Abstract
Light is an important factor for plant development and has serious effects on the growth, production and quality of potatoes. However, the physical and molecular mechanisms by which potato plantlets cope with different light qualities are not understood. In this study, the potato “Zhuanxinwu”, which is a germplasm potato resource with a high anthocyanin content, was used for physiological and transcriptome profiling analyses to uncover the different mechanisms that occur in response to blue, red and white light conditions, with the white light condition serving as the control. Multiple growth indexes, protective enzyme activity and metabolite accumulation were measured. The results indicated that white light promoted a shift in biomass allocation away from tubers to leaves to enhance dry leaf matter and reduce tuber fresh/dry weight relative to the effects of blue or red light. The leaf area and anthocyanin content values were greater for plants grown in blue light than those grown in white or red light, suggesting that combinations of different spectra were more conducive to regulating potato growth. A total of 2220 differentially expressed genes (DEGs) were found among the three samples, and the DEGs in the three comparison sets were analyzed. A total of 1180 and 984 DEGs were identified in the red light (Red) and blue light (Blue) conditions compared to the control condition, respectively, and 359 DEGs overlapped between the two comparison sets (Blue_vs_White and Red_vs_White). Interestingly, the 24 most common overlapped DEGs were involved in photosynthesis, respiration, and reactive oxygen species (ROS) scavenging. Of these DEGs, four genes involved in photosynthesis and two genes involved in pigment synthesis were highly expressed, implying that some genes could be implemented to cope with different light spectra by regulating the expression of DEGs involved in the corresponding metabolic pathways. In conclusion, our study characterizes physiological responses of potato to different light qualities and identifies potential pathways and candidate genes involved in these responses, thus providing a basis for further research on artificial light regulation of potato plant growth.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1410-0) contains supplementary material, which is available to authorized users.
Keywords: Potato, Different light spectra, RNA-seq, Physiological response, Differentially expressed genes
Introduction
Maximizing crop yields under abiotic and biotic stresses is an important strategy for improvement of the food supply. Many stress-related genes are induced following exposure to strong stress environments (Hancock et al. 2014; Ahuja et al. 2010; Basu et al. 2017). In general, many studies have shown high levels of stress-related physiological processes and genes involved in the regulation of plant survival under various abiotic stresses (Umezawa et al. 2006; Larkindale and Vierling 2008; Jamil et al. 2011; Basu et al. 2017). Although abiotic stresses under normal agronomic conditions rarely threaten plant death in temperate regions, mild stresses can also cause growth disturbances, leading to significant production losses (Skirycz et al. 2011). The initial response to those stresses is downregulation of genes involved in energy and protein metabolism accompanied by changes in plant growth to induce protective mechanisms (Cramer et al. 2011). Therefore, in an agricultural setting, growth cessation of crops can have an adverse effect on production, and characterization of physiological responses and growth changes under adverse conditions may serve as a good indicator for improved growth performance under mild stress conditions.
Light is one of the most important environmental factors for plant growth and development, and different light intensities and qualities can significantly affect plant growth and physiological metabolism. The different spectra of light strongly affect physiological and morphological changes in plant leaves (Hogewoning et al. 2010; Macedo et al. 2011). The absorption ratio of blue or red light is approximately 90% in plant leaves (Terashima et al. 2011). Thus, the amounts of blue and red light can strongly influence plant development and physiology (Parks 2003; McNellis and Deng 1995). Light intensity is also an important factor for plant growth and development. To adapt to various light environments, plants have formed diverse mechanisms to adjust the morphological and physiological characteristics of the leaves (Matsuda et al. 2016; Fan et al. 2013). The majority of previous studies have focused on the effects of different light intensities on plant growth and development under natural sunlight (Rocha et al. 2015; Ma et al. 2015; Fan et al. 2013). Conversely, little is known about the molecular basis of the growth and development of plants under red, blue and white light conditions.
The potato (Solanum tuberosum L.) is one of the top three important food crops worldwide and is second only to rice and wheat. With a global production of approximately 300 million tons (MT), potatoes are a key food crop for food security (Birch et al. 2012). In vitro culture and seedling detoxification are the main modes of in vitro potato propagation of the fine varieties, and inducing growth of potatoes by artificially regulating the light source is a prerequisite for cultivating strong seedlings and obtaining good plant varieties. Recently, the potato genome and transcriptome have been released, which have provided powerful resources for gene mapping, gene classification and functional annotation for functional analyses of potato responses to different artificial light qualities. However, the currently available transcriptomic data for potato plantlets under different light intensity conditions are insufficient, and some pathways involved in light regulation of potato growth have rarely been reported. In this study, RNA-seq data were generated from the potato cultivar ‘Zhuanxinwu’ and then used to examine transcriptome profiling and identify candidate genes involved in the regulation of potato growth under different light conditions. The growth of potatoes and the differentially expressed genes (DEGs) involved in their growth were analyzed to determine the possible molecular mechanisms by which potatoes respond to different light stresses. In total, 24 genes were differentially expressed in all three light conditions, and several potential photosynthesis- and carotenoid biosynthesis-related genes were identified. These genes will provide a basis for further study of how artificial light regulates and influences potato growth.
Materials and methods
Plant materials and light treatments
Potato plantlets of the cultivar ‘Zhuanxinwu’ were cultivated in vitro using MS medium (Murashige and Skoog 1962). The experiment was conducted at Jiangsu Vocational College of Agriculture and Forestry, China. Stem segments (10–15 mm in length with one leaf) were collected aseptically from potato plantlets in vitro and placed vertically in MS medium with 30 g L−1 of sucrose and 7.5 g L−1 of agar. After preculture for 3 days under fluorescent white lamps, the plantlets were placed under blue, white and red light conditions in vitro for 30 days. The control light source used wavelengths from 350 to 750 nm of a white fluorescent lamp (FL40D-EX/38, Huadian Co., China). The different spectral properties of the LED lights (the blue light peak at 440 nm and the red light peak at 630 nm) were examined using a spectroradiometer (SPIC-200, Everfine Co., Hangzhou, China). The photosynthetic photon flux density of each light was maintained at approximately 50 ± 5 µmol m−2 s−1, and the light treatments were conducted in an incubation room at a relative humidity of 75% under a 12-h photoperiod at 25 ± 2 °C. Ten bottles were used for each treatment, and each bottle contained 10 plantlets. Each experiment had three replicates. All samples from the two light treatment groups and one control group were collected, immediately frozen in liquid nitrogen and stored at − 80 °C.
Growth and physiological parameter measurements
After 30 days of the three in vitro light treatments, 10 plantlets from each treatment were randomly collected for measurement of their growth and physiological parameters. The leaf area, root length, stem length, and fresh and dry weight of each plant were examined as previously described (Ma et al. 2015). The chlorophyll (Chl) concentrations were determined by spectrophotometry according to Vincent and Nadeau (1983). The anthocyanin contents were determined using the method reported by Xu et al. (2015). The superoxide dismutase (SOD), catalase (CAT), and guaiacol peroxidase (POD) activities of the fresh leaves were measured by spectrophotometry using the method reported by Moradi and Ismail (2007) The sucrose acid invertase and sucrose neutral invertase activities were examined using the method described by Dai et al. (2016).
Total RNA extraction
All S. tuberosum ‘Zhuanxinwu’ samples from the three light treatment groups were collected after 30 days. Total RNA was extracted from the plant materials using the TRIzol reagent (Invitrogen Scientific, Inc., USA) according to the manufacturer’s instructions. The total RNA was digested using DNase I (TaKaRa, Dalian, China) to eliminate gDNA in each sample. The integrity and purity of the total RNA was measured using the NanoDrop 1000 micro ultraviolet visible spectrophotometer (Thermo Fisher Scientific, Inc., USA) and 1.5% agarose gel electrophoresis, respectively.
Library preparation and sequencing
Libraries were generated using the TruSeq RNA kit (Illumina). Approximately, 1.5 µg of total RNA from each sample was used to construct a cDNA library for subsequent Illumina sequencing. The mRNAs were purified and fragmented and then converted into cDNA for PCR amplification according to the Illumina RNA-seq protocol (Illumina, Inc., USA). Raw reads were generated using the Illumina Genome Analyzer II (Illumina) and Illumina HiSeq 2500 (Illumina) at the Beijing Genomics Institution (ShenZhen, China) according to the manufacturer’s recommendations. The RNA-seq raw data was deposited in the SRA databases of NCBI under BioProject PRJNA482047.
Read mapping and DEG screening
The FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used to filter the clean reads. The Illumina adapter sequences and the low-quality bases were removed using the fastx_clipper program and the fastq_quality_trimmer. All distinct clean reads were mapped to the potato (S. tuberosum) reference genome database (https://www.solgenomics.org/organism/Solanum_tuberosum/) using SOAPaligner/SOAP2 (http://soap.genomics.org.cn/). The gene expression levels of each sample were calculated using the fragments per kilobase of exon per million mapped fragments (FPKM) method (Mortazavi et al. 2008). The FPKM values were used to compare differences in gene expression among different light treatments. The DEGs were identified among pairwise comparisons of the three samples from the two light treatments (blue and red) and the control (white) condition using the edgeR package (Robinson et al. 2010). An absolute log2 fold change > 1 and false discovery rate (FDR) < 0.05 were used to identify the significance of differences in gene expression. Subsequently, the Blast2GO software (Conesa et al. 2005) was used to annotate the gene ontology (GO) classifications of the DEGs using a corrected p value < 0.05 as the screening cutoff. The GO enrichment contained three categories: molecular function, cellular component and biological process. KOBAS (Xie et al. 2011) was used to generate the enrichment analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways for the DEGs.
Quantitative real-time PCR (qPCR) verification of the DEGs
To confirm the confidence and reliability of the transcriptome sequencing data, the qPCR expression levels of six genes were compared with their corresponding expression levels from the sequencing data among the various sequencing samples. Gene-specific qPCR primers were designed using the Primer Express software (v3.0, Applied Biosystems) and are shown in Table S1. The elongation factor 1-α (ef1α) was selceted as an internal reference gene to normalize the expression values (Ma et al. 2016). The 20-µL PCR reaction contained 10 µL of SYBR Fast qPCR Mix (TaKaRa, Dalian, China), 2 µL of diluted cDNA, 1 µL each of the forward and reverse primers (10 µmol L−1), and 6 µL of ddH2O. The qPCR amplification was performed on a LightCycler 480 Real-Time PCR machine (Roche, Basel, Switzerland) under the following conditions: 95 °C for 3 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 40 s. The relative expression levels were calculated using the 2−(ΔΔCt) method (Livak and Schmittgen 2001), and each gene had three technical and three biological repeats.
Statistical analysis
The data are presented as the means ± standard errors (SEs). All statistical analyses were performed with the IBM Statistical Product and Service Solutions for Windows, Version 20.0 (IBM SPSS 20, Chicago, IL, USA). The data were analyzed for each treatment using one-way analysis of variance (ANOVA), and the significance of differences between the means was detected using Tukey’s test (p < 0.05).
Results and discussion
Effects of different light spectra (blue, white and red) on potato growth
Different light conditions influenced the vegetative growth of the potato plantlets (Fig. 1). Plantlets grown in the white light condition exhibited a shift in biomass allocation away from tubers to leaves corresponding to the blue or red light condition that indicated significantly enhanced leaf dry matter and reduced tuber fresh/dry weight. This shift caused a significantly reduced harvest index (Table 1), suggesting that the use of a single light did not significantly improve plant growth. After 30 days of incubation, different growth states were observed among the potato plantlets grown under the blue, white and red light conditions. As shown in Table 1, the average fresh weight (FW; mg), dry weight (DW; mg), chlorophyll a (Chla; µg g−1 DW), chlorophyll b (Chl b; µg g−1 DW) and carotene (Car; µg g−1 DW) values of the plants exposed to white light were 384.84 mg, 40.78 mg, 4.48 µg g−1 DW, 1.92 µg g−1 DW and 1.71 µg g−1 DW, respectively; these values were significantly greater than the values obtained for the plants exposed to the blue or red light (Table 1). However, the average leaf areas and anthocyanin contents of the plants exposed to blue light were approximately 86.06 mm2 and 40.55 µg g−1 DW, respectively, which were higher than the values obtained for the plants exposed to the white or red light. These results indicated the blue light might promote the growth and development of potato plantlets in vitro and suggested that some of its roles might be involved in the leaf area and pigment accumulation functions. Although blue light is harmful to living organisms, it can also increase reactive oxygen species (ROS) production to reduce oxidative stress (Hideg et al. 2013). When the plantlets were exposed to the blue light condition, the greatest average SOD, CAT and POD values were generated (340.83, 30.31, and 354.71 U/(g min), respectively). However, the sucrose acid invertase and sucrose neutral invertase contents in the leaves of plants grown under red light were 5.47 and 4.16 U[mg/(FW h)], respectively, which were significantly higher than the contents in the plants grown under blue or red light. These data confirm that the mechanism involved in blue light tolerance is based on increasing the activities of protected enzymes, such as SOD, CAT and POD, to promote plantlet survival (Hideg et al. 2013). These changes include leaf growth, chlorophyll synthesis and anthocyanin accumulation and indicate that blue light plays an important role in the formation of the specific qualities of the potato cultivar ‘Zhuanxinwu’. Therefore, these data suggest that multiple spectral combinations are more effective at artificially regulating potato growth.
Fig. 1.
Growth of potato plantlets under blue (A), white (B) and red (C) light quality conditions
Table 1.
Influence of light quality on the physiological responses of potato
| Parameter | Light quality | ||
|---|---|---|---|
| Blue light | Red light | White light | |
| Stem length (mm) | 45.00 ± 4.53c | 143.20 ± 20.97a | 69.00 ± 4.85b |
| Root length (mm) | 78.80 ± 12.43b | 80.40 ± 3.49b | 95.20 ± 3.14ab |
| Leaf area (mm2) | 86.06 ± 1.33a | 23.68 ± 0.30c | 75.46 ± 1.65b |
| Fresh weight (FW; mg) | 223.92 ± 44.77b | 341.76 ± 21.55ab | 384.84 ± 49.24ab |
| Dry weight (DW; mg) | 34.58 ± 7.00a | 32.10 ± 3.36a | 40.78 ± 4.30a |
| FW/DW (%) | 15.58 ± 0.69a | 9.41 ± 0.88c | 10.71 ± 0.26b |
| Chl a (µg g−1 DW) | 3.69 ± 0.17b | 2.85 ± 0.08c | 4.48 ± 0.12a |
| Chl b (µg g−1 DW) | 1.18 ± 0.09b | 0.96 ± 0.06c | 1.92 ± 0.16a |
| Car (µg g−1 DW) | 1.28 ± 0.08b | 1.01 ± 0.04c | 1.71 ± 0.10a |
| Anthocyanin (µg g−1 DW) | 40.55 ± 1.25a | 15.10 ± 1.50c | 29.60 ± 0.75b |
| Sucrose acid invertase {U[mg/(FW h)]} | 2.63 ± 0.11c | 5.47 ± 0.30a | 4.51 ± 0.19b |
| Sucrose neutral invertase {U[mg/(FW h)]} | 2.66 ± 0.10b | 4.16 ± 0.53a | 1.77 ± 0.16c |
| POD [U/(g min)] | 332.55 ± 30.55a | 178.01 ± 4.85b | 354.71 ± 14.63a |
| CAT [U/(g min)] | 15.62 ± 1.82b | 7.26 ± 0.74c | 30.31 ± 1.97a |
| SOD [U/(g min)] | 271.18 ± 15.35a | 316.5 ± 27.40a | 340.83 ± 37.54a |
Values followed by the same letter within a row do not significantly difference (by the LSD test, p = 0.05)
Sequencing data and DEG analysis
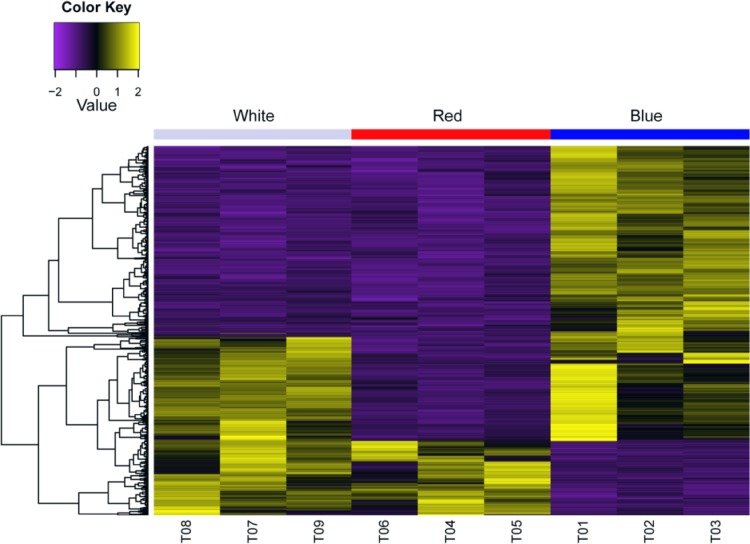
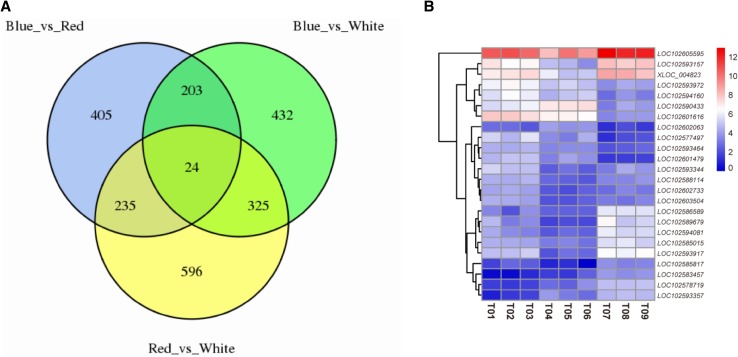
High-throughput sequencing, such as RNA-seq, has been used as an effective method to examine the transcriptional conditions of plants in different states, to compare differential expression profiles or differentially expressed genes, and to reveal the basis of plant molecular responses to different environments. In this study, potato leaf transcriptome data were obtained under three light conditions. Among the three transcriptome sequencing samples, a total of 22,484,237, 22,222,909 and 22,466,742 raw reads were generated from the corresponding red, blue, and white light cDNA libraries, respectively. Subsequently, these reads were filtered, and 60.18 Gb of clean bases were obtained; up to 78.28% of the clean reads could be mapped onto the published potato genome (Table 2 and Table S2). Then, the expression profiles of all transcripts were analyzed using pairwise comparisons of the three samples (Fig. 2 and Figure S1). Moreover, DEGs were identified among the different comparison groups [the data from the blue light-treated sample compared to the control (Blue_vs_White), the red light-treated sample compared to the control (Red_vs_White) and the blue light-treated sample compared to the red light-treated sample (Blue_vs_Red)]. A total of 2220 DEGs were identified, and 24 common overlapping DEGs were found among the different comparison data sets (Fig. 3 and Table S3). The Blue_vs_White comparison set contained 984 DEGs, of which 470 genes were upregulated DEGs and 514 genes were downregulated DEGs (Table 3). In the Red_vs_White comparison set, in which the red light was compared to the control condition, 1180 genes were differentially expressed, of which 773 genes were upregulated and 407 genes were downregulated. In addition, when the blue light-treated sample was compared to the red light-treated sample, 867 genes were differentially expressed, of which 177 genes were upregulated and 690 genes were downregulated (Table 3 and Table S3). The Red_vs_White comparison set had more DEGs than the Blue_vs_White or Blue_vs_Red comparison set, and those DEGs could be annotated using the Swiss-Prot, GO, KEGG, COG, KOG, Pfam, and nr databases. The annotation numbers varied from 491 to 1091 (Table S4).
Table 2.
Sample information and summary of sequencing reads
| Sample ID | Sample description | Read number | Base number | GC content | % ≥ Q30 |
|---|---|---|---|---|---|
| T01 | Blue light repeat 1 | 21,965,654 | 6,589,696,200 | 43.28 | 95.38 |
| T02 | Blue light repeat 2 | 22,222,909 | 6,666,872,700 | 44.63 | 95.26 |
| T03 | Blue light repeat 3 | 22,156,056 | 6,646,816,800 | 48.2 | 95.49 |
| T04 | Red light repeat 1 | 22,570,571 | 6,771,171,300 | 45.92 | 95.75 |
| T05 | Red light repeat 2 | 22,176,808 | 6,653,042,400 | 47.41 | 96.2 |
| T06 | Red light repeat 3 | 22,484,237 | 6,745,271,100 | 48.46 | 95.86 |
| T07 | White light repeat 1 | 22,362,037 | 6,708,611,100 | 47.42 | 96.18 |
| T08 | White light repeat 2 | 22,208,778 | 6,662,633,400 | 48.33 | 95.99 |
| T09 | White light repeat 3 | 22,466,742 | 6,740,022,600 | 47.5 | 95.77 |
Fig. 2.
Heat map of transcripts from potato plantlets under three light conditions
Fig. 3.
VNN map and clustering heat map of differential expression genes from potato plantlets. The VNN map (a) showed the differential expression genes from potato plantlets under three light conditions. The clustering heat map (b) showed the expression of overlapped differential expression genes from potato plantlets under three light conditions
Table 3.
Summary of DEGs from different comparison data sets
| Data set | Number of DEGs | Upregulation of DEGs | Downregulation of DEGs |
|---|---|---|---|
| Blue_vs_White | 984 | 470 | 514 |
| Red_vs_White | 1180 | 773 | 407 |
| Blue_vs_Red | 867 | 177 | 690 |
Functional annotation and analysis of DEGs
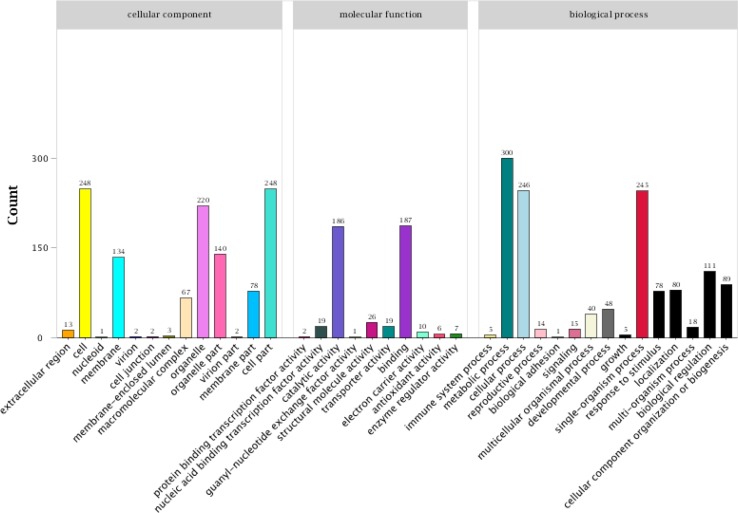
Mining differentially expressed genes is an important method for studies of transcriptome profiles under different states. Annotation of differential genes in different transcriptomes can be used to clarify which functional genes or metabolic pathways play important roles in transcriptional regulation. To investigate the functions of and highlight the metabolic pathways in which the DEGs were potentially involved during regulation of light tolerance, the GO classifications and KEGG pathway, annotations of genes encoding the identified DEGs from the three comparison sets (Blue_vs_White, Red_vs_White, and Blue_vs_Red) were analyzed (Fig. 4, Tables S4 and S5). These analyses identified 396, 1180 and 867 unique GO functional annotations from the Blue_vs_White, Red_vs_White, and Blue_vs_Red comparison groups, respectively. A total of 1295 genes were annotated in the biological process category, accounting for approximately 45.35% of the total GO functional annotation categories. The cellular component and molecular function categories contained 1098 and 462 genes, respectively (Fig. 4). Among those comparison data sets, many DEGs were significantly enriched in the following five GO categories: metabolic process, cell, cell part, cellular process and single organism (Fig. 4).
Fig. 4.
GO enrichment analysis of differential expression genes from potato plantlets
For KEGG pathway annotation of the DEGs, the data were retrieved from the published KOBAS database. A total of 249 DEGs from 4201 expressed genes were enriched in 54 pathways (Table S5). The KEGG pathway annotation terms for each intragroup analysis were compared to identify the top ten significantly enriched pathways (Table 4). The top ten enriched pathways contained 139 DEGs and included photosynthesis, carbon fixation in photosynthetic organisms, ribosome, glycolysis/gluconeogenesis, pentose phosphate pathway, photosynthesis-antenna proteins, oxidative phosphorylation, carotenoid biosynthesis, pyruvate metabolism, and glyoxylate and dicarboxylate metabolism (Table 4), which revealed that many genes were intensively enriched in metabolic and biosynthesis-related pathways. In current reports, transcript profiling and metabolic pathways have been analyzed for leaves from potato (S. tuberosum) plantlets treated with different light conditions. The expression profiling analysis indicated that genes involved in photosynthesis, photorespiration and carbohydrate-related processes were more drastically downregulated under the blue light condition than under the red light condition. In this study, we collected potato plantlets in vitro as the sequencing material to examine transcriptome changes under three different light conditions. Some significantly changed DEGs were identified and found to be associated with photosynthesis, secondary metabolism, pigment biosynthesis, and starch synthesis and transfer. The differences in the DEGs among these studies could be due to the use of different material properties, treatments and plant genetic backgrounds, which will require further data support.
Table 4.
Top ten enriched pathways of differential expression genes from potato plantlets
| Pathway | Ko_ID | DEGs | Expression genes |
|---|---|---|---|
| Photosynthesis | ko00195 | 36 | 86 |
| Carbon fixation in photosynthetic organisms | ko00710 | 23 | 86 |
| Ribosome | ko03010 | 15 | 286 |
| Glycolysis/gluconeogenesis | ko00010 | 12 | 129 |
| Pentose phosphate pathway | ko00030 | 11 | 58 |
| Photosynthesis—antenna proteins | ko00196 | 11 | 40 |
| Oxidative phosphorylation | ko00190 | 9 | 181 |
| Carotenoid biosynthesis | ko00906 | 8 | 39 |
| Pyruvate metabolism | ko00620 | 7 | 91 |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 7 | 45 |
RNA-seq data verification
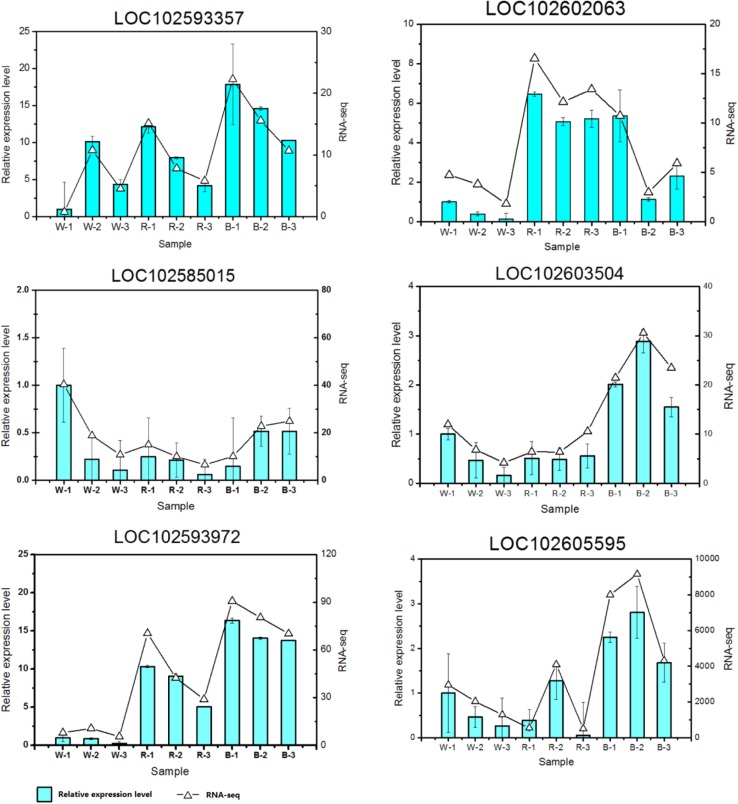
The accuracy of sequencing data is the basis of data utilization. With improvements in sequencing technology, high-throughput sequencing has gradually become a powerful means to detect gene expression. However, verification of sequencing data through qPCR is a key step in RNA-seq analyses. To confirm the data validity of the potato plantlet transcriptome under blue, white and red light conditions, six target genes were selected from the sequencing data to check the expression profiles using qPCR. The qPCR analyses of these selected genes and internal reference genes were performed for all sequencing samples using their qPCR primers (Table S1); the corresponding qPCR expression levels and RNA-seq values of the selected genes are shown using histograms in Fig. 5. Although the relative expression patterns of these selected genes were different from those obtained from the RNA-seq and qPCR data for all sequencing samples, the expression trends of these selected genes were similar to the data obtained using the two methods, which confirmed the reliability and accuracy of the RNA-seq results.
Fig. 5.
qPCR validations of RNA-seq results. The letters “B”, “R”, and “W” in the histogram represented the samples were treated under blue light, red light and white light, respectively
Analysis of genes related to photosynthesis and pigment biosynthesis
Potato production involves a series of complex biological processes mediated by both environmental factors and genes (Dinakar et al. 2012; Akula and Ravishankar 2011; Ahuja et al. 2010). In potatoes, the cell number and division of potato tubes are major events by which tubers bulk together with the associated starch and protein accumulation (Nazarian-Firouzabadi and Visser 2017; Van Harsselaar et al. 2017; Birch et al. 2012; Geigenberger 2003). Photoperiod signal perception and conduction in leaves initiates tuberization in the apical region of the underground potato tube. Dioxygenases catalyze oxygenation of polyunsaturated fatty acids (LOXs), such as linoleic and linolenic acids, which contributes to 9(S)-hydroperoxy linolenic acid (9(S)-HPOT) and 13(S)-hydroperoxy linolenic acid (13(S)-HPOT) accumulation. Then, these products are metabolized to tuberonic acid and tuberonic acid glucoside to induce tuber formation when the plantlets are placed in strong light conditions. Interestingly, some genes among the 24 common overlapped DEGs (Table S6) were involved in carotenoid biosynthesis (e.g., LOC102585015) and UDP-glucoronosyl and UDP-glucosyl transferase metabolism (LOC102593972), suggesting that these overlapped DEGs might have important functions in regulating carotenoid biosynthesis, transport and metabolism in response to light stress in potato plants. Moreover, the expression patterns of some genes were altered; those genes were involved in major biological processes or pathways (Table S6), such as posttranslational modification (LOC102593357), heat stress transcription factor (LOC102602063) and heat or oxidative stresses by the generation of ROS under light stress conditions. Light can mediate photosynthesis and carotenoid biosynthesis of potatoes and their signal transduction, and other related metabolic pathways change under different light intensity conditions, thereby altering the transcriptional responses of common overlapping genes related to photosynthesis and carotenoid biosynthesis. However, this mechanism needs to be further studied to explore light responses and the regulation of potato plantlets under different light conditions.
Conclusions
In this study, we examined the physiological responses of potato ‘Zhuanxinwu’ cultivar plantlets to three light wavelengths, including blue, white, and red light, and performed transcriptomic profiling. The growth and physiological parameter changes indicated that blue light reduced leaf fresh/dry matter, promoted expansion of the leaf area, improved chlorophyll synthesis and increased anthocyanin accumulation. These effects may play important roles in the formation of the specific quality of potato cultivar ‘Zhuanxinwu’, as suggested by multiple spectral combinations, and may provide a more effective method to regulate potato growth. The transcriptome analysis indicated that among the four comparison data sets containing 2220 DEGs, 24 common overlapped DEGs were involved in several potential photosynthesis and pigment biosynthesis pathways. This finding suggests that blue light plays an important role in the formation of the specific quality of the potato cultivar ‘Zhuanxinwu’. These findings yield new insights into the mechanisms by which potato plantlets respond to light quality and will help clarify the precise mechanisms related to light responses in potato plantlets. This work also opens the door for further studies on the effects of artificial light regulation on the yield and quality of potatoes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The study was funded by Grants from the National Natural Science Foundation of China (11674174), Jiangsu Vocational College of Agriculture and Forestry Research Project (2016kj002), Top-notch Academic Program Project of Jiangsu Higher Education Institutions (PPZY2015B173) and National Key R&D Program of China (2017YFB0403903).
Author contributions
ZX conceived the study and amended the manuscript. JX, ZY, YW and ZX performed experiments and data analysis. JX wrote the paper. All authors have read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare there is no conflict of interest.
References
- Ahuja I, de Vos RC, Bones AM, Hall RD. Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15(12):664–674. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Akula R, Ravishankar GA. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav. 2011;6(11):1720–1731. doi: 10.4161/psb.6.11.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Giri RK, Benazir I, Kumar S, Rajwanshi R, Dwivedi SK, Kumar G. Comprehensive physiological analyses and reactive oxygen species profiling in drought tolerant rice genotypes under salinity stress. Physiol Mol Biol Plants. 2017;23(4):837–850. doi: 10.1007/s12298-017-0477-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch PRJ, Bryan G, Fenton B, Gilroy EM, Hein I, Jones JT, Prashar A, Taylor MA, Torrance L, Toth IK. Crops that feed the world 8: potato: are the trends of increased global production sustainable? Food Secur. 2012;4(4):477–508. doi: 10.1007/s12571-012-0220-1. [DOI] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Fu M, Yang X, Chen Q. Ethylene inhibited sprouting of potato tubers by influencing the carbohydrate metabolism pathway. J Food Sci Technol. 2016;53(8):3166–3174. doi: 10.1007/s13197-016-2290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinakar C, Djilianov D, Bartels D. Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci. 2012;182:29–41. doi: 10.1016/j.plantsci.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Fan X-X, Xu Z-G, Liu X-Y, Tang C-M, Wang L-W, Han X-l. Effects of light intensity on the growth and leaf development of young tomato plants grown under a combination of red and blue light. Sci Hortic. 2013;153:50–55. doi: 10.1016/j.scienta.2013.01.017. [DOI] [Google Scholar]
- Geigenberger P. Regulation of sucrose to starch conversion in growing potato tubers. J Exp Bot. 2003;54(382):457–465. doi: 10.1093/jxb/erg074. [DOI] [PubMed] [Google Scholar]
- Hancock RD, Morris WL, Ducreux LJ, Morris JA, Usman M, Verrall SR, Fuller J, Simpson CG, Zhang R, Hedley PE, Taylor MA. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plant Cell Environ. 2014;37(2):439–450. doi: 10.1111/pce.12168. [DOI] [PubMed] [Google Scholar]
- Hideg E, Jansen MA, Strid A. UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 2013;18(2):107–115. doi: 10.1016/j.tplants.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Hogewoning SW, Trouwborst G, Maljaars H, Poorter H, van Ieperen W, Harbinson J. Blue light dose–responses of leaf photosynthesis, morphology, and chemical composition of Cucumis sativus grown under different combinations of red and blue light. J Exp Bot. 2010;61(11):3107–3117. doi: 10.1093/jxb/erq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil A, Riaz S, Ashraf M, Foolad MR. Gene expression profiling of plants under salt stress. Crit Rev Plant Sci. 2011;30(5):435–458. doi: 10.1080/07352689.2011.605739. [DOI] [Google Scholar]
- Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146(2):748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma X, Wang Y, Liu M, Xu J, Xu Z. Effects of green and red lights on the growth and morphogenesis of potato (Solanum tuberosum L.) plantlets in vitro. Sci Hortic. 2015;190:104–109. doi: 10.1016/j.scienta.2015.01.006. [DOI] [Google Scholar]
- Ma H, Cao X, Shi S, Li S, Gao J, Ma Y, Zhao Q, Chen Q. Genome-wide survey and expression analysis of the amino acid transporter superfamily in potato (Solanum tuberosum L.) Plant Physiol Biochem. 2016;107:164–177. doi: 10.1016/j.plaphy.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Macedo AF, Leal-Costa MV, Tavares ES, Lage CLS, Esquibel MA. The effect of light quality on leaf production and development of in vitro-cultured plants of Alternanthera brasiliana Kuntze. Environ Exp Bot. 2011;70(1):43–50. doi: 10.1016/j.envexpbot.2010.05.012. [DOI] [Google Scholar]
- Matsuda R, Yamano T, Murakami K, Fujiwara K. Effects of spectral distribution and photosynthetic photon flux density for overnight LED light irradiation on tomato seedling growth and leaf injury. Sci Hortic. 2016;198:363–369. doi: 10.1016/j.scienta.2015.11.045. [DOI] [Google Scholar]
- McNellis TW, Deng XW. Light control of seedling morphogenetic pattern. Plant Cell. 1995;7(11):1749–1761. doi: 10.1105/tpc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi F, Ismail AM. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot. 2007;99(6):1161–1173. doi: 10.1093/aob/mcm052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. 15 (3):473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
- Nazarian-Firouzabadi F, Visser RGF. Potato starch synthases: functions and relationships. Biochem Biophys Rep. 2017;10:7–16. doi: 10.1016/j.bbrep.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM. The red side of photomorphogenesis. Plant Physiol. 2003;133(4):1437–1444. doi: 10.1104/pp.103.029702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ABO, Honório SL, Messias CL, Otón M, Gómez PA. Effect of UV-C radiation and fluorescent light to control postharvest soft rot in potato seed tubers. Sci Hortic. 2015;181:174–181. doi: 10.1016/j.scienta.2014.10.045. [DOI] [Google Scholar]
- Skirycz A, Vandenbroucke K, Clauw P, Maleux K, De Meyer B, Dhondt S, Pucci A, Gonzalez N, Hoeberichts F, Tognetti VB, Galbiati M, Tonelli C, Van Breusegem F, Vuylsteke M, Inze D. Survival and growth of Arabidopsis plants given limited water are not equal. Nat Biotechnol. 2011;29(3):212–214. doi: 10.1038/nbt.1800. [DOI] [PubMed] [Google Scholar]
- Terashima I, Hanba YT, Tholen D, Niinemets U. Leaf functional anatomy in relation to photosynthesis. Plant Physiol. 2011;155(1):108–116. doi: 10.1104/pp.110.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol. 2006;17(2):113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Van Harsselaar JK, Lorenz J, Senning M, Sonnewald U, Sonnewald S. Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.) BMC Genom. 2017;18(1):37. doi: 10.1186/s12864-016-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent R, Nadeau D. A micromethod for the quantitation of cellular proteins in Percoll with the Coomassie brilliant blue dye-binding assay. Anal Biochem. 1983;135(2):355–362. doi: 10.1016/0003-2697(83)90696-6. [DOI] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(suppl_2):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Su X, Lim S, Griffin J, Carey E, Katz B, Tomich J, Smith JS, Wang W. Characterisation and stability of anthocyanins in purple-fleshed sweet potato P40. Food Chem. 2015;186:90–96. doi: 10.1016/j.foodchem.2014.08.123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.