Abstract
The Bsoft package is aimed at processing electron micrographs for the determination of the three-dimensional structures of biological specimens. Recent advances in hardware allow us to solve structures to near atomic resolution using single particle analysis (SPA). The Map Challenge offered me an opportunity to test the ability of Bsoft to produce reconstructions from cryo-electron micrographs at the best resolution. I also wanted to understand what needed to be done to work towards full automation with validation. Here, I present two cases for the Map Challenge using Bsoft: β-galactosidase and GroEL. I processed two independent subsets in each case with resolution-limited alignment. In both cases the reconstructions approached the expected resolution within a few iterations of alignment. I further validated the results by coherency-testing: i.e., that the reconstructions from real particles give better resolutions than reconstructions from the same number of aligned noise images. The key operations requiring attention for full automation are: particle picking, faster accurate alignment, proper mask generation, appropriate map sharpening, and understanding the amount of data needed to reach a desired resolution.
Keywords: single particle analysis, three-dimensional reconstruction, image processing, cryo-electron microscopy
Introduction
I developed Bsoft over the last 20 years with the help of others to process images of biological specimens (Heymann, 2001; Heymann, 2018; Heymann and Belnap, 2007; Heymann et al., 2008). The focus is mainly on single particle analysis (SPA), electron tomography and interpretation by segmentation and modeling. The algorithms for aligning single particles and calculating three-dimensional reconstructions have been extensively refined and used in many studies (Aksyuk et al., 2015; DiMattia et al., 2016; Heymann et al., 2003; Heymann et al., 2005; Keller et al., 2013; McHugh et al., 2014; Nemecek et al., 2012; Newcomb et al., 2017).
The Map Challenge (http://challenges.emdatabank.org/?q=2015_map_challenge) was conceived to gauge the state of single particle analysis (SPA), similar to the intent in the long-running CASP competition for structure prediction (Moult et al., 2018). As a first exercise, the organizers assembled seven data sets of already published structures. They challenged the 3DEM community to produce reconstructions comparable or better than the published ones.
As a software developer for SPA, the Map Challenge offered me an opportunity to test the newest version of Bsoft (Heymann, 2018). It is also a test of my understanding of single particle analysis and how to improve it. In particular, I tried to do as many of the operations as possible in an automated or semi-automated way. My ultimate goal is to have a fully automated processing workflow that removes the user bias from the analysis.
I selected two cases for processing: β-galactosidase (Bartesaghi et al., 2014) and GroEL (Vulovic et al., 2013). The first is an example of a rigid and pure protein that should not be difficult to reconstruct. The second is an artificially generated data set expected to give a good reconstruction. I achieved the expected resolution in both cases with proper validation, revealing some issues and complications as discussed below.
Validation
Bsoft allows the user to design processing in many different ways with considerable freedom in choosing parameters. It is therefore important for the user to follow good practices in processing the data. The recommended workflow in Bsoft starts with a very low-resolution reference map and splitting the data into two independent sets (see Figure 2 in (Heymann, 2018) for a general outline). Here, I selected two subsets of micrographs (β-galactosidase case) or particles (GroEL case) and processed them independently using resolution-limited alignment for each subset.
The goal of validation is to avoid issues that could influence the confidence we have in the final reconstruction. I considered the following biases with the corresponding remedies:
Initial reference bias: The initial reference map can influence the final reconstruction. I started with a highly low-pass (hard cutoff) filtered reference map (similar to the scheme in Scheres and Chen (Scheres and Chen, 2012)).
Mask bias: I used a reconstruction from one iteration as reference for the next. The map is typically masked to remove noise outside the particle envelope. I calculated a mask with a soft edge to avoid introducing high frequency terms that could compromise alignment. This is particularly important to preclude cross-talk between processing independent sets when using the same mask.
Noise bias: Most of the alignment information lies in the low frequency terms. I used a high resolution cutoff (effectively a hard low-pass filter) during alignment to minimize the effects of noise and improve the accuracy of the alignment. This resolution-limited alignment also has the advantage that any high frequency terms originating from a mask are excluded.
For validation, I used the following principles:
Independent sets: I split the micrographs (β-galactosidase) or particle images (GroEL) into two sets and proceeded to process them independently (Scheres and Chen, 2012). The maps from the two sets were used to calculate an “interset” FSC (Fourier shell correlation) curve (Harauz and van Heel, 1986).
Resolution-limited alignment: I aligned the particle images using resolution shells up to a high-resolution limit (Baker and Cheng, 1996). I chose to include only low frequency elements that are well represented (i.e., scored high, >0.8, in the previous FSC curve) in the reference maps, while excluding high frequency terms coming from noise or the mask. I calculate two maps from alternatingly selected particles within a set and calculate an “halfset” FSC curve. Any correlation beyond the high-resolution limit is taken as validation.
Coherence test: After the final alignment iteration, I calculated reconstructions from different numbers of randomly selected particle images, as well as from noise images aligned to the final map. With coherent particle images, the reconstructions should show much better resolution than those from the same number of noise images (Heymann, 2015).
Methods
Software
I did all of the processing using Bsoft 2.0 (Heymann, 2018). I used the following programs for specific steps in the SPA workflow (program names in italics): frame alignment: bseries; CTF (contrast transfer function) fitting: bctf; particle picking: bpick; global particle alignment: borient; local particle alignment: brefine; reconstruction: breconstruct; resolution estimation: bresolve; local resolution estimation: blocres; masking: bmask, beditimg, bfilter and bop; sharpening: bampweigh. The program bshow is an interactive display program used for checking CTF fits, particles picked, and reconstructions. I applied the CTF to reference projections during alignment, and corrected for it during reconstruction.
Masks
The mask used for the reference maps as well as for resolution estimation were calculated to avoid introducing high frequency components. First, I calculated a binary mask by thresholding the local variance map of the reference map (EMD_5995 for β-galactosidase and EMD_6422 for GroEL, http://www.emdatabank.org) and dilated it 2 times to close any holes (bmask). The mask was then smoothed with a 9x9x9 averaging kernel (bfilter) to soften the edges. The masks were used to remove background noise from the reference maps for alignment, as well as for estimating the resolution of reconstructions.
Computational setup
I ran many of the computationally intensive stages using the LSBR computer cluster (NIAMS, NIH) composed of a mixture of Macintosh Pro (multiple versions) and Linux (Fedora and Ubuntu, multiple versions) computers. The total number of cores were ~900, although availability varies because many of the computers are desktop machines in use during the day, and other users may also run jobs on the cluster. The cluster is managed with the Peach distributed processing system (Leong et al., 2005), allowing full usage as machines become available. The total estimated CPU usage for alignment was ~25600 hr (β-galactosidase, about 36 hr in real time, using ~700 cores) and ~2460 hr (GroEL, about 8 hr in real time, using ~300 cores). The difference lies in the sizes, symmetries and number of images processed, along the lines discussed in Heymann (Heymann, 2018). Reconstructions were calculated using 12 threads on a 2010 Macintosh Pro, with an estimated CPU usage for all iterations of ~50 hr (β-galactosidase, ~4 hr real time) and ~5 hr (GroEL, ~30 min real time).
Results
Case: β-galactosidase
The original micrographs were taken on a 300 kV FEI Titan Krios equipped with a Gatan K2 camera (Bartesaghi et al., 2014). The images were recorded as 509 dose-fractionated series (movies) (Table 1). Details of the command lines and scripts are provided in the Supplementary Material.
Table 1.
Data and processing report
| Parameter | β-Galactosidase | GroEL |
|---|---|---|
|
| ||
| Micrographs: | ||
| Number | 509 | - |
| Frames per micrograph | 5–40 | |
| Frame rate (/s) | 0.33–2.5 | |
| Dose per frame (e−/pixel) | 0.36–1.25† | |
| Accumulated dose (e−/Å2) | 15 – 33 | |
|
| ||
| CTF: Defocus range (μm) | 0.73–4.73 | 2.3–3.3 |
|
| ||
| Particles: | ||
| Picked/generated | 50139 | 10000 |
| Used in reconstruction | 19465* | 5088* |
|
| ||
| Reconstruction: | ||
| Symmetry | D2 | D7 |
| Resolution limit (Å) | 2.0 | 2.0 |
| Resolution estimate (Å, FSC0.143) | 3.4 | 4.3 |
| Sharpening | C-curve, LP 3Å# | C-curve, LP 3Å# |
Estimated from the image histograms
Selection based on projection matching cross-correlation
Amplitudes adjusted to the carbon electron scattering cross section and low-pass filtered to 3Å.
Preprocessing and particle picking
The preprocessing phase consisted of frame alignment, CTF determination, and particle picking. The frame alignment was done with bseries, which starts with a progressive alignment-averaging phase, followed by an iterative refinement (described in detail in (Heymann, 2018)). All frames were used and no dose weighting scheme was applied.
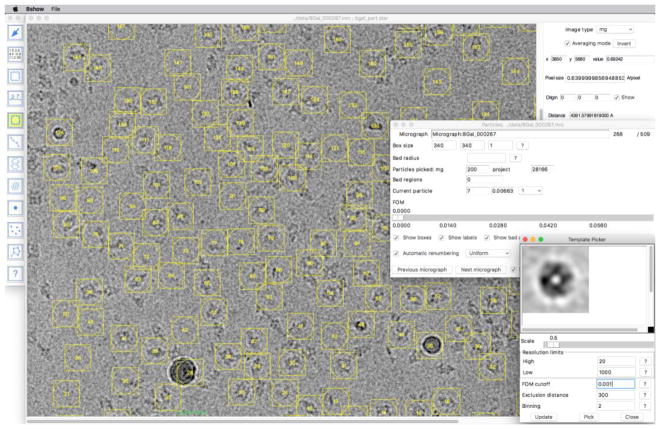
Initially, I picked a few particles in a micrograph and calculated a first template in bshow. I used this to find particles by cross-correlation (with a high resolution cutoff of 20 Å), cleaning out unwanted picks, and averaging the remainder for a new template. Once I was satisfied that it picked most acceptable particle candidates, I kept the template for automated picking (Figure 1).
Figure 1.
Particle images of β-galactosidase selected using the template picker in bshow. The panel in the bottom right shows the current particle average. This can be saved as a template or used for another round of particle picking using the parameters selected. The panel in the middle right shows the current particle parameters and statistics.
Next, I did the preprocessing phase fully automated using a script that distributed the jobs across the computational cluster with Peach (Leong et al., 2005). For each micrograph, the script sets up the parameter file (STAR format), aligns the frames, calculates a power spectrum, fits the CTF , and picks particles using the template.
I then checked the results of CTF fitting in bshow, adjusting the parameters when necessary. No micrographs were rejected because the Thon rings extended beyond 4 Å. I also checked the particle selections, deleting obvious erroneous picks such as ice or aggregates. This was the most intensive manual part of the processing and took about two days to complete for 509 micrographs.
Alignment and reconstruction
The first particle alignment used the published map produced from the data (Bartesaghi et al., 2014) (EMD_5995, http://www.emdatabank.org), limited to 60 Å. After the first global alignment with borient, I divided the micrographs into two sets that then constituted the independent sets for subsequent iterations. Particles were selected based on their FOM values (i.e., correlation coefficients from projection matching) and a reconstruction was calculated for each set (Figure 2a, iteration 1).
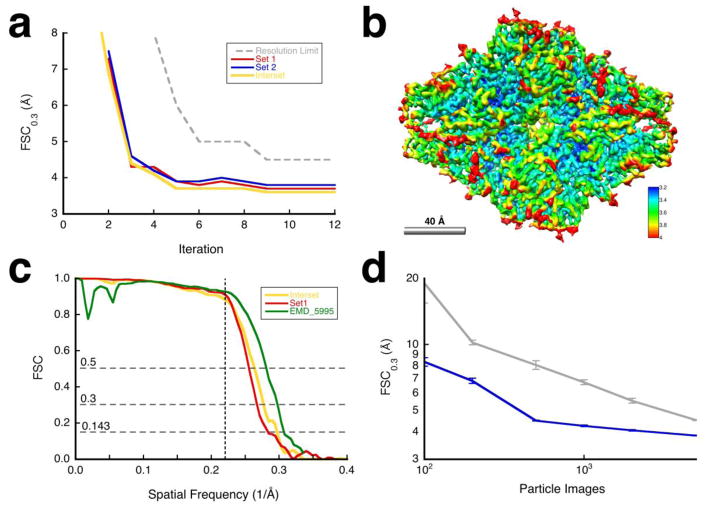
Figure 2.
Results for the β-galactosidase case. (a) Resolution estimates for the reconstructions for the different iterations: halfsets (blue/red) and interset (yellow). The dashed gray curve indicates the resolution limits used for alignment. I aligned the particles with a 60 Å limited reference in the first iteration to find global orientations (borient). During subsequent iterations, I refined the alignment, magnification and defocus for each particle (brefine). D2 symmetry was imposed on alignment and reconstruction. (b) Map sharpened against the carbon electron scattering cross section (bampweigh), colored according to the estimated local resolution (blocres, kernel = 26 Å, FSC0.3). (c) Comparison of the FSC curves for resolution limited alignment (halfset FSC for set 1 in red), independent sets (interset FSC in yellow), and relative to the published map (green) (Bartesaghi et al., 2014). The vertical dashed line indicates the resolution limit used in the final alignment (4.5 Å). (d) Validation by coherence: Resolution estimates with different numbers of randomly selected particles (blue) compared to aligned noise (gray) with error bars for 10 determinations at each point (Heymann, 2015).
I processed each set independently, using the masked reconstructed map from the previous iteration for the set as reference for the next and refined the previously determined orientations with brefine (Figure 2a). Within each set, two reconstructions were generated from alternatingly selected particles, masked, and used to calculate a halfset resolution. Similarly, the comparison between the reconstructions from the two independent sets gives an “interset” resolution. The alignment converged to a good solution within 5 iterations, with small but significant improvements afterwards (Figure 2a). I sharpened the final map against the carbon electron scattering cross section, producing a map (Figure 2b) that is very similar to the original map of Bartesaghi et al. (Bartesaghi et al., 2014).
Validation
Figure 2c shows an interset FSC curve (yellow line), an halfset FSC curve for set 1 (red line) and an FSC curve calculated against EMD_5995 (green line) (Bartesaghi et al., 2014). These curves are close to each other, with a resolution of 3.4 Å (at FSC = 0.143) from the interset curve, and a resolution of 3.6 Å (at FSC = 0.5) compared to the original map. The latter was reported to have a resolution of 3.2 Å (at FSC = 0.143, (Bartesaghi et al., 2014)).
To demonstrate coherence, I calculated reconstructions from different numbers of randomly chosen particles, and compared them to reconstructions from aligned noise images (Figure 2d). About five times fewer real particles are required to reach the same resolution as noise images, up to ~500 particles. Beyond that the curves converge, indicating an inherent limit in the data where it becomes indistinguishable from noise (and thus of dubious validity, as discussed in {Heymann, 2015 #4930}). Including more particle images would not improve the resolution significantly.
Case: GroEL
The individuals contributing the data set to the Map Challenge generated the particle images with the aim to simulate the conditions within an electron microscope, including noise and imposing transfer functions (Vulovic et al., 2013). Unfortunately, the original coordinates were incorrectly assumed to have D7 symmetry, and only orientations within one asymmetric unit were produced. The power spectra calculated from the images did not show the characteristic oscillations, precluding fitting the CTF. Therefore, I used the supplied parameters.
Alignment and reconstruction
For, the first particle alignment, I used a GroEL map from the EMDB (EMD_6422, http://www.emdatabank.org), limited to 40 Å. After alignment, I divided the particle images into two sets that then constituted the independent sets for subsequent iterations. Particles were selected based on their FOM values and a reconstruction was calculated for each set (Figure 3a, iteration 1).
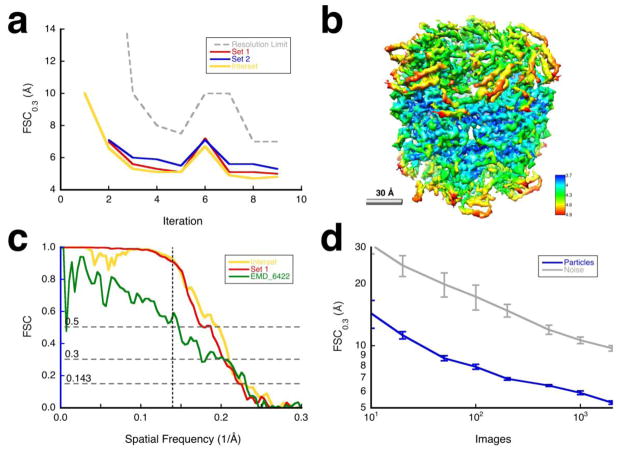
Figure 3.
Results for the GroEL case. (a) Resolution estimates for the reconstructions for the different iterations: halfsets (blue/red) and interset (yellow). The dashed gray curve indicates the resolution limits used for alignment. For iterations 1, 2 and 6, I aligned the particles globally (borient), while refining orientations, magnifications and defocus per particle during the other iterations (brefine). D7 symmetry was imposed during alignment and reconstruction. (b) Map sharpened against the carbon electron scattering cross section (bampweigh), colored according to the estimated local resolution (blocres, kernel = 28Å, FSC0.3). (c) Comparison of the FSC curves for resolution limited alignment (halfset FSC for set 1 in red), independent sets (interset FSC in yellow), and relative to the published map (green) (Vulovic et al., 2013). The vertical dashed line indicates the resolution limit used in the final alignment (7 Å). (d) Validation by coherence: Resolution estimates with different numbers of randomly selected particles (blue) compared to aligned noise (gray) with error bars for 10 determinations at each point (Heymann, 2015).
I subsequently processed each set independently, using the masked reconstructed map from the previous iteration for the set as reference for the next (Figure 3a). Within each set, two reconstructions were generated from alternatingly selected particles, masked, and used to calculate a halfset resolution. An interset resolution was calculated between reconstructions from the two independent particle sets. Iterations 1, 2 and 6 were globally aligned with borient, while the other iterations were run using brefine. The alignment reached a good solution within 5 iterations (Figure 3a). It is possible that the initial global searches misaligned some particles due to the low resolution of the reference, and that may degrade the reconstruction. I therefore did a global alignment at iteration 6 to potentially correct bad particle orientations. The slight improvement with local refinement afterwards could be attributed to correcting some orientations. The final reconstruction is shown in Figure 3b.
Validation
Figure 3c shows the FSC curves for the GroEL images (corresponding to those in Figure 2c for β-galactosidase). The curves indicate poor correlation even at lower frequencies, partly due to how the images were generated. The imposition of D7 symmetry further lowers the correspondence to EMD_6422 (green curve). Even with these limitations, the final map (Figure 3b) appears reasonable with identifiable secondary structure elements.
The coherence of the particle images decreases monotonically with the number of images, with a resolution always better than for noise images (Figure 3d). The apparent absence of an inflection point as is seen for the β-galactosidase case (Figure 2d) is likely due to the synthetic nature of the data.
Discussion
The Map Challenge presented data sets with already published targets. The question was therefore whether the target could be reproduced. I chose the β-galactosidase case (Bartesaghi et al., 2014) as a relatively small molecule with low symmetry, a good test for using Bsoft. I also chose the GroEL case (Vulovic et al., 2013) because it is a synthetic data set that should yield good results. In both cases I achieved the best result with current protocols. In the following I discuss what I learnt through this process.
Picking β-galactosidase particles
The new direct detectors generate large numbers of micrographs, making manual particle picking a laborious task. However, automated picking has not advanced to the point where I am satisfied with the outcome as compared to manual picking. The current state of the art is to overpick particles and trust the downstream selection to identify the best particle images.
I automatically picked β-galactosidase particle images with some manual cleanup as described in the Results section. It is evident from Figure 1 that many of the images picked are clearly not the particle of interest and would be omitted during manual picking. Yet, the final map is close enough to the target to suggest that selection of particles after alignment (~40%) worked to some extent (Figure 2b,c). However, the published target was also reconstructed from automatically selected particle images (Bartesaghi et al., 2014). Therefore, I don’t really know if manual picking would have produced a better map.
Synthetic images
It is difficult to generate synthetic images that have statistical characteristics comparable to real micrographs. Nevertheless, we base much of our software development on synthetic cases where we know the correct orientations and statistical parameters. The GroEL case represents another attempt at realistic simulation (Vulovic et al., 2013). Unfortunately, the symmetry of the molecule was not correctly considered in generating the images for the Map Challenge and I could not fit the CTF to the particle power spectra. Given these limitations, the resultant lower correlation with respect to the target is reasonable (Figure 3c).
Particle alignment
I used existing maps, EMD_5995 (Bartesaghi et al., 2014) and EMD_6422, for the initial references. The use of a correct reference map, even low-pass filtered, helps the processing to reach convergence quickly, resulting in valid reconstructions within a few iterations (Figures 2 and 3). An advantage here is the relative simplicity of the specimens (little contamination in the β-galactosidase case and synthetic images in the GroEL case). In more realistic cases, the presence of contaminants (“junk”) poses serious problems for particle picking and alignment programs. In those cases, tens of iterations are typically required to reach convergence.
An important part of the approach I use in Bsoft is to incorporate two validation concepts: resolution-limited alignment and independent sets (Heymann, 2018). In both cases the results show no overfitting by comparing the interset FSC with the FSC calculated against the structure-generated reference (Figures 2c and 3c). This is further corroborated by analyzing the coherency of the reconstructions compared to noise (Figures 2d and 3d) (Heymann, 2015).
What is required for automation?
One of my goals was to ascertain the state of automation in SPA with Bsoft. One persistent issue in SPA is how to automatically pick good particle images. The intuition is that if we only pick good images, we should get the best possible reconstructions. An open question is how much do bad particle images affect the final reconstructions. Presumably, bad images contribute randomly to the map, thus representing additional noise beyond that expected from the background. The optimal approach may center around a certain fraction of good or acceptable images. This obviously varies with the nature of the specimen.
The alignment of particle images can be automated in most current software packages. However, there are some crucial issues that need to be addressed before we can fully automate the process. The most critical is how to treat the reference maps. In most cases the map is masked to remove extraneous noise. It is important to have a soft-edged mask to avoid high frequency components. This can however still introduce shape elements that influence alignment and violate the independency of sets separated before processing. With the resolution-limited approach in Bsoft, any influence of high frequency elements is eliminated.
A second issue is that the reference map is often sharpened. This may affect some alignment algorithms but not others. In Bsoft, the cross-correlation calculated in borient covers a frequency band, where the relative amplitudes in the individual resolution shells matter. Conversely, each resolution shell is normalized (i.e., the intensities divided by their sum) in brefine, making it insensitive to reference map sharpening. How the reference map is handled may therefore affect the outcome in an algorithm-dependent manner. In the current state-of-the-art, the user decides how to do both masking and sharpening. It should be possible to incorporate these steps in a fully automated workflow.
Finally, the current efficiency of the programs can be significantly improved by eliminating redundant calculations and optimizing others. For example, the majority of workflows include several instances of forward and backward Fourier transformation. Another example is that the global search strategies often over-sample the search grid. In a better design the need for these could be reduced. This will however require a better understanding of both the theory and what operations are unnecessarily repeated during processing.
Future map challenges
This Map Challenge was a first attempt to assess the state of SPA. Considerable freedom was given to the participants to choose from several cases and process the data. A more defined challenge would present only one or two cases, preferably without known targets.
Supplementary Material
Acknowledgments
I thank the Map Challenge organizing committee for their efforts to advance single particle analysis: Bridget Carragher, Wah Chiu, Cathy Lawson, Jose-Maria Carazo, Wen Jiang, John Rubinstein, Peter Rosenthal, Fei Sun, Janet Vonck, and Ardan Patwardhan. This work was supported by the Intramural Research Program of the National Institute for Arthritis, Musculoskeletal and Skin Diseases, NIH.
Abbreviations
- SPA
Single particle analysis
- CTF
Contrast transfer function
- FSC
Fourier shell correlation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aksyuk AA, Newcomb WW, Cheng N, Winkler DC, Fontana J, et al. Subassemblies and asymmetry in assembly of herpes simplex virus procapsid. MBio. 2015;6:e01525–01515. doi: 10.1128/mBio.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci U S A. 2014;111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Cheng N, Huang R, Heymann JB, et al. The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly. Structure. 2016;24:1068–1080. doi: 10.1016/j.str.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Heymann JB. Bsoft: image and molecular processing in electron microscopy. J Struct Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- Heymann JB. Validation of 3D EM Reconstructions: The Phantom in the Noise. AIMS Biophys. 2015;2:21–35. doi: 10.3934/biophy.2015.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB. Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 2018;27:159–171. doi: 10.1002/pro.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol. 2008;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, et al. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Iwasaki K, Yim YI, Cheng N, Belnap DM, et al. Visualization of the binding of Hsc70 ATPase to clathrin baskets: implications for an uncoating mechanism. J Biol Chem. 2005;280:7156–7161. doi: 10.1074/jbc.M411712200. [DOI] [PubMed] [Google Scholar]
- Keller PW, Huang RK, England MR, Waki K, Cheng N, et al. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol. 2013;87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong PA, Heymann JB, Jensen GJ. Peach: a simple Perl-based system for distributed computation and its application to cryo-EM data processing. Structure. 2005;13:505–511. doi: 10.1016/j.str.2005.01.015. [DOI] [PubMed] [Google Scholar]
- McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, et al. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 2014;33:1896–1911. doi: 10.15252/embj.201488566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins. 2018;86(Suppl 1):7–15. doi: 10.1002/prot.25415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB. Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage φ6 procapsid. J Virol. 2012;86:11616–11624. doi: 10.1128/JVI.01347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Fontana J, Winkler DC, Cheng N, Heymann JB, et al. The Primary Enveloped Virion of Herpes Simplex Virus 1: Its Role in Nuclear Egress. MBio. 2017:8. doi: 10.1128/mBio.00825-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulovic M, Ravelli RB, van Vliet LJ, Koster AJ, Lazic I, et al. Image formation modeling in cryo-electron microscopy. J Struct Biol. 2013;183:19–32. doi: 10.1016/j.jsb.2013.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.