Abstract
Sodium channelopathies are a common genetic cause of paroxysmal disorders of the brain, peripheral nervous system, muscle, and heart. The phenotypes produced depend on a combination of the channel affected and its functional consequence; unfortunately, for missense variants, the latter is not clinically available. However we will show that the location of a missense sodium channel variant can be used as a surrogate for functional studies. We present data from epilepsy to illustrate clinical and treatment implications of sodium channel variants, the relationship between function, location and treatment response, then generalize this to other sodium channelopathies.
INTRODUCTION
Voltage-gated sodium channels have been implicated in numerous inherited paroxysmal disorders of the nervous system, muscle, and heart. Our goal is to provide a framework that helps neurologists understand the clinical and treatment implications of sodium channel variants the encounter in clinical practice. This will be accomplished through our objectives of (i) recognizing the relationship between location of a missense sodium channel gene variant and its effect on channel function, and (ii) categorizing clinical phenotype based on functional effect of a variant. The relationship between location, function and treatment response is also discussed. These interactions can be illustrated by the sodium channelopathies seen in people with epilepsy but generalize beyond that disorder.
Multiple mutations in genes that encode α-subunits sodium channels within the central nervous system have been associated with early onset epileptic encephalopathies and certain autosomal dominant epilepsy syndromes. The α-subunits protein folds into four homologous domains1. Each domain consists of six transmembrane segments (termed S1 through S6) connected by small extracellular and intracellular linkers identified by the transmembrane segments they separate. The four domains are linked by large intracellular loops. The transmembrane S4 segment is the main voltage sensor that triggers activation. The adjacent region transmembrane segment S4 and S5 (the S4–5) acts as an electromechanical couple translating depolarization to channel opening. Studies examining the effects of scorpion toxins have shown that the S3–4 segment plays an important role in the movement of S4 during inactivation. The pore through which sodium ions transverse the membrane consists of portions of transmembrane segments S5 and S6 and loop connecting S5–6. The sodium channel α-subunit genes SCN1A, SCN2A, and SCN8A are those most frequently associated with epilepsy.
Kanai noted that missense mutations in the pore of SCN1A were associated with more severe epilepsy phenotypes2. Using a larger database, Zuberi and colleagues demonstrated that epilepsy-associated missense SCN1A variants occurred most commonly in the pore region and voltage sensor than in other locations3. Meng and colleagues demonstrated that variants in the pore were more likely to be associated with a severe epilepsy phenotype and noted that these are functionally most often associated with a loss of channel function. In contrast, variants in the voltage sensor region (which they defined as the S3–4, S4 and S4–5 regions) were unlikely to demonstrate a complete loss of function4.
Although SCN1A, SCN2A, and SCN8A genes are highly conserved, the main epilepsy-associated functional consequences of SCN1A variants differ from those of SCN2A and SCN8A5. At the extreme, nonsense and frameshift mutations typically lead to loss of function and are common in epilepsy associated with SCN1A, accounting for about half of the cases of SCN1A-associated early onset epileptic encephalopathies (EOEE). In addition, many of the missense variants in SCN1A that cause EOEE result from complete or partial loss of channel function4. In contrast, functional consequences of missense variants in SCN2A and SCN8A are different, with variants associated with EOEE demonstrating electrophysiological changes that most often result in a gain of function6–8.
The differing physiological effects can have therapeutic implications. SCN2A variants associated with early onset epileptic encephalopathies respond to sodium channel modulating antiepileptic medications. In contrast, when epilepsy is seen with loss-of-function SCN2A variants, the seizures have a later onset and tend to respond more poorly to sodium channel agents8. Similarly, loss-of-function variants in SCN1A respond poorly to sodium channel modulators9. It is unknown if these functional differences are reflected in different distribution of epilepsy-associated SCN2A and SCN8A missense variants as well. To examine this question, we compared the distribution of SCN1A, SCN2A, and SCN8A variants associated with epilepsy and compared the types of neuronal sodium channel gene variants in people with epilepsy and other neurological disorders.
DISTRIBUTION OF VARIANTS IN CENTRAL NERVOUS SYSTEM SODIUM CHANNEL GENES
Pathogenic and potentially pathogenic missense variants in SCN1A, SCN2A, and SCN8A were identified from the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk; accessed 3/22/16) and from the open access Database of Chromosomal Imbalance and Phenotype in Humans (DECIPHER) v9.14 data (http://decipher.sanger.ac.uk; accessed 4/28/17). Additional pathogenic variants in SCN8A were also identified from www.scn8a.net (accessed 2/17/17) and in SCN8A and SCN2A from recent publications. After review of literature associated with each variant, only variants that could be classified as likely pathogenic or pathogenic for epilepsy based on criteria provided by the American College of Medical Genetics10 were included in the epilepsy dataset. Some variants were identified in people with neurological disorders other than epilepsy. These were examined separately. If a variant was identified but the presence or absence of seizures could not be determined for the phenotype provided, it was excluded. A comparative dataset of sodium channel variants from 60,706 people free of severe pediatric disease was obtained from the Exome Aggregate Consortium (ExAC; http://exac.broadinstitute.org;11).
Domains in the voltage-gated sodium channel amino acid sequence were defined from the SWISS-PROT database (last accessed 12/6/17) and then grouped according to approximate functional domains based on previously published groupings4. The pore region was defined as segments S5, S5-S5, S6; the voltage sensor region (VSR) as S4 and its associated linkers (S3–S4 and S4–S5). Other transmembrane segments and their linking regions were grouped (TMO) and the intracellular loops linking domains I–IV were grouped together (Loops). The N-terminus (N) and C-terminus (C) were grouped separately. Thus 6 different regions (VSR, pore, TMO, loops, N, and C) were compared.
Multiple different amino acid substitutions were seen at some locations but the amino acid location was only included once during statistical analysis. Because the number of amino acids differed between each region, the relative frequency of variants was calculated by dividing the number of variants per amino acid in a particular segment by the number of variants per amino acid in the entire protein. This approach permitted a comparison between different channels despite differences in the overall number of variants identified between the channels. This method is similar to that reported elsewhere3, 5. A 2 × 6 Fisher’s Exact Test was used to compare the distribution of variants across the entire channel. A 2 × 2 Fisher’s Exact Test or chi square test (based on the number of variants) was used to compare specific regions within the channels. The p-values listed are based on Fisher’s Exact Test unless noted otherwise.
The number of missense, non-sense, frameshift and splice site variants in SCN1A, SCN2A and SCN8A as well as their associated phenotypes are summarized in Table 1. Of the pathogenic or likely pathogenic epilepsy-associated missense variants identified, 386 SCN1A variants occurred at unique amino acid locations within NaV1.1, 102 of the 116 epilepsy-associated SCN2A variants occurred at unique amino acid locations within NaV1.2, and 42 of the 46 epilepsy-associated SCN8A variants occurred at unique amino acid locations within NaV1.6. All nine SCN1A variants and all three SCN8A variants that did not have epilepsy as part of their phenotype occurred at unique locations as did nine of 11 SCN2A variants. An additional six novel variants in SCN2A were identified from exome studies in people with autism16, 17; however, detailed information about their phenotypes was not available to ascertain whether participants had concomitant epilepsy. These were excluded from our analysis. In the population-based ExAC database, 372 variants at 328 unique locations in SCN1A; 287 variants at 253 unique locations in SCN2A, and 249 variants at 219 unique locations in SCN8A were identified.
Table 1.
Comparisons of Functional Effect, Clinical Phenotype and Variant Type in SCN1A, SCN2A and SCN8A Channelopathies
| PHENOTYPE | ||||||
|---|---|---|---|---|---|---|
| Variant type | Effect on Channel Function |
EOEE | B(F) NIS (SCN2A/8A) or GEFS + (SCN1A) |
Epilepsy (other) |
Other; No epilepsy |
Number of variants |
| SCN2A/8A | ||||||
| Missense | Gain, loss or mixed: dependent on location | 65 % (n=113) | 15 % (n=26) | 12 % (n=22) | 8 % (n=14) | 175 |
| Nonsense/frameshift/splice site | Complete loss | 0 | 0 | 33 % (n=15) | 67% (n=30) | 45 |
| SCN1A | ||||||
| Missense | Gain, loss or mixed: dependent on location | 83 % (n=434) | 9 % (46) | 6 % (n=34) | 2 % (n=9) | 523 |
| Nonsense/frameshift/splice site | Complete loss | 93 % (n=476) | 1 % (n=7) | 6 % (n=28) | 0 | 511 |
Abbreviations: EOEE = early onset epileptic encephalopathy as associated syndromes; B(F)NIS = Benign (familial) neonatal/infantile seizures; GEFS+ = Genetic Epilepsy with Febrile Seizures Plus; Other; No epilepsy, includes developmental delay, schizophrenia, autism spectrum disorders, ataxia and familial hemiplegic migraine with no history of seizures.
Epilepsy-Associated SCN1A, SCN2A and SCN8A variants
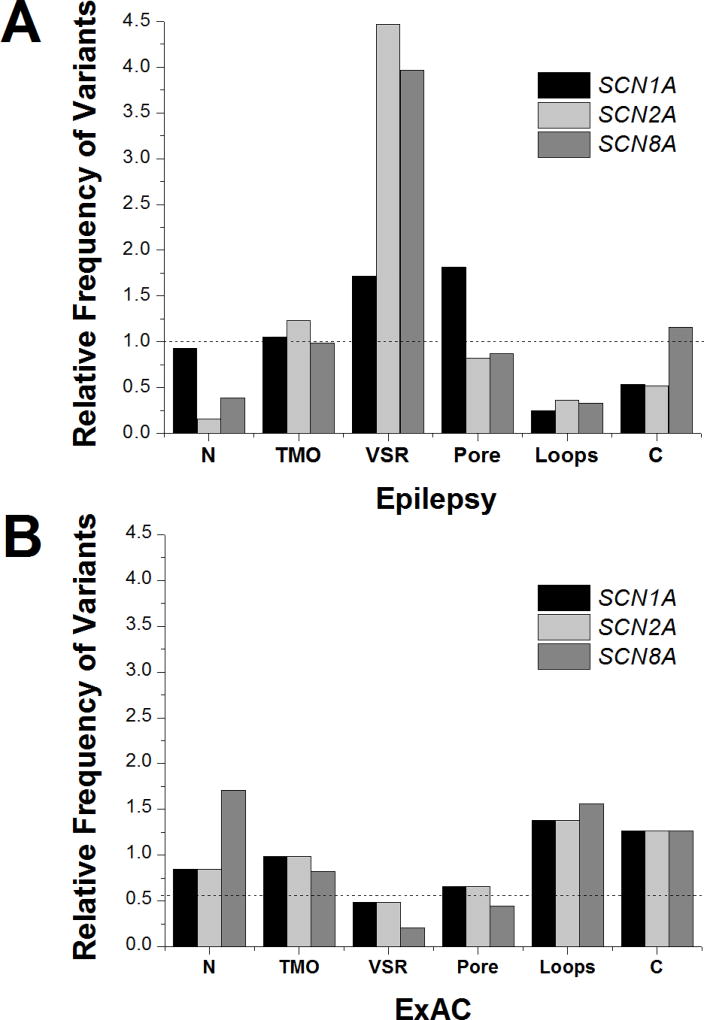
The distribution of epilepsy-associated variants for each gene are shown in Figure 1A. There were no statistical differences between the distribution of epilepsy variants between SCN2A and SCN8A (p=0.28); therefore, these were combined for subsequent comparisons. The distribution of epilepsy-associated variants in SCN1A was significantly different than in SCN2A/SCN8A (p<0.0001; Chi-square). The differences were driven by differences in the VSR and pore. Consistent with prior observations, the VSR and pore were the predominant locations in SCN1A to harbor variants pathogenic for epilepsy. For SCN1A, these regions both had an almost 2-fold enrichment of epilepsy-associated variants compared to the protein as a whole (1.7-fold increase in VSR; 1.8-fold increase in the pore). In contrast, while epilepsy-associated pathogenic variants in the VSR region were common in SCN2A/8A (4.0-fold increase; p <0.0001 compared with SCN1A), variants in the pore were slightly less likely to occur in SCN2A/8A compared to the genes in their entirety (0.97-fold decrease; p <0.001 compared with SCN1A).
FIGURE 1.
Distribution of (A) epilepsy-associated missense variants and (B) population-based missense variants in SCN1A, SCN2A and SCN8A. The dashed line represents the variant frequency over the entire protein.
Overall, there was relative sparing of the intracellular loops for all genes (0.29-fold decrease in SCN1A and 0.40-fold decrease in SCN2A/8A) and this distribution was not different between the genes (p = 0.26). However, within the intracellular loop that connects domains III and IV there is a region that acts as the inactivation gate18. This loop also has been identified as a common region for prolonged QT syndrome-associated variants in SCN5A19. Comparing the epilepsy and population based databases, this loop has significantly more epilepsy-associated variants than the other intracellular loops for both SCN1A (p < 0.01) and SCN2A/8A (p<0.001).
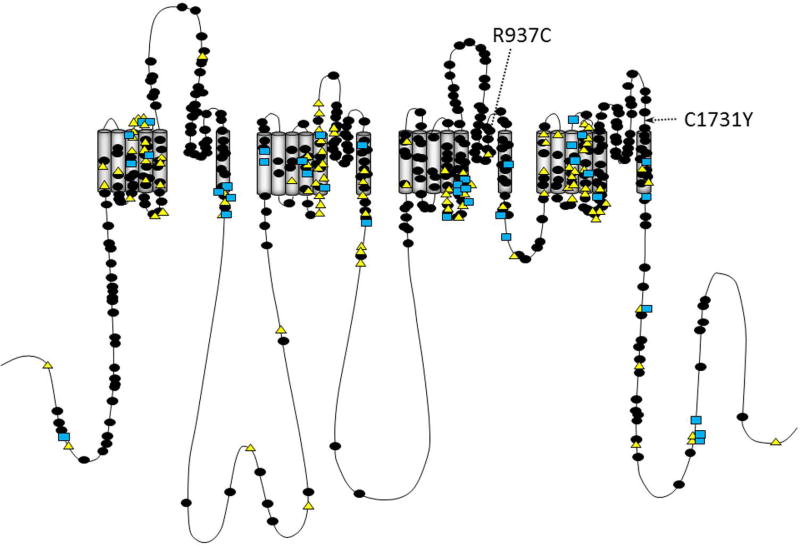
The distribution of variants within the segments of the pore and VSR is illustrated in Figure 2. The most notable difference between the pore regions for the sodium channels occurred in the extracellular S5–6 loop; for SCN1A, 123 of the 185 (66%) of the epilepsy-associated variants located in the pore occurred in the S5–6 segment. In contrast, only seven of the thirty-four (21%) epilepsy-associated pore variants in SCN2A/8A occurred in the S5–6 region (Odds Ratio (OR) 9.1; p<0.0001; Confidence Interval (CI) 4.2–20.2; Chi-Square). The frequency of epilepsy-associated variants in S4–5 was somewhat more common in SCN2A/8A compared with SCN1A (OR 2.1; 95% CI 1.1–3.9; p =0.02, Chi Square).
FIGURE 2.
Predicted transmembrane topology of NaV showing the location of epilepsy-associated variants in SCN1A (filled circles), SCN2A (triangles) and SCN8A (squares). Location of variants examined here are identified by the arrows.
Population-Based SCN1A, SCN2A and SCN8A variants
The distribution of variants identified from the ExAC database are shown in Figure 1B. As was the case with the epilepsy-associated variants, there were no statistical differences between the distribution of epilepsy variants between SCN2A and SCN8A (p=0.27; Fisher’s Exact Test); therefore, these were combined for subsequent comparisons. In contrast to the epilepsy-associated variants, the overall distribution of variants within ExAC did not differ between SCN1A and SCN2A/8A (p=0.24). With the exception of the pore region, the distribution of variants in ExAC was largely the inverse of that seen with epilepsy associated variants. Regions with relative sparing in the epilepsy group were overrepresented in the population-based dataset and vice versa. For example, in the ExAC database the intracellular loops had a relative variant rate of 1.4 for both SCN1A and SCN2A/8A and VSR had a relative variant rate of 0.54 for SCN1A and 0.30 for SCN2A/8A when compared to the proteins as a whole. However, variants in the pore regions were infrequent in both SCN1A (0.66-fold decrease) and SCN2A/8A (0.58-fold decrease) within the ExAC database.
Non-epilepsy associated pathological SCN2A and SCN8A missense variants
An additional 11 missense variants (at nine unique locations) in SCN2A and three missense variants in SCN8A were identified in people with intellectual disabilities who did not have concurrent epilepsy; these were absent from the ExAC database. Although the number of variants is small, the variants seen in people with developmental disabilities without epilepsy tended to be more common in the pore (OR 4.3; CI 1.5–13; p = 0.009) than in other regions when compared to epilepsy-associated variants in SCN2A/8A.
FUNCTIONAL EFFECTS OF NON-EPILEPSY ASSOCIATED PATHOLOGICAL MISSENSE VARIANTS IN THE PORE REGION
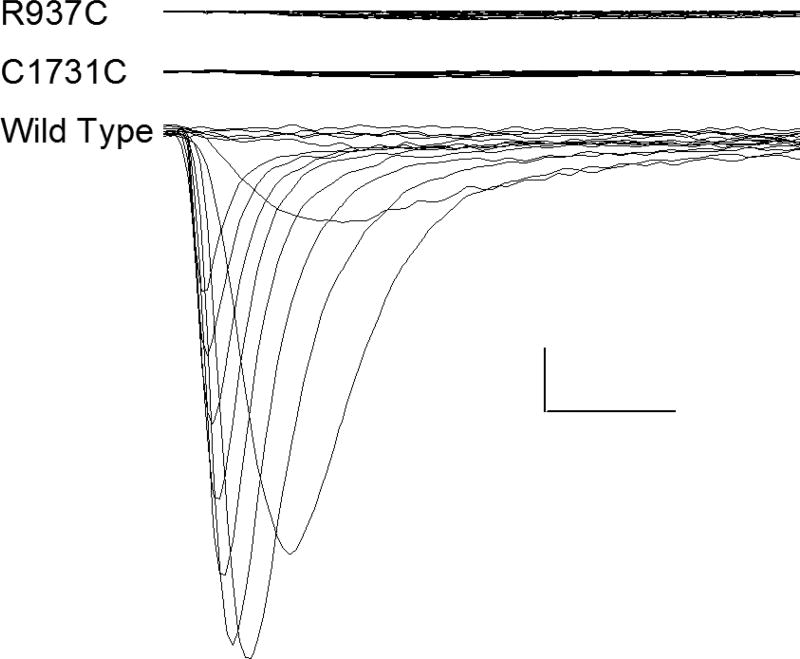
Frameshift, splice site, and nonsense variants typically result in a loss of function and these have also been reported in SCN2A and SCN8A. In contrast to SCN1A (where all reported frameshift, nonsense, and splice-site variants have epilepsy as part of the phenotype; Table 1), these types of variants in SCN2A/8A are not associated with epilepsy in the majority of cases (30 of 45; p <0.0001). For those who did have epilepsy, the onset of seizures tended to occur at over one year of age8. The non-epileptic phenotypes associated with frameshift, nonsense, and splice-site variants in SCN2A and SCN8A included developmental delay, autism, dysmorphic appearance, and schizophrenia. While the effects of missense variants (in SCN1A, SCN2A, and SCN8A) seen in people with epilepsy have been well studied, less is known about the effects of missense variants seen in people with developmental disorders without epilepsy. To examine the relationship between location, function, gene, and phenotype we selected two SCN2A variants (R937C and C1731Y) for in vitro functional evaluation. These variants were selected because they were located in the S5–6 region of the pore and were identified in people with developmental delay without epilepsy. These two factors would predict a loss of channel function. The functional studies were done using methods previously described by others (see Acknowledgments)12–15. NaV1.2 currents in cells transfected with wild-type or mutant cDNAs were recorded and representative traces are shown in Figure 3. Not all cells that were successfully transfected with the beta subunits (which contained the markers that identified transfected cells) also appeared to be transfected with adequate amounts of cDNA for the alpha subunit (which produces the Nav1.2 current). The percentage of tested cells exhibiting quantifiable sodium currents (defined as peak current ≤−400 pA) was significantly lower for R937C and C1731Y as compared to WT channels (WT, 30%, n = 53; R937C, 0%, n = 45, p < 0.0001; C1731Y, 0%, n = 40, p <0.0001; Wilcoxon Rank Sum Test). Of the sixteen cells transfected with WT cDNA which had suitable macroscopic currents, the mean current density was −136 ± 33 pA/pF. Similar to R937C and C1731Y, none of the non-transfected cells produced a quantifiable current. Thus, these pore-region SCN2A variants did result in the predicted loss of function.
FIGURE 3.
NaV1.2 activation currents in cells transfected with wild-type (WT), R937C, and C1731Y plasmids. The R937C and C1731Y variants have been reported in children without epilepsy. The location of these non-epilepsy associated variants are identified in Figure 2 by the arrows. Currents were activated by voltage steps to between −80 and +60 mV from a holding potential of −120 mV. Calibration bars 200 pA (vertical), 1 ms (horizontal). To ensure that transfection or mutagenesis errors were not the cause of the low currents for R937C and C1731Y, transfections were done with separate preparations of DNA, and transfections were repeated three to four times.
DISCUSSION
Sodium channelopathies are a common cause of early onset epileptic encephalopathies and some self-limited epilepsy syndromes. Here we demonstrated that epilepsy-associated missense variants mutations SCN2A and SCN8A tend to occur in the main voltage sensor and adjacent linking regions while variants in the pore region are underrepresented in epilepsy. In particular, the S5–6 segment in SCN2A and SCN8A are largely spared from epilepsy-associated mutations. In contrast, epilepsy-associated SCN1A variants occur preferentially from the voltage sensor region and associated linkers and throughout the entire pore. In contrast to SCN2A and SCN8A, the most common area for SCN1A-associated epilepsy is in the S5–6 region of the pore. Few variants in the voltage sensor region or pore are seen in the population-based ExAC database for any of these sodium channels indicating an evolutionary disadvantage for mutations in the pore regardless of the sodium channel gene. This implies that severe childhood onset disorders other than epilepsy may result from SCN2A and SCN8A variants in the pore region. Severe developmental disability is one such possibility. SCN2A is one of the most commonly identified gene in exome sequencing studies examining the genetic contribution to neurodevelopmental disorders16, 17, 20, 21.
Functional difference may underlie the varied clinical presentations and differences in distribution of the epilepsy-associated sodium channelopathies. The majority of people with SCN2A variants predicted to produce loss of channel function (nonsense, frameshift, and splice-site variants) do not have epilepsy8. In addition, missense variants in people with SCN2A variants seen in non-epileptic individuals with developmental delay result in loss of channel function. Ben-Shalom and colleagues demonstrated that autism-associated SCN2A variants in the pore region resulted in loss of channel function22. Our findings support their observations. Similarly, two SCN8A missense variants seen in people with developmental disabilities without epilepsy have been studied and these variants also produced non-functional channels23. These observations support the notion that loss-of-function SCN2A and SCN8A variants are more commonly associated with conditions other than epilepsy. In contrast, all variants in SCN1A which result in complete loss-of-function result in epilepsy4. Gain-of-function variants in SCN1A, SCN2A, and SCN8A are also associated with epilepsy4, 7, 8.
In addition to differing clinical phenotypes, functional consequences of SCN1A, SCN2A, and SCN8A gene variants have important therapeutic implications. Early onset epileptic encephalopathies associated with pathogenic SCN1A variants are thought to be poorly responsive to antiepileptic treatments that modulate sodium channel function. In contrast, many patients with SCN2A and SCN8A-associated early onset epileptic encephalopathies tend to respond better to anticonvulsant medications and the associated missense variants have shown a gain-of-function in in vitro functional assessments6. Wolff and colleagues8 recently showed that there are important phenotype and treatment response differences in people with gain-of-function SCN2A variants compared to those with loss-of-function variants. Gain-of-function variants tended to present with earlier onset epilepsy (< 3 months of age) and had a better response to sodium channel modulating anti-epileptic treatments. In contrast, when epilepsy was present in people with loss-of-function variants, the seizures had a later age of onset and were often exacerbated by treatment with sodium channel modulators. Although these functional effects have significant implications for personalization of therapy for sodium-channel epileptic encephalopathies, at present there are numerous obstacles to widespread use of in vitro functional studies for missense variants in clinical care24.
The different distribution of epilepsy-associated variants in SCN2A/8A compared to SCN1A provides an opportunity to aid in selection of therapies without laborious functional studies. Loss-of-function missense variants often localize to the pore region (especially the S5–6 region); while variants with a gain-of-function (or mixed loss- and gain-of-function) are seen in the voltage sensor (S4) and adjacent linkers. This suggests that certain locations of variants within the resultant protein can be a surrogate when more detailed functional studies are not available. Sodium channel blocking drugs should be avoided in pore-region variants but may be useful for some VSR variants. Even a small subset of people with SCN1A mutations could respond favorably to sodium channel modulation, provided the variant is in a region that predicts gain- of-function (e.g. in S4–5). Unfortunately, the functional consequences of variants outside of the pore and S4–5 are less predictable. This approach may also have treatment implications beyond epilepsy. Behavioral disorders can be seen in people with autism and developmental delay and sodium channel modulators such as lamotrigine are potential treatment options. Avoiding these agents in children with SCN2A or SCN8A variants without epilepsy may be sensible.
The association between functional consequences, clinical phenotype, and location of variant generalizes to sodium channelopathies outside of the central nervous system as well (Table 3)5. For example, cardiac conduction disorders associated with SCN5A variants depend upon the physiological effect of the variant. Loss of function variants are associated with Brugada Syndrome and gain of function variants are associated with Prolonged QT Syndrome Type 3. The distribution of prolonged QT associated variants shares similarities with the epilepsy-associated gain of function SCN2A and SCN8A variants, with many variants occurring in the VSR and a there is a relative sparing of the S5–6 region in the pore25. In addition, variants in the DIII–DIV are vastly over-represented in prolonged QT syndrome. SCN5A variants associated with Brugada Syndrome are more widely distributed within the transmembrane regions; however, analogous to SCN1A variants in epilepsy, SCN5A variants located within the pore region are associated with a more complete loss of function and a more severe clinical phenotype26. Similarly, gain of function variants in SCN4A are associated with a number of paroxysmal neuromuscular conditions (such as hyperkalemic periodic paralysis, hypokalemic periodic paralysis, and paramyotonia congenita) and in SCN9A are associated with paroxysmal pain disorders (such as paroxysmal extreme pain disorder and inherited erythromelalgia). The majority gain of function variants in SCN4A occur within the VSR and pore with relative sparing of S5–627. In addition to the VSR, disorders associated with gain of function variants in SCN9A are also common in the DIII–IV linker. In contrast to the other sodium channels, loss of function variants in SCN4A and SCN9A are seen primarily when both alleles have a loss of function variant. Taken all together, the clinical phenotype of sodium channelopathies associated with variants in the VSR and DIII–IV linking differ from those seen with variants in the S5–6 portion of the pore.
A limitation to this approach is that the pore and associated electromechanical coupling regions have not been clearly delineated. There is evidence that the intracellular portions of the transmembrane segments S5 and S6 contribute to the coupling of depolarization to channel opening. However, a comprehensive analysis of these regions in neuronal sodium channels is not available. The epilepsy-associated SCN2A and SCN8A variants in S5 and S6 are more common toward the intracellular part of the these transmembrane regions; however, better definition of this area and characterization of the functional consequences of variants in the region are needed before functional effects can be inferred from the location of a particular variant in the proximal S5 and distal S6 areas.
Symptoms produced by sodium channel variants depend on a combination of channels affected and the functional consequence of the mutation on channel function. This difference is reflected in the distribution of missense variants within the channels. In SCN2A and SCN8A epilepsy-associated variants are clustered in the voltage sensor region, and spare the pore region; especially the S5–6 segment; and are frequently associated with a gain-of-function; while epilepsy-associated SCN1A variants common in both the voltage sensor region and the pore and result from loss-of-function, gain-of-function, or mixed changes. In contrast, loss-of-function variants SCN2A and SCN8A variants are more often seen with neurological disorders other than epilepsy; as a result, few variants in the pore are seen in people with epilepsy caused by SCN2A or SCN8A mutations. This distinction has important clinical implications. Epilepsy resulting from variants in the pore region may respond poorly to sodium channel blocking antiepileptic medications while those in the voltage sensor region may respond more favorably to these agents. Location of the variant could also be important for drug studies in people with sodium channelopathies because it provides a simple way of focusing on an even more homogeneous subject population.
Table 2.
Simplified Framework for Functional Effect, Location, and Phenotype of Variants in Sodium Channelopathies*
| SCN1A | SCN2A | SCN4A | SCN5A | SCN8A | SCN9A | |
|---|---|---|---|---|---|---|
| Predominant Location of gene | Central nervous system | Central nervous system | Muscle | Heart | Central nervous system | Dorsal Root Ganglion |
| LOSS OF FUNCTION | ||||||
| Type of variant | Nonsense and Frameshift more than missense | Nonsense and Frameshift more than missense | Few reports in literature | Missense more than Nonsense | Missense more than Frameshift | Nonsense more than missense |
| Common location of Missense variants | P > VSR | P | Few reports in literature | Distributed throughout TM | P | P (S5–6) |
| Prototypic phenotype | EOEE | ASD, DD with or without later onset seizures | Normal (het) Congenital Myopathy (hom / biallelic) | Brugada Syndrome | DD, ataxia | Normal (het) Congenital indifference to pain (hom / biallelic) |
| GAIN OF FUNCTION** | ||||||
| Type of variant | Missense | Missense | Missense | Missense | Missense | Missense |
| Common location of missense variants | VSR | VSR; sparing of S5–6 | VSR > P with sparing of S5–6 and S6 | DIII–DIV linker, VSR, S6 | VSR; sparing of S5–6 | DIII–DIV linker, VSR > P with sparing of S5–6 |
| Prototypic phenotype | EOEE > GEFS+ | EOEE> B(F)NIS | Paroxysmal neuromuscular disorders | Prolonged QT Syndrome 3 | EOEE | Paroxysmal pain disorders |
There is considerable molecular and clinical heterogeneity of the variants. The table lists the most frequent associations but this is not exclusive. This has been compiled from multiple sources5, 19, 26, 27, 29–32;
Many missense variants in VSR have complicated functional effects with mixed electrophysiological changes but most exhibit some gain of function properties.
Abbreviations: As in Table 1, in addition, DD = Developmental delay; ASD= Autism spectrum disorders; P = pore (S5, S5–6, S6); VSR= voltage sensor region (S3–4, S4; S4–5); TMO= transmembrane; het= heterozygous, hom=homozygous.
Acknowledgments
We would like to thank Dr. Alfred L. George, Jr., for providing, SCN2A-WT, pIRES-CD8-hβ1 and pIRES-EGFP-hβ2 as well as for teaching his electrophysiological techniques and methodology We thank Mr. Yonggen Song for technical assistance. This study makes use of data generated by the DECIPHER community28. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for DECIPHER was provided by the Wellcome Trust. Those who carried out the original analysis and collection of the data generated by the DECIPHER community bear no responsibility for the further analysis or interpretation. The authors would like to thank Anna Byars for editorial assistance. This work was supported by NIH grant NINDS R01 NS062756. This source was not involved in study design, data collection, analysis or interpretation, or in the writing of the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
Study conception and design: KDH, TMB
Data acquisition and analysis: KDH, TMB, PSH
Drafting manuscript and figures. KDH
POTENTIAL CONFLICTS OF INTEREST
Nothing to report
References
- 1.Catterall WA. Voltage-gated sodium channels at 60: structure, function and pathophysiology. The Journal of physiology. 2012 Jun 1;590(11):2577–89. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanai K, Hirose S, Oguni H, et al. Effect of localization of missense mutations in SCN1A on epilepsy phenotype severity. Neurology. 2004 Jul 27;63(2):329–34. doi: 10.1212/01.wnl.0000129829.31179.5b. [DOI] [PubMed] [Google Scholar]
- 3.Zuberi SM, Brunklaus A, Birch R, Reavey E, Duncan J, Forbes GH. Genotype-phenotype associations in SCN1A-related epilepsies. Neurology. 2011 Feb 15;76(7):594–600. doi: 10.1212/WNL.0b013e31820c309b. [DOI] [PubMed] [Google Scholar]
- 4.Meng H, Xu HQ, Yu L, et al. The SCN1A mutation database: updating information and analysis of the relationships among genotype, functional alteration, and phenotype. Human mutation. 2015 Jun;36(6):573–80. doi: 10.1002/humu.22782. [DOI] [PubMed] [Google Scholar]
- 5.Brunklaus A, Ellis R, Reavey E, Semsarian C, Zuberi SM. Genotype phenotype associations across the voltage-gated sodium channel family. Journal of medical genetics. 2014 Oct;51(10):650–8. doi: 10.1136/jmedgenet-2014-102608. [DOI] [PubMed] [Google Scholar]
- 6.Meisler MH, Helman G, Hammer MF, et al. SCN8A encephalopathy: Research progress and prospects. Epilepsia. 2016 Jul;57(7):1027–35. doi: 10.1111/epi.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagnon JL, Barker BS, Hounshell JA, et al. Pathogenic mechanism of recurrent mutations of SCN8A in epileptic encephalopathy. Annals of clinical and translational neurology. 2016 Feb;3(2):114–23. doi: 10.1002/acn3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolff M, Johannesen KM, Hedrich UBS, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017 May 1;140(5):1316–36. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 9.Liao WP, Shi YW, Long YS, et al. Partial epilepsy with antecedent febrile seizures and seizure aggravation by antiepileptic drugs: associated with loss of function of Na(v) 1.1. Epilepsia. 2010 Sep;51(9):1669–78. doi: 10.1111/j.1528-1167.2010.02645.x. [DOI] [PubMed] [Google Scholar]
- 10.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemke JR, Riesch E, Scheurenbrand T, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012 Aug;53(8):1387–98. doi: 10.1111/j.1528-1167.2012.03516.x. [DOI] [PubMed] [Google Scholar]
- 12.Lossin C, Rhodes TH, Desai RR, et al. Epilepsy-associated dysfunction in the voltage-gated neuronal sodium channel SCN1A. J Neurosci. 2003 Dec 10;23(36):11289–95. doi: 10.1523/JNEUROSCI.23-36-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002 Jun 13;34(6):877–84. doi: 10.1016/s0896-6273(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 14.Misra SN, Kahlig KM, George AL., Jr Impaired NaV1.2 function and reduced cell surface expression in benign familial neonatal-infantile seizures. Epilepsia. 2008 Sep;49(9):1535–45. doi: 10.1111/j.1528-1167.2008.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohmori I, Kahlig KM, Rhodes TH, Wang DW, George AL., Jr Nonfunctional SCN1A is common in severe myoclonic epilepsy of infancy. Epilepsia. 2006 Oct;47(10):1636–42. doi: 10.1111/j.1528-1167.2006.00643.x. [DOI] [PubMed] [Google Scholar]
- 16.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014 Nov 13;515(7526):209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014 Nov 13;515(7526):216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahern CA, Payandeh J, Bosmans F, Chanda B. The hitchhiker's guide to the voltage-gated sodium channel galaxy. The Journal of general physiology. 2016 Jan;147(1):1–24. doi: 10.1085/jgp.201511492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapplinger JD, Giudicessi JR, Ye D, et al. Enhanced Classification of Brugada Syndrome-Associated and Long-QT Syndrome-Associated Genetic Variants in the SCN5A-Encoded Na(v)1.5 Cardiac Sodium Channel. Circ Cardiovasc Genet. 2015 Aug;8(4):582–95. doi: 10.1161/CIRCGENETICS.114.000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deciphering Developmental Disorders S. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015 Mar 12;519(7542):223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Gama AM, Pochareddy S, Li M, et al. Targeted DNA Sequencing from Autism Spectrum Disorder Brains Implicates Multiple Genetic Mechanisms. Neuron. 2015 Dec 2;88(5):910–7. doi: 10.1016/j.neuron.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Shalom R, Keeshen CM, Berrios KN, An JY, Sanders SJ, Bender KJ. Opposing Effects on NaV1.2 Function Underlie Differences Between SCN2A Variants Observed in Individuals With Autism Spectrum Disorder or Infantile Seizures. Biological psychiatry. 2017 Aug 1;82(3):224–32. doi: 10.1016/j.biopsych.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagnon JL, Barker BS, Ottolini M, et al. Loss-of-function variants of SCN8A in intellectual disability without seizures. Neurology Genetics. 2017 Aug;3(4):e170. doi: 10.1212/NXG.0000000000000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George AL., Jr Lessons learned from genetic testing for channelopathies. Lancet neurology. 2014 Nov;13(11):1068–70. doi: 10.1016/S1474-4422(14)70123-1. [DOI] [PubMed] [Google Scholar]
- 25.Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010 Jan;7(1):33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meregalli PG, Tan HL, Probst V, et al. Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm. 2009 Mar;6(3):341–8. doi: 10.1016/j.hrthm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Yoshinaga H, Sakoda S, Shibata T, et al. Phenotypic variability in childhood of skeletal muscle sodium channelopathies. Pediatric neurology. 2015 May;52(5):504–8. doi: 10.1016/j.pediatrneurol.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. American journal of human genetics. 2009 Apr;84(4):524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett DL, Woods CG. Painful and painless channelopathies. Lancet neurology. 2014 Jun;13(6):587–99. doi: 10.1016/S1474-4422(14)70024-9. [DOI] [PubMed] [Google Scholar]
- 30.Jurkat-Rott K, Holzherr B, Fauler M, Lehmann-Horn F. Sodium channelopathies of skeletal muscle result from gain or loss of function. Pflugers Arch. 2010 Jul;460(2):239–48. doi: 10.1007/s00424-010-0814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaharieva IT, Thor MG, Oates EC, et al. Loss-of-function mutations in SCN4A cause severe foetal hypokinesia or 'classical' congenital myopathy. Brain. 2016 Mar;139(Pt 3):674–91. doi: 10.1093/brain/awv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lampert A, O'Reilly AO, Reeh P, Leffler A. Sodium channelopathies and pain. Pflugers Arch. 2010 Jul;460(2):249–63. doi: 10.1007/s00424-009-0779-3. [DOI] [PubMed] [Google Scholar]