Abstract
Background
This study was conducted to identify whether the presence of circulating tumor DNA (ctDNA) in plasma before treatment with EGFR‐tyrosine kinase inhibitors (TKIs) is associated with clinical outcomes.
Methods
Fifty‐seven pairs of tissues and plasma samples were obtained from patients with NSCLC adenocarcinoma harboring activating EGFR mutations before the administration of EGFR‐TKI treatment. ctDNA mutation was identified using the PANAMutyper EGFR mutation kit. Both qualitative and quantitative analyzes of the data were performed.
Results
Concordance rates with tissue biopsy were 40.4% and 59.6% for the qualitative and quantitative methods, respectively. Bone metastasis showed a statistically significant correlation with ctDNA detection (odds ratio 3.985, 95% confidence interval [CI] 1.027–15.457; P = 0.046). Progression‐free survival (PFS) was significantly shorter in the group detected with ctDNA than in the undetected ctDNA group (median PFS 9.8 vs. 20.7 months; hazard ratio [HR] 2.30, 95% CI 1.202–4.385; P = 0.012). Detection of ctDNA before treatment with EGFR‐TKIs (HR 2.388, 95% CI 1.138–5.014; P = 0.021) and extra‐thoracic lymph node metastasis (HR 13.533, 95% CI 2.474–68.747; P = 0.002) were independently associated with PFS. Six of 11 patients (45.5%) monitored by serial sampling showed a dynamic change in ctDNA prior to disease progression.
Conclusion
Quantitative testing can increase the sensitivity of the ctDNA detection test. Patients with detectable ctDNA had significantly shorter PFS after receiving EGFR‐TKIs than those with undetectable ctDNA. Tumor burden may be associated with plasma ctDNA detection. A shorter PFS was associated with detection of ctDNA and extra‐thoracic lymph node metastasis. Dynamic changes in the ctDNA level may help predict clinical outcomes.
Keywords: Circulating tumor DNA, EGFR‐tyrosine kinase inhibitor (TKI), non‐small cell lung cancer, progression‐free survival, sensitivity
Introduction
Current practice for the treatment of patients with advanced‐stage non‐small cell lung cancer (NSCLC) has become increasingly dependent on identifying the presence of genetic mutations.1, 2, 3 EGFR mutations can be detected in tissues extracted through surgery or other invasive procedures. In clinical situations, diagnosis and mutation testing should be performed simultaneously with tissue samples or cytology. However, a significant number of patients do not have enough tissue,4 do not have a lesion available for biopsy, or refuse to undergo a repeat biopsy. Complications from intrathoracic biopsy have been reported in approximately 17% of patients.5 Because of the heterogeneity of NSCLC, a biopsy may need to be performed at multiple sites, which is frequently not realistic given the poor performance status of advanced‐stage lung cancer patients.6, 7, 8, 9, 10
Given these limitations, alternative methods for analyzing the EGFR mutation status of NSCLC have been investigated. Non‐invasive EGFR mutation detection methods based on plasma or serum show great potential. Circulating tumor DNA (ctDNA) in the plasma can be used to detect EGFR mutations in NSCLC patients, providing a level of information similar to that of tumor tissue biopsies.11 Dynamic changes in ctDNA EGFR mutation status are associated with clinical outcomes of EGFR‐tyrosine kinase inhibitor (EGFR‐TKI) treatment, such as disease progression and progression‐free survival (PFS).12, 13
This study aimed to measure whether the presence of ctDNA in plasma before treatment with EGFR‐tyrosine kinase inhibitors (TKIs) is associated with PFS. A qualitative and quantitative analysis was performed on the same samples and the results were compared.
Methods
Patients
Fifty‐seven patients with NSCLC adenocarcinoma harboring activating EGFR mutations (exon 19 deletion and L858R mutation) at the Asan Medical Center, Seoul, Korea between January 2014 and December 2016 were enrolled in the study. E19 deletions and L858R mutations were detected in all patients via tissue biopsy and treated with EGFR‐TKIs. Plasma samples were collected from patients before treatment with first‐generation EGFR‐TKIs (IRB No. 2016‐0692).
Patient data were collected from medical records and radiological images to assess baseline information, tumor response, and PFS. Never smokers were defined as having smoked < 100 cigarettes in their lifetime. PFS was measured from the first day of treatment with EGFR‐TKIs until tumor progression or the last follow‐up date.
EGFR mutation analysis for tissue and plasma
We used direct Sanger sequencing or peptide nucleic acid (PNA)‐mediated PCR clamping assay (PNAClamp EGFR Mutation Detection kit, PANAGENE Inc., Daejeon, Korea) to detect EGFR mutations in tissue. The PNA clamping probe binds to wild‐type DNA and suppresses amplification. Only mutant‐type DNA is selectively amplified.
The PANAMutyper EGFR kit (PANAGENE Inc.) was used to detect mutations in plasma. PANAMutyper is a technology that integrates PNAClamp and PANA RealTyper. PANA RealTyper is Multiplex Melting Curve Analysis using a fluorescence‐labeled PNA probe. Fluorescent‐labeled PNA probes only fluoresce when bound to the target sequence. PNA probes with a unique melting temperature (Tm) according to the nucleotide sequence are denatured from the target DNA at the Tm as the temperature increases and the fluorescence signal decreases. Genotyping of the target DNA is possible by analyzing the temperature at which the signal decreases (Tm). This method could increase sensitivity and allow the detection of ctDNA in plasma. Currently, the PANAMutyper EGFR kit is provided as a qualitative assay, but in this study, quantitative analysis was performed in the same manner. Quantitative results were converted into reactions according to the linearity equation and provided as semi‐quantitative indices. The linearity of PANAMutyper R EGFR was evaluated by serial dilution of mutant EGFR standard materials spiked with human plasma wild type for EGFR. With this method < 1 copies/reaction and > 1000 copies/reaction are displayed as 1 and > 1000, respectively. The specificity of liquid biopsy for most ctDNA detection methods is close to 100%, and according to data provided by the laboratory, the specificity of the method used for ctDNA detection in this study was also 100%.
Statistical analysis
Fisher's exact and Wilcoxon rank‐sum tests were used for categorical and continuous variables. The concordance rate was assessed by comparing the match between primary tissue and ctDNA sequencing results. Multiple logistic regression analysis was used to analyze risk factors associated with ctDNA detection and disease progression. The Kaplan–Meier method was used to estimate PFS distribution, and the Cox proportional hazards model was used to investigate the effect of specified risk factors on disease progression. Results were considered statistically significant at a P value of < 0.05. All analyses were performed using SPSS version 22.0 (IBM Corp., Armonk, NY, USA).
Ethics statement
The Asan Medical Center institutional review board approved the study (No. 2016‐1228). We received informed consent from all patients for study participation and genetic analysis.
Results
Patient characteristics
Baseline examination of EGFR mutations was performed in all patients using matched tumor tissues and plasma samples prior to treatment with first‐generation EGFR‐TKIs. The majority of patients were women (39/57, 68.4%), never smokers (41/57, 71.9%), and had extrathoracic metastatic diseases (M1b) (34/57, 59.6%). Most patients received gefitinib (55/57 96.5%) as first‐line treatment. The most common mutation identified was an E19 deletion; T790M was not detected before treatment. Treatment responses were observed in 98.2% of patients, including partial responses (44/57, 77.2%) and stable disease (12/57, 21.1%) (Table 1). The median PFS was 14.9 months (95% confidence interval [CI] 7.55–22.25). During a median follow‐up period of 23.3 months (interquartile range 14.3–29.0), disease progression and death occurred in 40 (70.2%) and 9 (15.8%) patients, respectively. All deaths were related to disease progression.
Table 1.
Baseline patient and tumor characteristics
| Characteristics | All patients (n = 57) |
|---|---|
| Age (years) (mean ± SD) | 57 ± 10.6 |
| Gender, female (%) | 39 (68.4) |
| Smoking status | |
| Never | 41 (71.9) |
| Ever | 16 (28.1) |
| ECOG performance status | |
| 0 | 6 (10.5) |
| 1 | 49 (86) |
| 2 | 2 (3.5) |
| EGFR mutation by tissue biopsy | |
| Exon 19 del | 39 (68.4) |
| L858R | 18 (31.6) |
| Stage | |
| M0/< M1a | 23 (40.4) |
| M1b | 34 (59.6) |
| TKI | |
| Gefitinib | 55 (96.5) |
| Erlotinib | 1 (1.8) |
| Afatinib | 1 (1.8) |
| Best response | |
| Partial response | 44 (77.2) |
| Stable disease | 12 (21.1) |
| Progressive disease | 1 (1.8) |
Unless otherwise stated, data are presented as number (%).ECOG, Eastern Cooperative Oncology Group; SD, standard deviation; TKI, tyrosine kinase inhibitor.
The two ctDNA EGFR mutation testing methods were compared for concordance with the tumor tissue. The concordance rates were 40.4% and 59.6% for the qualitative and quantitative methods, respectively (Table 2). Individual patient results are reported in Table S1. Receiver operating characteristic curve analysis was performed by quantitative test to predict the concordance rate between the qualitative analysis and tissue results. The area under the receiver operating characteristic curve was 0.804 (95% CI 0.686–0.922; P < 0.001) and the cutoff value was 1.5 copies/reaction (sensitivity 68.2%, specificity 71.4%). When the samples were dichotomized based on the cutoff value, the concordance rates between ctDNA qualitative analysis and the tissue results were 28.1% in < 1.5 copies/reaction and 100% in ≥ 1.5 copies/reaction.
Table 2.
Comparison of plasma and tissue samples according to qualitative (PANAMutyper) and quantitative (PANAgene‐SQI) ctDNA tests
| Tissue EGFR mutation | ||||
|---|---|---|---|---|
| Testing method | E19del | L858R | Total patients | |
| ctDNA EGFR mutation (PANAMutyper) | E19del | 14 (35.9%) | 0 | 14 |
| L858R | 0 | 9 (50%) | 9 | |
| Wild | 25 | 9 | 34 | |
| Total patients | 39 | 18 | 57 | |
| ctDNA EGFR mutation(PANAgene‐SQI) | E19del | 21 (53.8%) | (1) | 21 |
| L858R | 0 | 13 (72.2%) | 13 | |
| Wild | 18 | 5 | 23 | |
| Total patients | 39 | 18 | 57 | |
E19del, exon 19 deletion; ctDNA, circulating tumor DNA.
At the time of diagnosis, there was a statistically significant correlation between bone metastasis and ctDNA detection (Table 3). The ability to detect ctDNA using the qualitative test did not have a statistically significant effect on PFS (median PFS: ctDNA detected 11.5 vs. ctDNA undetected group 13.5 months, hazard ratio [HR] 1.417, 95% CI 0.80–2.52; P = 0.234) (Fig 1a). However, in the quantitative test results, PFS was statistically significantly shorter in the ctDNA detected than in the ctDNA undetected group (median PFS 9.8 vs. 20.7 months, respectively, HR 2.30, 95% CI 1.202–4.385; P = 0.012) (Fig 1b). Detection of ctDNA before treatment with EGFR‐TKIs (HR 2.388, 95% CI 1.138–5.014) and extrathoracic lymph node (LN) metastasis (HR 13.533, 95% CI 2.474–68.747) were independently associated with PFS (Table 4).
Table 3.
Multivariate analysis of risk factors for ctDNA detection
| Multivariable analysis | ||
|---|---|---|
| Variable | OR (95% CI) | P |
| Age | 6.182 (0.481–79.444) | 0.691 |
| Gender (male) | 7.291 (0.734–72.435) | 0.090 |
| Smoking | 5.928 (0.552–63.367) | 0.142 |
| Metastasis to pleural effusion | 0.657 (0.085–5.077) | 0.469 |
| Pericardial metastasis | 0.227 (0.0–241.538) | 0.955 |
| Metastatic pleural nodules | 0.648 (0.100–4.189) | 0.833 |
| Contralateral lung metastasis | 1.216 (0.225–6.567) | 0.598 |
| Bone metastasis | 3.985 (1.027–15.457) | 0.046 |
| Extrathoracic lymph nodes metastasis | 11.533 (2.200–60.469) | 0.257 |
| Brain metastasis | 0.448 (0.096–2.490) | 0.467 |
| Adrenal metastasis | 7.482 (0.363–154.317) | 0.125 |
| Liver metastasis | — | 0.999 |
CI, confidence interval; ctDNA, circulating tumor DNA; OR, odds ratio.
Figure 1.
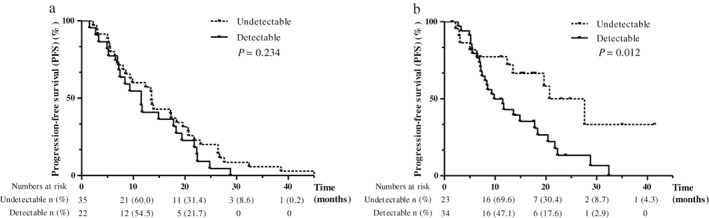
Kaplan–Meier survival curves of progression‐free survival of patients to circulating tumor DNA (ctDNA) EGFR mutation. (a) Qualitative and (b) quantitative analysis of ctDNA.
Table 4.
Univariate and multivariate analysis of prognostic factors for progression‐free survival
| Univariate analysis | Multivariable analysis | |||
|---|---|---|---|---|
| Variable | HR* (95% CI) | P | HR* (95% CI) | P |
| Age | 0.978 (0.948–1.010) | 0.174 | 0.970 (0.939–1.002) | 0.070 |
| Gender | 0.797 (0.400–1.589) | 0.520 | 0.881 (0.433–1.789) | 0.774 |
| ctDNA (+) | 2.483 (1.196–5.156) | 0.015 | 2.388 (1.138–5.014) | 0.021 |
| Smoking | 1.452 (0.716–2.947) | 0.314 | ||
| Histologic differentiation | 2.981 (1.018–8.731) | 0.088 | ||
| M1b metastasis | 2.453 (1.159–5.194) | 0.019 | 1.843 (0.836–4.063) | 0.128 |
| Metastasis to pleural effusion | 0.854 (0.258–2.834) | 0.797 | ||
| Pericardial metastasis | 1.129 (0.152–8.358) | 0.905 | ||
| Pleural nodules | 0.290 (0.112–0.748) | 0.010 | ||
| Contralateral lung metastasis | 0.898 (0.423–1.906) | 0.778 | ||
| Bone metastasis | 1.769 (0.935–3.346) | 0.080 | 0.761 (0.339–1.706) | 0.783 |
| Extrathoracic lymph nodes metastasis | 10.731(2.258–51.004) | 0.003 | 13.533 (2.474–68.747) | 0.002 |
| Brain metastasis | 1.140 (0.593–2.194) | 0.694 | ||
| Adrenal metastasis | 2.155 (0.814–5.708) | 0.122 | ||
| Liver metastasis | 2.386 (0.822–6.923) | 0.110 | ||
| Other sites (metastasis) | 3.133 (0.414–23.730) | 0.269 | ||
CI, confidence interval; ctDNA, circulating tumor DNA; HR, hazard ratio.
Dynamic changes in EGFR mutations were analyzed in 11 patients using serial sampling. In four patients, ctDNA mutations were unable to be detected from diagnosis to disease progression. In one patient, there was no change in the level of ctDNA. Dynamic changes in the ctDNA level were observed in the remaining six patients (Fig 2). Negative conversion of detectable ctDNA after EGFR‐TKI administration was found in 45.5% (5/11) of patients (Fig 2b–f), and increased ctDNA levels before or after disease progression were observed in 54.5% (6/11). T790M mutation was detected in 36.4% (4/11) of patients before or after disease progression (Fig 2c–f).
Figure 2.
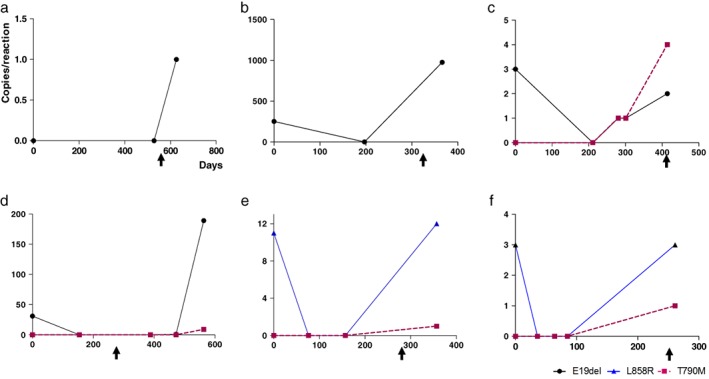
(a–f) Serial monitoring in six patients detected with circulating tumor DNA after EGFR‐tyrosine kinase inhibitor administration. Arrows indicate disease progression.
Discussion
Although ctDNA is now actively studied, numerous challenges need to be overcome before ctDNA can be used as a routine method of detection in a clinical setting. Although the specificity of liquid biopsy is close to 100% regardless of the technique used, the sensitivity is lower and varies by the method used.13, 14, 15, 16 When very few copies of ctDNA are available, it is not always detectable in the peripheral blood, thus ctDNA analysis results might be false negative. Therefore, one major challenge is low sensitivity. The concordance rate of ctDNA depends on which method is used;17, 18, 19, 20 however, few studies have performed both quantitative and qualitative analysis of ctDNA using the same sample. In this study, the detection rate of ctDNA was higher by quantitative than by qualitative analysis. To analyze the cause of these differences, we individually identified all results of the registered patients (Table S1). When the data were divided into two groups according to the amount of ctDNA detected by quantitative analysis, the detection rate of ctDNA by the qualitative test was higher in the group with > 1.5 copies/reaction. This discrepancy may be attributable to several factors. Because pre‐analysis conditions undoubtedly play a crucial role,21 slight differences in plasma DNA extraction and quantification methods in each laboratory may affect the detection rate of ctDNA. Moreover, the detection rate could vary depending on the threshold setting.22, 23
In current clinical practice, only qualitative analysis of ctDNA is performed. This study highlights the potential for sensitivity enhancement by using quantitative analysis, as we found that additional quantitative analysis can increase the sensitivity of the ctDNA detection test.
Several studies have shown that high ctDNA copy numbers are associated with shorter overall survival and PFS.24, 25, 26 In our study, detection of ctDNA by qualitative analysis was not statistically significant to PFS, but detection of ctDNA by quantitative analysis yielded a statistically significantly shorter PFS. For these reasons, quantitative analysis of ctDNA may be more pertinent.
Although some studies have found no correlation between ctDNA levels and tumor burden,24, 27 most studies have suggested that detection of ctDNA is significantly associated with tumor burden.28, 29, 30, 31 In this study, bone metastasis was associated with detectable ctDNA, supporting the higher detection rate of ctDNA in patients with a more extensive tumor burden.
This study showed a pattern of decreased ctDNA levels after treatment that increased before or after tumor progression, which mirrored results found in other studies.32, 33 In one patient, increased ctDNA levels and T790M mutation appeared much earlier than tumor progression, as assessed by imaging (Fig 2c). These results indicate that quantitative analysis of ctDNA can be used as a marker of therapeutic response or the development of resistance. Although we achieved meaningful results, a limitation of this study was the small patient population.
An attractive feature of ctDNA is that it contains DNA mutations found in primary and metastatic lesions,34, 35, 36 which may be a way to overcome problems caused by tumor heterogeneity. More studies should be performed to validate the potential of ctDNA for routine clinical use, and precise medicine is the next step in the treatment of NSCLC.
Quantitative testing can increase the sensitivity of ctDNA detection. Patients with detectable ctDNA had significantly shorter PFS after receiving EGFR‐TKI than patients with undetectable ctDNA. Tumor burden may be associated with plasma ctDNA. A shorter PFS was associated with detection of ctDNA and extrathoracic LN metastasis. Dynamic changes in the ctDNA level may help predict clinical outcomes.
Disclosure
No authors report any conflict of interest.
Supporting information
Table S1 Comparison between qualitative and quantitative analysis of circulating tumor DNA (ctDNA) EGFR mutation.
Acknowledgments
This study was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (HI15C0516).
References
- 1. Nana‐Sinkam SP, Powell CA. Molecular biology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence‐based clinical practice guidelines. Chest 2013; 143 (Suppl 5): e30S–e9S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang YS, Choi CM, Lee JC. Mechanisms of epidermal growth factor receptor tyrosine kinase inhibitor resistance and strategies to overcome resistance in lung adenocarcinoma. Tuberc Respir Dis 2016; 79: 248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park JY, Jang SH. Epidemiology of lung cancer in Korea: Recent trends. Tuberc Respir Dis 2016; 79: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barlesi F, Mazieres J, Merlio JP et al Routine molecular profiling of patients with advanced non‐small‐cell lung cancer: Results of a 1‐year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016; 387: 1415–26. [DOI] [PubMed] [Google Scholar]
- 5. Overman MJ, Modak J, Kopetz S et al Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J Clin Oncol 2013; 31: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piotrowska Z, Niederst MJ, Karlovich CA et al Heterogeneity underlies the emergence of EGFRT790 wild‐type clones following treatment of T790M‐positive cancers with a third‐generation EGFR inhibitor. Cancer Discov 2015; 5: 713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sequist LV, Waltman BA, Dias‐Santagata D et al Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3 (75): 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suda K, Murakami I, Katayama T et al Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res 2010; 16: 5489–98. [DOI] [PubMed] [Google Scholar]
- 9. Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: Implications for targeted therapeutics. Br J Cancer 2013; 108: 479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weber B, Meldgaard P, Hager H et al Detection of EGFR mutations in plasma and biopsies from non‐small cell lung cancer patients by allele‐specific PCR assays. BMC Cancer 2014; 14: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diaz LA Jr, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol 2014; 32: 579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim CG, Shim HS, Hong MH et al Detection of activating and acquired resistant mutation in plasma from EGFR‐mutated NSCLC patients by peptide nucleic acid (PNA) clamping‐assisted fluorescence melting curve analysis. Oncotarget 2017; 8: 65111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mok T, Wu YL, Lee JS et al Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first‐line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015; 21: 3196–203. [DOI] [PubMed] [Google Scholar]
- 14. Douillard JY, Ostoros G, Cobo M et al First‐line gefitinib in Caucasian EGFR mutation‐positive NSCLC patients: A phase‐IV, open‐label, single‐arm study. Br J Cancer 2014; 110: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sundaresan TK, Sequist LV, Heymach JV et al Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood‐based analyses. Clin Cancer Res 2016; 22: 1103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karlovich C, Goldman JW, Sun JM et al Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of Rociletinib (CO‐1686). Clin Cancer Res 2016; 22: 2386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taniguchi K, Uchida J, Nishino K et al Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011; 17: 7808–15. [DOI] [PubMed] [Google Scholar]
- 18. Yam I, Lam DC, Chan K et al EGFR array: Uses in the detection of plasma EGFR mutations in non‐small cell lung cancer patients. J Thorac Oncol 2012; 7: 1131–40. [DOI] [PubMed] [Google Scholar]
- 19. Kim HR, Lee SY, Hyun DS et al Detection of EGFR mutations in circulating free DNA by PNA‐mediated PCR clamping. J Exp Clin Cancer Res 2013; 32: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thress KS, Brant R, Carr TH et al EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross‐platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015; 90: 509–15. [DOI] [PubMed] [Google Scholar]
- 21. Vendrell JA, Mau‐Them FT, Béganton B, Godreuil S, Coopman P, Solassol J. Circulating cell free tumor DNA detection as a routine tool for lung cancer patient management. Int J Mol Sci 2017; 18: pii:E264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high‐throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 2011; 12: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shu Y, Wu X, Tong X et al Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep 2017; 7: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nygaard AD, Holdgaard PC, Spindler KL, Pallisgaard N, Jakobsen A. The correlation between cell‐free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 2014; 110: 363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alegre E, Fusco JP, Restituto P et al Total and mutated EGFR quantification in cell‐free DNA from non‐small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumour Biol 2016; 37: 13687–94. [DOI] [PubMed] [Google Scholar]
- 26. Tissot C, Toffart AC, Villar S et al Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J 2015; 46: 1773–80. [DOI] [PubMed] [Google Scholar]
- 27. García‐Saenz JA, Ayllón P, Laig M et al Tumor burden monitoring using cell‐free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 2017; 17: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oxnard GR, Paweletz CP, Kuang Y et al Noninvasive detection of response and resistance in EGFR‐mutant lung cancer using quantitative next‐generation genotyping of cell‐free plasma DNA. Clin Cancer Res 2014; 20: 1698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newman AM, Bratman SV, To J et al An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20: 548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yanagita M, Redig AJ, Paweletz CP et al A prospective evaluation of circulating tumor cells and cell‐free DNA in EGFR‐mutant non‐small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res 2016; 22: 6010–20. [DOI] [PubMed] [Google Scholar]
- 31. Pérez‐Ramírez C, Cañadas‐Garre M, Robles AI, Molina MÁ, Faus‐Dáder MJ, Calleja‐Hernández MÁ. Liquid biopsy in early stage lung cancer. Transl Lung Cancer Res 2016; 5: 517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Forshew T, Murtaza M, Parkinson C et al Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4: 136ra68. [DOI] [PubMed] [Google Scholar]
- 33. Pérez‐Callejo D, Romero A, Provencio M, Torrente M. Liquid biopsy based biomarkers in non‐small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res 2016; 5: 455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yi X, Ma J, Guan Y, Chen R, Yang L, Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer 2017; 140: 2642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan KC, Jiang P, Zheng YW et al Cancer genome scanning in plasma: Detection of tumor‐associated copy number aberrations, single‐nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013; 59: 211–24. [DOI] [PubMed] [Google Scholar]
- 36. De Mattos‐Arruda L, Weigelt B, Cortes J et al Capturing intra‐tumor genetic heterogeneity by de novo mutation profiling of circulating cell‐free tumor DNA: A proof‐of‐principle. Ann Oncol 2014; 25: 1729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Comparison between qualitative and quantitative analysis of circulating tumor DNA (ctDNA) EGFR mutation.


