hydrostatic pressure. The latter mode of switching is often fully reversible and allows the protein to be probed under native buffer conditions. It requires that the volume of the protein in the unfolded state is substantially lower than in the folded state.7–11 Analogous to pressure-jump fluorescence experiments,12 the temporal change in the NMR spectrum following a step change in pressure contains kinetic information.4,13
We recently developed a pressure-jump apparatus capable of reliably switching the hydrostatic pressure in an NMR sample cell between 1 bar and up to 2.8 kbar within a few milliseconds,14 and demonstrated it for ubiquitin, whose folding mechanism has been the subject of much experimental and computational work.15–22 Although wild-type ubiquitin requires more than 5 kbar to unfold,23 mutations that increase the volume difference between the folded and unfolded state reduce the pressure needed to unfold the protein.24–25 Indeed, the V17A/V26A ubiquitin mutant (VA2-ubiquitin) lowered the mid-point of pressure-induced unfolding to 1.4 kbar, while retaining the native structure.14, 22
At 2.5 kbar, 25 °C, the 15N-1H heteronuclear single quantum correlation (HSQC) NMR spectrum of VA2-ubiquitin shows the protein is ca 94% unfolded, yielding narrow chemical shift dispersion and sharp resonances; the remaining 6% retains its natively folded spectrum. Immediately after dropping the pressure to 1 bar, the 15N shifts of the protein remain close to those of the pressure-denatured state, but also show evidence of exchange broadening (Figure S1).14
For a two-state folder, jumping from high pressure (denaturing) to 1 bar (native) conditions triggers a folding process that is manifested as a changing admixture of unfolded and folded proteins. For multi-state folders, intermediates will also become populated and can exhibit molten globule characteristics with correspondingly broad NMR resonances.26–27 We demonstrate that the ensemble-averaged chemical shifts for such an admixture can be monitored stroboscopically28,29 as a function of folding time, and permits determination of the folding intermediate chemical shifts.
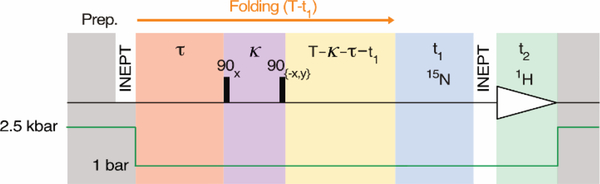
The NMR experiment (Figure 1) is a modification of the regular gradient-enhanced heteronuclear single quantum correlation (HSQC) pulse scheme30 synchronized with the pressure jump, which includes a stroboscopic element to probe the resonance frequency during folding. During the preparation period (grey), 1H magnetization is first transferred to in-phase 15Nz by means of a refocused INEPT element at high pressure, where the efficiency of such transfers is very high. This transfer is immediately followed by the opening of a hydraulic valve that drops the pressure in the sample cell to 1 bar, initiating the folding process. Following a duration τ (Figure 1, peach), the 15N chemical shift frequency is encoded “stroboscopically” by the 90x-κ−90-x pulse pair (purple) which modulates the 15Nz magnetization, and thereby the spectral intensity, Mx(τ), in the HSQC spectrum, by cos(ωNκ), where ωN is the 15N angular offset frequency, averaged over duration κ and over all molecules that contribute to the final 2D HSQC spectrum (Figure 2A). Similarly, a 90x-κ−90y pulse pair is used to generate a second HSQC spectrum whose intensity, My(τ), is modulated by sin(ωNκ) (Figure 2B). The ratio of intensities observed in these two spectra therefore equals tan(ωNκ), thereby yielding <ωN> at time τ, where the brackets denote the ensemble-averaged value. Linear weighting of contributions to <ωN> from folded, unfolded or intermediate states of the protein requires that κ(ΔωN) « π, where ΔωN represents the angular chemical shift difference between them.
Figure 1.
Pressure-jump pseudo-3D NMR pulse scheme used for stroboscopic measurement of 15N chemical shifts during protein folding. For details, see Figure S2.
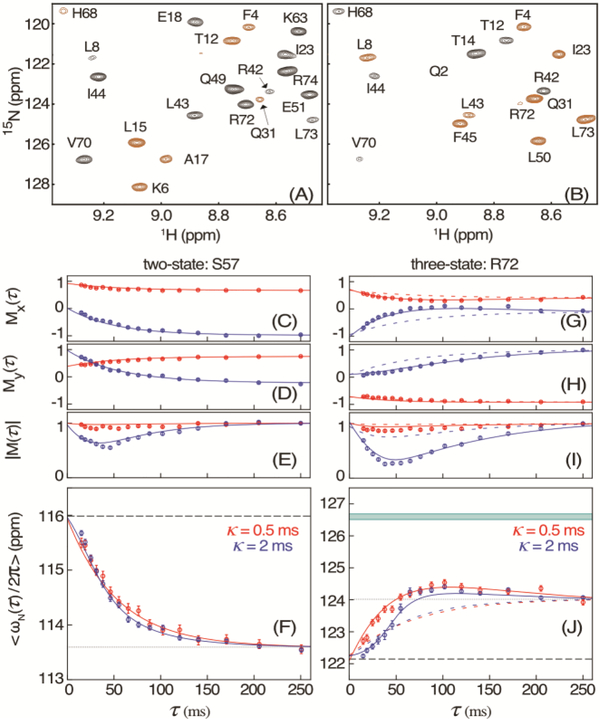
Figure 2.
Measurement of 15N chemical shifts during VA2-ubiquitin folding. (A, B) Small region of the HSQC spectrum, recorded with the scheme of Figure 1 for τ = 10 ms and κ = 2 ms, modulated by (A) cos(ωNκ) and (B) sin(ωNκ). Extension of the 50-ms t1 time domain to 75 ms by the NUS reconstruction program SMILE34 was used to slightly increase both resolution and sensitivity. Negative intensity is shown in brown. For full spectra, see Figure S3. (C) Mx(τ) and (D) My(τ) magnetization of residue S57 after evolution for κ = 0.5 ms (red) and κ = 2 ms (blue), and (E) the corresponding magnitude |M(τ)|, with these values normalized to |M(τ)| = 1 at τ = 248 ms. Solid colored lines correspond to the values predicted for a two-state folding model with ρ = 14.3 s−1 and known chemical shifts14 of the unfolded (F, dashed line) and folded (F, dotted) states at 1 bar. (F) Apparent average chemical shift <ωN/2π> at time τ after the pressure drop. (G-J) Analogous to (C-F) but for residue R72 with solid lines corresponding to a three-state model with ku→i = 13.2 s−1; ku→f = 6.5 s−1; and ki→f = 12.0 s−1, and colored dashed lines for the two-state model. The band at 126.6 ppm in (J) depicts the fitted 15N shift of the intermediate state. The full set of residues is shown in Figure S4. The error bars in (C-E) and (G-I) indicate the RMS noise in the Mx and My spectra, scaled by the normalization factor of |M|. Error bars in (F) and (J) indicate the uncertainty in the average chemical shift derived from the RMS noise in the Mx and My spectra
For a two-state folder at time t after the pressure is dropped to 1 bar, the probability that the angular resonance frequency of any given nucleus switches from unfolded ωU to folded ωF during a short interval dt equals ρe-ρtdt, where ρ is the protein folding rate. Thus, at time τ a fraction e−ρτ resonates with frequency ωU and the remainder, (1 - e−ρτ), with ωF. During the κ interval, the corresponding magnetization vectors accumulate phases of ωUκ and ωFκ, respectively, of which the fractions cos(ωUκ) and cos(ωFκ) are returned to the +z axis by the subsequent 90-x pulse, or sin(ωUκ) and sin(ωFκ) for the second experiment where a 90y pulse terminates the stroboscopic observation. Only proteins that are natively folded at the start of the subsequent 15N evolution period contribute to the resonance intensity of the folded protein in the HSQC spectrum. While this requirement is met for all proteins in the (1 - e−ρτ) folded fraction, only a fraction 1 - e−ρ(Τ−τ) of the unfolded protein fraction (e-ρτ) will switch to the folded state between its stroboscopic encoding and the start of 15N t1 evolution. We also must account for the small fraction, A, of the protein that does not unfold at 2.5 kbar (A≈6%).
In the limit where ρκ « 1, the amplitude of the observed, folded protein magnetization is then given by
| (1) |
and an analogous expression for My(τ) in which sin substitutes for cos. Depending on the values of ωUκ, the amplitude of the HSQC signal can be positive or negative (Figure 2A, B; Figure S3).
The weighted average frequency is given by
| (2) |
up to the addition of an integer multiple of 2π/κ, analogous to aliasing in traditional Fourier sampling. For a two-state folder with small chemical shift difference (ωU-ωF) and ρT » 1, <ω(τ)> simply becomes the population-weighted average frequency, decaying exponentially from [(1-A)ωU + AωF] to ωF when τ is incremented from 0 to T (Figure 2F). When κ « π(ΔωN)−1 is not satisfied, the signal magnitude exhibits a minimum in |M(τ)| = √[Mx(τ)2 + My(τ)2] where
| (3) |
with fractional amplitude cos[(ωU −ωF)τmin/2] relative to |M(Τ)|. Note that ωU at 1 bar can be measured at high precision from a separate pressure-jump HSQC spectrum (Figure S1),14 and ωF is known from the folded protein HSQC spectrum. Therefore, for the 2-state folding model, Mx(τ), My(τ), |M(τ)|, and <ω(τ)> can be calculated for any value of κ (Figures 2 and S4), and then be compared to experimental data to test validity of the model. The use of large κ values subsequently becomes advantageous, as the minimum of |M(τ)| becomes deeper, and its position better defined. As seen from eq 3, for a 2-state folder, the position of the minimum should be the same for all residues.
Experiments were carried out at 600 MHz 1H frequency, for a 0.3 mM 15N labeled VA2ubiquitin sample, pH 6.4, using a Bruker Instruments cryogenic probehead with a z-axis pulsed field gradient accessory. A 2.7-mm ID zirconia sample cell (Daedalus Inc.) was used, rated to withstand pressures of up to 3 kbar.31 Two data sets were collected, using κ = 0.5 ms and κ = 2 ms. For κ = 0.5 ms, <ω(τ)> can be determined uniquely from the ratio of Mx(τ) and My(τ) over a bandwidth of 2 kHz, which suffices to cover the entire range of backbone amide 15N chemical shifts in VA2-ubiquitin. The precision, ε, at which <ω(τ)>= θ /κ can be determined from My(τ) / Mx(τ) is limited by the uncertainty in θ, which is determined by the root-mean-square noise, N, in these spectra:
| (4) |
In practice, we obtain |M(τ)| / N ratios of ca 40:1 when using 2 scans per free induction decay (FID). This corresponds to an uncertainty of ε(τ) ≈ 50 rad/s, or 0.13 ppm for κ = 0.5 ms. As seen from eq (4), longer κ durations reduce the uncertainty in <ω(τ)>; however, (ωU - ωF)κ « π may no longer be satisfied, causing a loss in |M(τ)| when τ approaches τmin, which may offset the gain from the larger denominator.
We have previously shown that VA2-ubiquitin folds with comparable efficiency via two parallel pathways: one with a single barrier, and one with two barriers, giving rise to a meta-stable, short-lived intermediate that was identified through an upfield shifted resonance of L50-Cδ2H3.14 However, in the above analysis of M(τ), three-state folding will be indistinguishable from two-state if the resonance frequency of the intermediate ωI equals that of the folded state ωF, as applies for example for S57 (Figure 2; Figure S4). By contrast, many other residues deviate strongly from simple two-state behavior. For example, <ω(τ)> of R72 initially increases from its unfolded value of 122 ppm to downfield of 124 ppm, before returning to its folded value of 123.9 ppm (Figure 2J). The latter behavior points to a chemical shift value of the intermediate that is downfield of 124 ppm. Considering the folding process is dominated by the sum of two- and three-state pathways, fitting the predicted <ω(τ)> to the data (SI, text) yields the chemical shifts, ωI, of the intermediate in the three-state pathway (Figure 3B; Table S1).
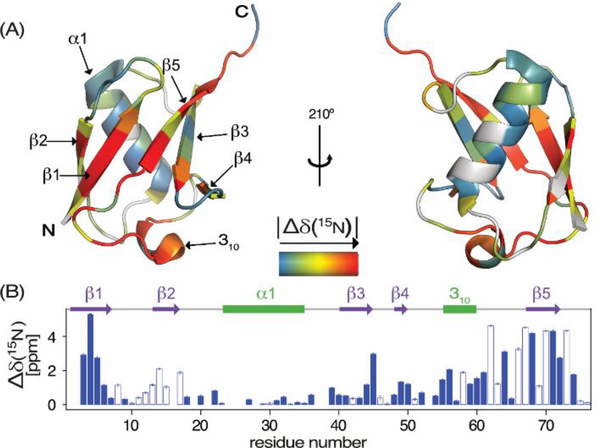
Figure 3.
15N chemical shift differences, |Δδ(15N)| = |δI – δF|, between the meta-stable intermediate and natively folded VA2-ubiquitin. (A) Color coded on a backbone ribbon structure. Residues for which Δδ(15N) could not be determined are shown in grey. (B) Δδ(15N) as a function of residue number, with negative values depicted as open bars.
Evaluation of ωF and ωI shows that the largest differences between them are located in strands β5, the loop preceding β5, strand β1, and the last few residues of β3 (Figure 3). Remarkably, deviations from 2-state folding extend to at least residue R74, indicating that this residue must be structured in the intermediate state, even while in the native protein it is significantly disordered. Remaining residuals in the 3-state fits (Figure 2J; Figure S4) could point to low fractional population of additional intermediates, but also could result from small non-idealities in the pressure profile, or from the change in temperature following the adiabatic expansion associated with the pressure drop.
We have shown that it is possible to determine 15N chemical shifts of short-lived protein folding intermediates. We anticipate that the method can be readily extended to the measurement of 13C′ and 13Cα frequencies, provided the folding process is not more complex than three-state. These chemical shifts then will provide an alternate pathway to studying the structure of such intermediates, analogous to the probing of chemical shifts of invisible structural states by relaxation dispersion NMR.32–33
Supplementary Material
ACKNOWLEDGMENT
We thank J. Ying.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Data fitting procedure and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Hoeltzli SD; Frieden C, Stopped-flow NMR spectroscopy - Real-time unfolding studies of 6-F19-tryptophan-labeled E. Coli dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 9318–9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koide S; Dyson HJ; Wright PE, Characterization of a folding intermediate of apoplastocyanin trapped by proline isomerization. Biochemistry 1993, 32, 12299–12310. [DOI] [PubMed] [Google Scholar]
- 3.Balbach J; Forge V; Vannuland NAJ; Winder SL; Hore PJ; Dobson CM, Following protein folding in real-time using NMR spectroscopy. Nat. Struct. Biol 1995, 2, 865–870. [DOI] [PubMed] [Google Scholar]
- 4.Roche J; Dellarole M; Caro JA; Norberto DR; Garcia AE; Garcia-Moreno B; Roumestand C; Royer CA, Effect of Internal Cavities on Folding Rates and Routes Revealed by Real-Time Pressure-Jump NMR Spectroscopy. J. Am. Chem. Soc 2013, 135, 14610–14618. [DOI] [PubMed] [Google Scholar]
- 5.Schlepckow K; Wirmer J; Bachmann A; Kiefhaber T; Schwalbe H, Conserved folding pathways of alpha-lactalbumin and lysozyme revealed by kinetic CD, fluorescence, NMR, and interrupted refolding experiments. J. Mol. Biol 2008, 378, 686–698. [DOI] [PubMed] [Google Scholar]
- 6.Schanda P; Forge V; Brutscher B, Protein folding and unfolding studied at atomic resolution by fast two-dimensional NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 11257–11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royer CA; Hinck AP; Loh SN; Prehoda KE; Peng XD; Jonas J; Markley JL, Effects of Amino-Acid Substitutions on the Pressure Denaturation of Staphylococcal Nuclease as Monitored by Fluorescence and Nuclear-Magnetic-Resonance Spectroscopy. Biochemistry 1993, 32, 5222–5232. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi T; Yamada H; Akasaka K, Thermodynamics of Unfolding of Ribonuclease-a under High-Pressure - a Study by Proton Nmr. J. Mol. Biol 1995, 250, 689–694. [DOI] [PubMed] [Google Scholar]
- 9.Kitahara R; Hata K; Li H; Williamson MP; Akasaka K, Pressure-induced chemical shifts as probes for conformational fluctuations in proteins. Prog. Nucl. Magn. Reson. Spectrosc 2013, 71, 35–58. [DOI] [PubMed] [Google Scholar]
- 10.Urbauer JL; Ehrhardt MR; Bieber RJ; Flynn PF; Wand AJ, High-resolution triple-resonance NMR spectroscopy of a novel calmodulin peptide complex at kilobar pressures. J. Am. Chem. Soc 1996, 118, 11329–11330. [Google Scholar]
- 11.Inoue K; Yamada H; Akasaka K; Hermann C; Kremer W; Maurer T; Doker R; Kalbitzer HR, Pressure-induced local unfolding of the Ras binding domain of RalGDS. Nature Structural Biology 2000, 7, 547–550. [DOI] [PubMed] [Google Scholar]
- 12.Dumont C; Emilsson T; Gruebele M, Reaching the protein folding speed limit with large, sub-microsecond pressure jumps. Nat. Methods 2009, 6, 515–U70. [DOI] [PubMed] [Google Scholar]
- 13.Kremer W; Arnold M; Munte CE; Hartl R; Erlach MB; Koehler J; Meier A; Kalbitzer HR, Pulsed Pressure Perturbations, an Extra Dimension in NMR Spectroscopy of Proteins. J. Am. Chem. Soc 2011, 133, 13646–13651. [DOI] [PubMed] [Google Scholar]
- 14.Charlier C; Alderson TR; Courtney JM; Ying J; Anfinrud P; Bax A, Study of protein folding under native conditions by rapidly switching the hydrostatic pressure inside an NMR sample cell. Proc. Natl. Acad. Sci. USA 2018, 115, E4169–E4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briggs MS; Roder H, Early Hydrogen-Bonding Events in the Folding Reaction of Ubiquitin. Proc. Natl. Acad. Sci. U. S. A 1992, 89, 2017–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khorasanizadeh S; Peters ID; Roder H, Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nat. Struct. Biol 1996, 3, 193–205. [DOI] [PubMed] [Google Scholar]
- 17.Krantz BA; Sosnick TR, Distinguishing between two-state and three-state models for ubiquitin folding. Biochemistry 2000, 39, 11696–11701. [DOI] [PubMed] [Google Scholar]
- 18.Jackson SE, Ubiquitin: a small protein folding paradigm. Org. Biomol. Chem 2006, 4, 1845–1853. [DOI] [PubMed] [Google Scholar]
- 19.Went HM; Benitez-Cardoza CG; Jackson SE, Is an intermediate state populated on the folding pathway of ubiquitin? FEBS Lett. 2004, 567, 333–338. [DOI] [PubMed] [Google Scholar]
- 20.Piana S; Lindorff-Larsen K; Shaw DE, Atomic-level description of ubiquitin folding. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 5915–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy G; Thirumalai D, Collapse Precedes Folding in Denaturant-Dependent Assembly of Ubiquitin. J. Phys. Chem. B 2017, 121, 995–1009. [DOI] [PubMed] [Google Scholar]
- 22.Alderson TR; Charlier C; Torchia DA; Anfinrud P; Bax A, Monitoring Hydrogen Exchange During Protein Folding by Fast Pressure Jump NMR Spectroscopy. J. Am. Chem. Soc 2017, 139, 11036–11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herberhold H; Winter R, Temperature- and pressure-induced unfolding and refolding of ubiquitin: A static and kinetic Fourier transform infrared spectroscopy study. Biochemistry 2002, 41, 2396–2401. [DOI] [PubMed] [Google Scholar]
- 24.Roche J; Caro JA; Norberto DR; Barthe P; Roumestand C; Schlessman JL; Garcia AE; Garcia-Moreno E B; Royer CA, Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 6945–6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roche J; Dellarole M; Caro JA; Guca E; Norberto DR; Yang YS; Garcia AE; Roumestand C; Garcia-Moreno B; Royer CA, Remodeling of the Folding Free Energy Landscape of Staphylococcal Nuclease by Cavity-Creating Mutations. Biochemistry 2012, 51, 9535–9546. [DOI] [PubMed] [Google Scholar]
- 26.Ptitsyn OB, Molten globule and protein folding. Adv. Prot. Chem, 1995, 47, 83–229. [DOI] [PubMed] [Google Scholar]
- 27.Balbach J; Forge V; Lau WS; Jones JA; VanNuland NAJ; Dobson CM, Detection of residue contacts in a protein folding intermediate. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 7182–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupce E; Freeman R, SPEED: single-point evaluation of the evolution dimension. Magn. Reson. Chem 2007, 45 (9), 711–713. [Google Scholar]
- 29.Hansen AL; Bruschweiler R, Absolute Minimal Sampling in High-Dimensional NMR Spectroscopy. Angew. Chem. Int. Ed 2016, 55, 14169–14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kay LE; Keifer P; Saarinen T, Pure Absorption Gradient Enhanced Heteronuclear Single Quantum Correlation Spectroscopy with Improved Sensitivity. J. Am. Chem. Soc 1992, 114, 10663–10665. [Google Scholar]
- 31.Peterson RW; Wand AJ, Self-contained high-pressure cell, apparatus, and procedure for the preparation of encapsulated proteins dissolved in low viscosity fluids for nuclear magnetic resonance spectroscopy. Rev. Sci. Instrum. 2005, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekhar A; Kay LE, NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 12867–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neudecker P; Robustelli P; Cavalli A; Walsh P; Lundstroem P; Zarrine-Afsar A; Sharpe S; Vendruscolo M; Kay LE, Structure of an Intermediate State in Protein Folding and Aggregation. Science 2012, 33, 362–366. [DOI] [PubMed] [Google Scholar]
- 34.Ying J; Delaglio F; Torchia DA; Bax A, Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J. Biomol. NMR 2017, 68, 101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.