Abstract
Graphene oxide (GO) sheets have been used successfully as a supporting substrate film in several recent cryogenic electron-microscopy (cryo-EM) studies of challenging biological macromolecules. However, difficulties in preparing GO-covered holey carbon EM grids have limited their widespread use. Here, we report a simple and robust method for covering holey carbon EM grids with GO sheets and demonstrate that these grids can be used for high-resolution single particle cryo-EM. GO substrates adhere macromolecules, allowing cryo-EM grid preparation with lower specimen concentrations and provide partial protection from the air-water interface. Additionally, the signal of the GO lattice beneath the frozen-hydrated specimen can be discerned in many motion-corrected micrographs, providing a high-resolution fiducial for evaluating beam-induced motion correction.
Introduction
Recent technological breakthroughs have made single particle cryogenic electron microscopy (cryo-EM) a versatile and routine method for structure determination of macromolecules at high-resolution (Bai et al., 2015; Cheng, 2015). With automated data acquisition (Mastronarde, 2005; Suloway et al., 2005) enabled by stable high-end electron microscopes equipped with direct electron detection cameras and streamlined image processing software (Kimanius et al., 2016; Punjani et al., 2017), structure determination by single particle cryo-EM has never been easier. What remains more or less unchanged is the plunge freezing technique (Dubochet et al., 1988), which works well for many samples, particularly structurally stable ones. For fragile complexes, however, preparing good frozen-hydrated cryo-EM grids with intact and monodispersed particles is often a challenging task and a main bottleneck to cryo-EM structure analysis. It has been suggested that exposing protein samples to an air-water interface during plunge freezing can damage fragile protein complexes or induce preferred orientations in thin vitreous ice (Dubochet et al., 1988; Glaeser and Han, 2017; Glaeser, 2018). Additionally, some samples may prefer to stick to the carbon matrix of holey grids instead of being suspended in the vitreous ice spanning the holes ( Zhao et al., 2015; Snijder et al., 2017). A common approach to mitigate these problems is to apply a continuous thin layer of substrate to the holey carbon grid, making the distribution of particles in the holes more even and holding protein samples away from the air-water interface (Williams and Glaeser, 1972; Russo and Passmore, 2014; Han et al., 2016; Glaeser, 2018).
Amorphous carbon is the most commonly used substrate (Grassucci et al., 2007), but it adds significant background noise to particle images, limiting its use to relatively large particles such as ribosomes (Gao et al., 2007). Other substrates include monolayer sheets of graphene (Pantelic et al., 2012; Russo and Passmore, 2014) and two-dimensional crystals of streptavidin (Wang et al., 2008; Han et al., 2017). More recently, thin sheets of graphene oxide (GO) were introduced as a substrate (Pantelic et al., 2010). In two subsequent studies, GO coated grids were used on challenging macromolecular targets to ameliorate a tendency to aggregate at high concentrations and a bad preferred orientation problem, respectively (Bokori-Brown et al., 2016; Boland et al., 2017).
Compared to other options, GO sheets are nearly electron transparent, hydrophilic enough to adhere macromolecules from dilute solutions, inexpensive to purchase or synthesize, and amenable to functionalization (Pantelic et al., 2010; Chen et al., 2012). However, obtaining EM grids evenly covered with one or several layers of GO sheets has not been easy. In our experience, our attempts to use the previously reported drop-casting method (Pantelic et al., 2010) mostly produced grids with irregular coverage of GO sheets over the holes. Typically, fewer than twenty percent of holes were covered by one or a few layers of GO sheets, while the majority of the grids were either covered with multi-sheet aggregates or lacked GO entirely. Because we regularly need to screen tens of cryo-EM grids before finding conditions suitable for automated data acquisition and high-resolution structure determination, reproducibility and ease of manufacture are key considerations for any substrate.
To improve the usable area on GO covered grids, we established a simple and robust surface assembly procedure for evenly covering holey carbon EM grids with one to very few layers of GO sheets. These GO-covered grids are suitable for high-resolution single particle cryo-EM studies of biological macromolecules. We prepared frozen-hydrated archaeal 20S proteasomes using such GO-covered Quantifoil EM grids and collect a dataset resulting in a 2.5Å resolution 3D reconstruction, comparable to our best previous results in freestanding vitreous ice (Li et al., 2013; Zheng et al., 2017). We also used cryogenic electron tomography experiments (cryo-ET) to confirm that particles are concentrated close to the GO sheet, with about 90% of particles closer to the GO substrate than the air-water interface. In addition, lattice images of the graphene oxide film recorded together with frozen-hydrated 20S proteasome particles provide information for evaluating beam-induced motion correction.
Fabricating GO-covered holey carbon cryo-EM grids
Ideally, a GO covered grid would be completely covered in a single monolayer GO sheet without any wrinkled regions or GO aggregates. Commercial GO are available in the form of small flakes. Direct application of an aqueous suspension of GO flakes to a glow-discharged EM grid (drop-casting) (Pantelic et al., 2010) tends to leave many regions of the grid uncovered and deposits high-contrast multi-flake aggregates over many others. Initially we speculated that our commercial GO suspension might have deteriorated with age, so we used bath sonication to break up weakly aggregated sheets followed by centrifugation to isolate mostly large single GO flakes. This treatment reduces the presence of aggregates but does not greatly improve coverage uniformity.
GO surface is sufficiently hydrophobic to be enriched at air-water interfaces (Kim et al., 2010). We used this property to assemble a thin, mostly continuous film of GO flakes at the surface of a dish of pure water. By draining the water, the assembled GO film is then slowly lowered onto submerged holey carbon EM grids with their holey carbon film sides facing up (Figure 1A). In agreement with previous reports (Kim et al., 2010; Cote et al., 2011), we found that methanol-dispersed GO flakes spread and float easily on a pure water subphase. Once completely dried, GO flakes stably adhere to holey carbon films and remain adhered during blotting. The boundaries of individual GO flake on EM grids can be directly observed in the transmission electron microscope (TEM) (Figure 1B).
Figure 1. EM grids can be evenly coated with GO sheets using surface assembly.
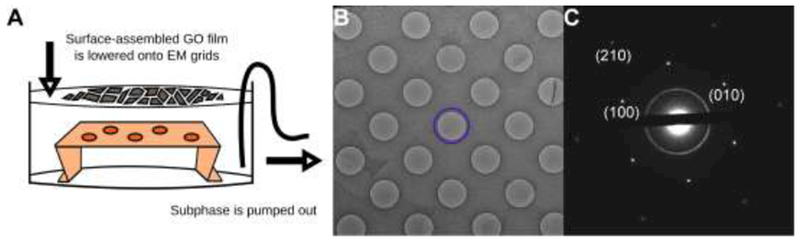
A. Schematic of the apparatus used for surface assembly of thin GO films and subsequent deposition onto holey carbon EM grids. B. Low magnification image of a holey carbon (Quantifoil) grid covered with a thin film of GO sheets. A hole covered by a single GO sheet is circled in blue. C. Electron diffraction pattern of the hole in B. Without a scrolled edge, it is hard to discern by image contrast alone whether a hole has a GO sheet spanning it. Instead, the diffraction pattern shows unambiguously the hexagonal lattice of each GO sheet.
According to the vendors, such as Sigma-Aldrich, their GO flakes are predominantly monolayer sheet, based on atomic force microscope height measurements. Selected area electron diffraction patterns taken from a single flake shows a single hexagonal lattice of Bragg peaks (Figure 1C), suggesting that such flake either contains a single layer of GO sheet, or multiple sheets stacking together coherently as a thin three-dimensional (3D) crystal. Because GO is exfoliated through chemical oxidation of graphite, it possible that some flakes contain more than one layer of GO sheet coherently stack together. We did not attempt to experimentally determine how many layers of GO sheets are coherently stacked together in each flake (Meyer et al., 2007), since the flakes are sufficiently thin that we did not observe any noticeable influence of image contrast. As a result, we assume that most of flakes contain a single layer of GO sheet. Diffraction patterns with sharp high-order spots indicates a long-range periodicity of the GO lattice over the hole (Figure 1C). While GO grids can acquire surface contaminants which render them less hydrophilic, they can be cleaned without damage by brief glow-discharge in air (five to ten seconds) immediately before use. A detailed protocol for fabricating GO grids by surface assembly is provided in the supplementary methods.
Suspensions of GO sheets can vary significantly in the lateral sheet size and degree of oxidation. The cryo-EM experiments in this study were performed on grids coated with GO sheets from a commercial source (Sigma-Aldrich). We have also recently produced grids with home-made suspensions of GO with larger sheet size. These GO suspensions were made according to an established protocol (Marcano et al., 2010) . Larger GO sheets coat grids more evenly and produce fewer high-contrast ‘edges’ in images, increasing the efficiency of data collection. Even so, surface assembly is robust to these variations and works well with both commercially-available GO suspensions and home-made GO sheets. The only important parameter to optimize with our protocol is the amount of GO applied to the surface. We typically make one or two test grids and ensure satisfactory coverage by screening in a transmission electron microscope before producing a large batch of grids.
Single particle cryo-EM of archaeal 20S proteasome on a GO grid
Using the archaeal 20S proteasome as a test specimen, we evaluated the practicality of using GO grids for high-resolution single particle cryo-EM. As previously reported, the commonly used plunge freezing procedure works well for GO grids, though we have found that longer blotting times (10-30 seconds) are often preferred. For the 20S proteasome, achieving an optimal particle distribution required a specimen concentration approximately ten times lower with GO (0.05 mg/mL) than without (0.5 mg/mL) (Figure 2A, Supplementary Figure 1A). By cryo-ET, we confirmed that 20S proteasome particles directly adhere to the GO substrate, as almost 90% of particles were closer to the GO substrate than the air-water interface (Figure 2B, Supplementary Figure 1C-D). Based on the locations of the few proteasomes and gold nanoparticle fiducials not adhered to the GO face, we estimate the thickness of the vitreous ice in this hole to be about 65 nm, while the deviation in the Z-location of 3D template-matched 20S proteasome particles is only about 5 nm. This suggests that 20S proteasomes on GO are protected from the air-water interface and lie on a common plane, a clear benefit when estimating micrograph defocus.
Figure 2. High-resolution single-particle cryo-EM on GO grids.
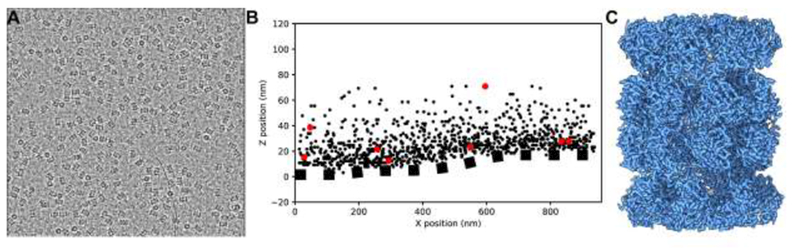
A. Micrograph of 20S proteasome particles over a GO support film. The GO film adds minimal background contrast. The specimen was applied at a concentration 0.05mg/mL, approximately ten times less than we typically use for this specimen. B. 3D localization of 20S particles from cryogenic electron tomography (cryo-ET). This tomogram was taken from a different GO grid than the one we used for the single-particle results: the 20S proteasome concentration here was higher and the GO substrate was far thicker (5-8 sheets). These conditions enabled us to identify how densely the GO surface could be coated with macromolecular specimen and to discern the position of the GO layer within low-resolution tomogram. Fiducial markers and points across the GO substrate surface were picked manually, while 20S proteasome particles were located in the tomogram using template matching. The GO is not flat but ripples and bends, as shown by the dotted line fitted by a spline to the points manually picked on the GO surface. The maximal distance between a picked proteasome particle and the GO substrate surface was 66nm, bounding the relative position of the air-water interface. 862 out of 969 picked proteasome particles (88.9%) are closer to the GO substrate than the air-water interface, strongly suggesting a direct interaction. All tomographic processing was performed using the IMOD software suite (Mastronarde, 2005). C. 3D reconstruction of the 20S proteasome on a GO support resolved at 2.5Å.
To compute a 3D reconstruction by single-particle analysis, we used the SerialEM automated data acquisition procedure to collect 740 dose-fractionated stacks. When choosing areas for data acquisition, we did not attempt to distinguish if holes were covered by GO or not. In this dataset, approximately 73% of the collected images were of high-quality, judged from the particle distribution and lack of numerous high-contrast GO edges. 10% of micrographs had thick crystalline ice quality likely unrelated to the GO, 10% of the holes are missing the GO support, and 7% of the holes had suboptimal coverage (usually too many overlapping sheets). Beam-induced motion was corrected using MotionCor2 (Zheng et a1., 2017).
For 3D reconstruction, we included only particles collected from holes covered with GO sheets and did not attempt to computationally remove the periodic graphene lattice, as might be done when using other crystalline lattice supports such as streptavidin crystal grids (Wang et al., 2008; Han et al., 2016). From this dataset, we determined a 3D reconstruction of the 20S proteasome at a resolution of 2.5Å (Figure 2C, Supplementary Figure 1E). The density map is visually indistinguishable in quality from our previously reported map of the 20S proteasome in freestanding vitreous ice but used fewer particles in the final reconstruction (117,578 particles on GO vs. 187,011 in freestanding vitreous ice (Zheng et al., 2017). This suggests that despite the added background contrast and the micrographs lost due to defects in the GO grid fabrication, GO poses no significant barrier to achieving high-resolution reconstructions.
Evaluating correction of beam-induced motion
GO sheet typically maintains a long-range periodicity (Figure 1C) that can tolerate the amount of electron beam radiation used to image the biological sample (Supplementary Figure 2). For a typical image recorded as a movie stack of subframes, only weak spots are seen in the sum image before motion correction (Figure 3A). These peaks can also be observed in short three-frame averages during the exposure, suggesting that the GO lattice is not significantly deteriorated by radiation damage but is primarily blurred by beam-induced motion (Supplementary Figure 2). After full motion correction with MotionCor2 (Zheng et al., 2017), the power spectrum show one complete set of hexagonally arranged peaks around 2.1Å resolution, beyond the physical Nyquist frequency of the K2 camera (2.43Å at the selected magnification) (Figure 3D). These peaks signal the first order reflections of the underlying GO lattice, with each hexagonal set of peaks corresponding to a single GO sheet. Reflections are typically very sharp, with an average half width of 10 Fourier pixels, suggesting that the vitrified GO lattice covering the image area maintains a long-range periodicity.
Figure 3. Evaluation of motion correction using GO peak intensities.
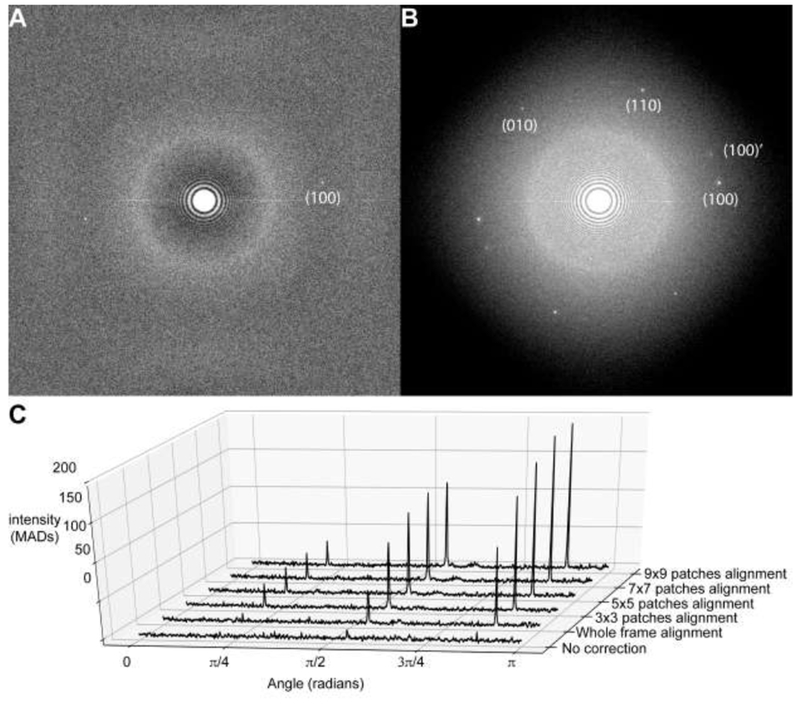
Power spectra from the image in Figure 2A before (A.) and after (B.) motion correction. Power spectra were calculated by averaging the periodograms of overlapping 512×512 pixel windows using a script in Python. C. GO peak height increases as the number of local motion patch trajectories calculated by MotionCor2 is increased. To generate 1D radial profiles showing the GO peaks with respect to the background, the aforementioned power spectrum was transformed into polar coordinates by cubic spline interpolation and a radial band of intensities from 2.0Å to 2.2Å was extracted. The maximum intensity component in this band was taken for each angle sampled in the half-circle 0 to π. This was done because the actual spatial frequency of the GO peak deviated from the expected position at 2.13Å, likely due to residual specimen tilt or uncorrected anisotropy in the magnification system (Zhao et al., 2015a). To compare 1D profiles between different motion correction schemes, the intensities in each 1D profile were scaled according to the median absolute deviation (MAD), which is a robust measure of scale. All calculations were performed in python with the scientific computing library scipy, with code available on request.
In MotionCor2 (Zheng et al., 2017), global motions are corrected by iteratively refining translational shifts for each frame, while nonuniform local motions are corrected by fitting numerous trajectories of local motions to a time-variant polynomial function. This polynomial estimates the instantaneous shift for any location in the image at any time point in the exposure, allowing local motions to be corrected smoothly at every image pixel. The number of image patches where local motion trajectories are measured is a free parameter in MotionCor2: a coarse grid of patches (i.e. 3×3 patches) will provide fewer measurements, but they should have high signal-to-noise ratio (SNR), while a finer grid (9×9 patches) will provide a finer measurement of localized motion, but at the expense of increased computation and a lower per-patch SNR.
GO peaks are nearly absent in the power spectra before motion correction, but they are readily visualized after global motion correction (Figure 3A and C). Applying local motion correction, we observe a trend where increasing the number of patches significantly increases GO peak intensity. For this particular image, using 7×7 patches increased the peak intensities approximately 200% with respect to the global alignment, while 9×9 patches provided no additional significant increase. While the intensities of all three distinct GO lattice peaks increase with better motion correction, the amount of improvement is different for each. We speculate that this could occur if the image still contains uncorrected motion, such as that occurring within each frame and if this motion were orthogonal to the direction of the foreshortened peaks.
Recently, Han et al (2017) observed a correlation between the resolutions of streptavidin 2D crystal and of the reconstruction determined from particles absorbed to the streptavidin substrate (Han et al., 2017) and proposed the use of peak intensity from a 2D streptavidin crystal to evaluate cryo-EM image quality. Resolution of high-resolution peaks from streptavidin crystal are sensitive indicators of specimen damage incurred during vitrification or exposure to the electron beam, and can thus be used to optimize vitrification conditions or to exclude images of damaged specimen.
Compared with a protein crystal lattice, GO is tolerant to vitrification and electron radiation damage, maintaining long-range order throughout the exposure (Supplementary figure 2). Beam-induced motion is the primary factor that weakens GO reflections during an exposure. The intensity and sharpness of the reflections after motion correction could be directly correlated with the quality of the correction.
To use resolutions or peak intensities of substrate crystals to evaluate corrections of local motions, the larger unit cell of streptavidin lattice is more advantageous. However, unlike GO lattice whose crystallinity is usually stable and can be verified by electron diffractions, crystallinity of streptavidin 2D crystals varies and image unbending can also improve the resolution (Henderson et al., 1986), making it necessary to separate resolution improvement by unbending or motion correction.
Conclusions and discussion
We have established a simple and robust procedure for covering holey carbon EM grids with thin films of GO sheets and have demonstrated the utility of the resultant grids for determining 3D reconstructions of macromolecules at high-resolution. We also showed that GO peaks in the image power spectrum are sensitive fiducial markers for the quality of the beam-induced motion correction. By simplifying their production, we anticipate that our method will make the benefits of GO grids more widely accessible.
Nevertheless, we believe the GO grids described here are only one step towards a more universal substrate for high-resolution single-particle cryo-EM. In the course of testing this protocol, we have attempted to use GO grids in several challenging, on-going cryo-EM projects. In some cases, macromolecular particles were bound and concentrated onto the GO surface in a native state like we report here for the 20S proteasome (Supplementary Figure 3). In other cases, particles were not visibly bound to the GO or appeared denatured (data not shown). This could occur if the proteins have no affinity for GO, as they would then have no protection from the air-water interface. There have also been reports that GO itself can destabilize protein structure and compromise enzymatic activity (Bai et al., 2017). In either case, an obvious solution is to functionalize the GO surface, increasing its affinity for interactions that preserve protein structure while passivating residual non-oxidized hydrophobic domains that may promote denaturation. GO has abundant epoxide groups on its surface that are amenable to such functionalization. We are actively working along these lines to improve the applicability of GO grids to fragile specimens.
Even if proteins interact favorably with GO and are protected from the air-water interface, they can still denature when directly exposed to filter paper during blotting. To protect GO-bound proteins from possible deleterious filter paper interactions, we can apply sample to the back side of the grid. During blotting, the grid bars should help prevent the filter paper from directly contacting the GO-bound proteins (Supplementary Figure 4). This method was originally used by electron crystallographers who called it ‘back-side injection’ (Gyobu et al., 2004). We have found that long blotting times (20-30s) are needed to get appropriately thin vitreous ice with back side application.
If a protein specimen is properly adhered to GO grids, we expect that imaging conditions should not be worse than if no substrate is used, and may be better. Particles bound to a common surface should have locally-correlated heights, improving the accuracy of defocus estimation. We noticed that the GO peaks in the power spectrum do not follow precise hexagonal symmetry, and that the lattice spacing for each of the three peaks vary slightly around 2.1 Å. This can be caused either by anisotropic magnification of the microscope or a slight tilt of the specimen, or a combination of the both. If the anisotropic magnification of the microscope is precisely calibrated, it is possible to derive the tilt angle and orientation of the specimen, facilitating a better local defocus determination. At a resolution of ~2.5Å, we did not notice any influence of the underlying GO lattice on particle alignment or classification for the 20S proteasome reconstruction. We therefore did not attempt to computationally remove the underlying GO lattice. It may become necessary to do so if the resolution of a reconstruction is beyond the GO lattice space, lattice space, ~2.1 Å.
Finally, we showed that subtle improvements in beam-induced motion correction can be detected by examining the intensity of GO peaks present in the power spectra of differently motion corrected micrographs. Note that we did not use GO peak intensity as an objective function to directly optimize subframe alignment, in which the motions between consecutive subframes larger than the unit cell of the GO lattice (2.13Å) could degenerate subframe registration solutions (i.e., aliasing). However, it is possible to optimize the motion correction by maximizing the GO peak intensity as an objective function. This is because the residual local motions after whole-frame rigid body motion correction are relative small (Supplementary Figure 1B in Zheng et al, 2017), thus a displacement between two consecutive subframes is likely much less than an angstrom and registration of shifts larger than the GO lattice unit cell can be excluded. If the periodic GO signal is used only to locally refine the initial registration trajectory estimated from aperiodic low-resolution image features (as MotionCor2 already does), deleterious artifacts of overfitting will be unlikely, permitting faithful recovery of the highest resolution features captured in the dose-fractionated electron micrographs.
Supplementary Material
Supplemental Figure 1: Cryo-EM of proteasome particles prepared using GO covered grid
A. Image of a hole partially covered with a GO. Fewer proteasomes are observed in the upper right-hand corner where the GO support is absent. The additional background contrast added by the GO substrate is barely perceptible. B-D. Projections through Z-slices of the tomogram summarized in Figure 2B. The slices depict B. the air-water interface (58-80 nm away from the lowest apparent point of the GO substrate), C. the higher part of the GO surface where bound proteasomes are observed (23-39nm), and D., the lower part of the GO surface where bound proteasomes are observed (8-23nm). E. Gold-standard FSC curves for the 20S proteasome, calculated from independently reconstructed half-maps with a spherical mask and with a tight mask.
Supplemental Figure 2: GO peak intensity is sensitive to initial beam-induce motion but not electron radiation
From the dose-fractionated stack used in Figure 3, we calculated power spectra from running three-subframe averages of without motion correction. Each row shows the 1D radial profile of a power spectrum calculated from an averaged image of three consecutive subframes. The 1D radial profile was calculated identically to those in Figure 3C. Two of three GO peaks are visible in most three-frame average images. The third peak is too weak to be detected from power spectrum of individual frames. While the peaks are not detected at the beginning of the exposure when beam-induced motion is greatest, they are not obviously reduced at the end of the exposure. The color scale represents the MAD.
Supplemental Figure 3: Image of a 170kDa complex on GO
Region of a cryo-EM image of a substrate-bound E. coli HptG complex (expected molecular weight ~170kDa) weakly crosslinked with bissulfosuccinimidyl suberate and bound to a GO grid. This particle is about 4 times smaller than the 20S proteasome and was never observed in vitreous ice without a support film, even at very high concentrations. On GO grids, individual particles are apparent when the specimen is applied at 1OOnM, suggesting that the GO is binding and concentrating the complex onto its surface. For this experiment, we used a GO grid coated with a homemade GO suspension. The specimen was applied to a GO grid, blotted for 4s on a Mark IV Vitrobot (100% humidity, blot force 0), and plunged into liquid ethane. The image was collected on a 200kV Talos Arctica equipped with a Gatan K2 camera (1.15Å/pixel, 60e- total exposure, 0.2s/frame, 12s exposure) and motion corrected with MotionCor2 (Zheng et al; 2017). The defocus of the image is estimated to be 2.4 microns (Zhang, 2016).
Supplemental Figure 4: Schematic representation of the back-side application technique.
Supplemental Movie 1: Tomogram of the 20S proteasome on a GO grid
Relative heights are given in nanometers with respect to the graphene oxide substrate. Few 20S proteasomes particles are bound at the air-water interface. The graphene oxide substrate has ripples and bends but primarily consists of upper and lower ‘plateaus’ that are both completely saturated with 20S proteasome particles. Some particles are observed in the vitreous ice layer, possibly interacting with the GO-bound layer of particles. The GO substrate itself is significantly thicker than would be ideal for single-particle cryo-EM, but for this tomography experiment the wrinkles of the multiple sheets unanbiguously signal the beginning of the GO substrate.
Acknowledgement
This work is supported in part by NIH grants R01GM082893, R01GM098672, R01HL134183, P50GM082250, P01GM111126, and 1S10OD020054 to Y.C. and NIH U54 CA209891 and UCSF Program for Breakthrough Biomedical Research to D.A.A. D.A.A and Y.C. are Investigators of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bai XC, McMullan G, and Scheres SH (2015). How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 40, 49–57. [DOI] [PubMed] [Google Scholar]
- Bokori-Brown M, Martin TG, Naylor CE, Basak AK, Titball RW, and Savva CG (2016). Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nat Commun 7, 11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland A, Martin TG, Zhang Z, Yang J, Bai XC, Chang L, Scheres SH, and Barford D (2017). Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat Struct Mol Biol 24, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Feng H, and Li J (2012). Graphene oxide: preparation, functionalization, and electrochemical applications. Chem Rev 112, 6027–6053. [DOI] [PubMed] [Google Scholar]
- Cheng Y (2015). Single-Particle Cryo-EM at Crystallographic Resolution. Cell 161, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote LJ, Kim J, Tung VC, Luo JY, Kim F, and Huang JX (2011). Graphene oxide as surfactant sheets. Pure Appl Chem 83, 95–110. [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, and Schultz P (1988). Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21, 129–228. [DOI] [PubMed] [Google Scholar]
- Glaeser RM (2018). Proteins, interfaces, and cryo-EM grids. Current opininon in colloid and interface science https://doi.Org/10.1016/j.cocis.2017.12.009. [DOI] [PMC free article] [PubMed]
- Glaeser RM, and Han BG (2017). Opinion: hazards faced by macromolecules when confined to thin aqueous films. Biophys Rep 3, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BG, Watson Z, Cate JH, and Glaeser RM (2017). Monolayer-crystal streptavidin support films provide an internal standard of cryo-EM image quality. J Struct Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BG, Watson Z, Kang H, Pulk A, Downing KH, Cate J, and Glaeser RM (2016). Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromolecules. J Struct Biol 195, 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, and Zemlin F (1986). Structure of purple membrane from halobacterium halobium: recording, measurement and evaluation of electron micrographs at 3.5 Å resolution. ultramicroscopy 19, 147–178. [Google Scholar]
- Kim J, Cote LJ, Kim F, Yuan W, Shull KR, and Huang JX (2010). Graphene Oxide Sheets at Interfaces. J Am Chem Soc 132, 8180–8186. [DOI] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, and Cheng Y (2013). Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcano DC, Kosynkin DV, Berlin JM, Sinitskii A, Sun Z, Slesarev A, Alemany LB, Lu W, and Tour JM (2010). Improved synthesis of graphene oxide. ACS Nano 4, 4806–4814. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Meyer JC, Geim AK, Katsnelson MI, Novoselov KS, Booth TJ, and Roth S (2007). The structure of suspended graphene sheets. Nature 446, 60–63. [DOI] [PubMed] [Google Scholar]
- Pantelic RS, Meyer JC, Kaiser U, Baumeister W, and Plitzko JM (2010). Graphene oxide: a substrate for optimizing preparations of frozen-hydrated samples. J Struct Biol 170, 152–156. [DOI] [PubMed] [Google Scholar]
- Pantelic RS, Meyer JC, Kaiser U, and Stahlberg H (2012). The application of graphene as a sample support in transimission electron microscopy. Solid State Communications 152, 1375–1382. [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Russo CJ, and Passmore LA (2014). Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas. Nat Methods 11, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Borst AJ, Dosey A, Walls AC, Burrell A, Reddy VS, Kollman JM, and Veesler D (2017). Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J Struct Biol 198, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Wang L, Ounjai P, and Sigworth FJ (2008). Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection. J Struct Biol 164, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RC, and Glaeser RM (1972). Ultrathin carbon support films for electron microscopy. Science 175, 1000–1001. [DOI] [PubMed] [Google Scholar]
- Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, and Brunger AT (2015). Mechanistic insights into the recycling machine of the SNARE complex. Nature 518, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Cryo-EM of proteasome particles prepared using GO covered grid
A. Image of a hole partially covered with a GO. Fewer proteasomes are observed in the upper right-hand corner where the GO support is absent. The additional background contrast added by the GO substrate is barely perceptible. B-D. Projections through Z-slices of the tomogram summarized in Figure 2B. The slices depict B. the air-water interface (58-80 nm away from the lowest apparent point of the GO substrate), C. the higher part of the GO surface where bound proteasomes are observed (23-39nm), and D., the lower part of the GO surface where bound proteasomes are observed (8-23nm). E. Gold-standard FSC curves for the 20S proteasome, calculated from independently reconstructed half-maps with a spherical mask and with a tight mask.
Supplemental Figure 2: GO peak intensity is sensitive to initial beam-induce motion but not electron radiation
From the dose-fractionated stack used in Figure 3, we calculated power spectra from running three-subframe averages of without motion correction. Each row shows the 1D radial profile of a power spectrum calculated from an averaged image of three consecutive subframes. The 1D radial profile was calculated identically to those in Figure 3C. Two of three GO peaks are visible in most three-frame average images. The third peak is too weak to be detected from power spectrum of individual frames. While the peaks are not detected at the beginning of the exposure when beam-induced motion is greatest, they are not obviously reduced at the end of the exposure. The color scale represents the MAD.
Supplemental Figure 3: Image of a 170kDa complex on GO
Region of a cryo-EM image of a substrate-bound E. coli HptG complex (expected molecular weight ~170kDa) weakly crosslinked with bissulfosuccinimidyl suberate and bound to a GO grid. This particle is about 4 times smaller than the 20S proteasome and was never observed in vitreous ice without a support film, even at very high concentrations. On GO grids, individual particles are apparent when the specimen is applied at 1OOnM, suggesting that the GO is binding and concentrating the complex onto its surface. For this experiment, we used a GO grid coated with a homemade GO suspension. The specimen was applied to a GO grid, blotted for 4s on a Mark IV Vitrobot (100% humidity, blot force 0), and plunged into liquid ethane. The image was collected on a 200kV Talos Arctica equipped with a Gatan K2 camera (1.15Å/pixel, 60e- total exposure, 0.2s/frame, 12s exposure) and motion corrected with MotionCor2 (Zheng et al; 2017). The defocus of the image is estimated to be 2.4 microns (Zhang, 2016).
Supplemental Figure 4: Schematic representation of the back-side application technique.
Supplemental Movie 1: Tomogram of the 20S proteasome on a GO grid
Relative heights are given in nanometers with respect to the graphene oxide substrate. Few 20S proteasomes particles are bound at the air-water interface. The graphene oxide substrate has ripples and bends but primarily consists of upper and lower ‘plateaus’ that are both completely saturated with 20S proteasome particles. Some particles are observed in the vitreous ice layer, possibly interacting with the GO-bound layer of particles. The GO substrate itself is significantly thicker than would be ideal for single-particle cryo-EM, but for this tomography experiment the wrinkles of the multiple sheets unanbiguously signal the beginning of the GO substrate.


